Abstract
The RNA polymerase III (pol III) type III promoters U6 and 7SK are routinely used to express short hairpin RNA (shRNA) molecules from a DNA construct. In this study, we identified, characterised and compared the porcine 7SK promoter in porcine (homologous) and non-porcine (heterologous) derived cell lines. The porcine 7SK small nuclear RNA (snRNA) was identified by alignment with known sequences and further characterisation of the upstream regions determined the presence of typical RNA pol III sequence motifs. The porcine 7SK (po7SK) promoter was cloned and a one-step PCR strategy used to construct shRNA expression cassettes. The 7SK promoter activity was quantified by knockdown of the exogenous reporter gene encoding the enhanced green fluorescent protein (EGFP). Results indicated the po7SK promoter was functional in both homologous and heterologous cells lines. The identification and characterisation of the porcine RNA pol III promoter will contribute to the area of RNAi delivery and further develop our understanding of RNA promoter structure and function.
Keywords: RNAi, 7SK, U6, pol III, shRNA
INTRODUCTION
RNA polymerase (pol) III type III promoters are the most commonly used promoters for the expression of short hairpin RNA (shRNA; Amarzguioui et al, 2005). Type III promoters such as U6 and 7SK are often preferred because they naturally express small, highly-abundant non-coding RNA transcripts referred to as small-nuclear RNAs (snRNAs). Type III promoters do not contain intragenic control regions, the promoter region is located entirely upstream of the transcription start site (+1) and transcription termination occurs at a defined tract of 4-5 thymidines (Geiduschek and Kassavetis, 2001). The pol III type III promoters contain sequence motifs located within regions referred to as the enhancer and core region (Schramm and Hernandez, 2002). The enhancer region consists of an octamer motif (OCT) and an SphI Post-octamer Homology domain (SPH), while a proximal sequence element (PSE) and a TATA-like element form the core region (Schramm and Hernandez, 2002). The mouse U6 and human U6/H1 promoters are the most commonly used promoters for shRNA expression. Promoter activity may vary considerably for the family of U6 promoters, such as the five human U6 promoters which all have varied transcriptional efficiencies (Domitrovich and Kunkel, 2003).
More recently, researchers have identified the human 7SK promoter as the most efficient pol III promoter for the expression of short hairpin RNAs (shRNAs) in vivo, surpassing the activity of the human U6-1 promoter (Koper-Emde et al, 2004). Numerous 7SK pseudo-genes are known to exist, but only a single 7SK promoter is known to be present in the human (Murphy et al, 1987), bovine (Lambeth et al, 2006), mouse (Moon and Krause, 1991) and chicken (Bannister et al, 2007) genomes.
With a large portion of the porcine genome sequence currently available and the conserved nature of the 7SK snRNAs (Humphries et al, 1987), we sought to identify, characterise and compare the porcine 7SK (po7SK) promoter for promoter efficiency relative to various known RNA pol III type III promoters.
MATERIALS AND METHODS
Cell culture and transfection
Swine testis (ST) cells (ATCC, CRL-1746) were maintained in Eagles Minimal Essential Media (EMEM) with 1% (w/v) sodium pyruvate and 0.25% (w/v) lactalbumin hydrolysate (LAH). Baby hamster kidney (BHK; ATCC, CCL-10) were grown and maintained in Basal Medium Eagle (BME) and 10% (w/v) tryptose phosphate broth (TBP). African green monkey kidney cells (VERO; ATCC, CCL-81) were grown and maintained in EMEM. All growth media used contained additional supplements including; 2 mM glutamine, 10 mM HEPES, 10% (v/v) fetal calf serum (FCS) with the addition of penicillin (100 U/ml) and streptomycin (100 μg/ml). Cells were maintained in a 5% (v/v) CO2 atmosphere at 37°C. Cells were harvested by washing twice in phosphate buffered saline-A (PBSA), followed by the addition of 0.25% (v/v) PBSA diluted trypsin. All cell lines were transfected at approximately 80-90% confluencey in 8-well chamber slides (Nunc) for fluorescence microscopy, 24-well plates for flow cytometry analysis or 25 cm2 culture flask for RNA isolation. Cells for microscopy and flow cytometry analysis were transfected with 0.5 μg of each plasmid using Lipofectamine 2000 (Invitrogen) according to manufactures instructions. When the cells were to be used for RNA isolation, 10 μg of plasmid DNA and 25 μl of Lipofectamine 2000 reagent was used.
Isolation of the porcine 7SK promoter
The po7SK promoter was PCR amplified from pig genomic DNA (Wizard Genomic DNA purification kit, Promega) using PCR platinum supermix (Invitrogen) and primers; po7SK-F (5′-GGCTCAGAGCCAGGAGAAAA AC-3′) and po7SK-R (5′-GAGGGCCTGAGGAAGGC CGC-3′). The 436 bp amplified PCR product was cloned into pGEM®-T Easy vector system (Promega), sequenced confirmed and named pGEM-po7SK.
The sequence was compared using mega-Basic Local Alignment Search Tool (mega-BLAST; Altschul et al, 1990). The putative po7SK promoter was deposited into GenBank under the accession number EU39883.
Construction of the porcine 7SK shRNA expression vectors
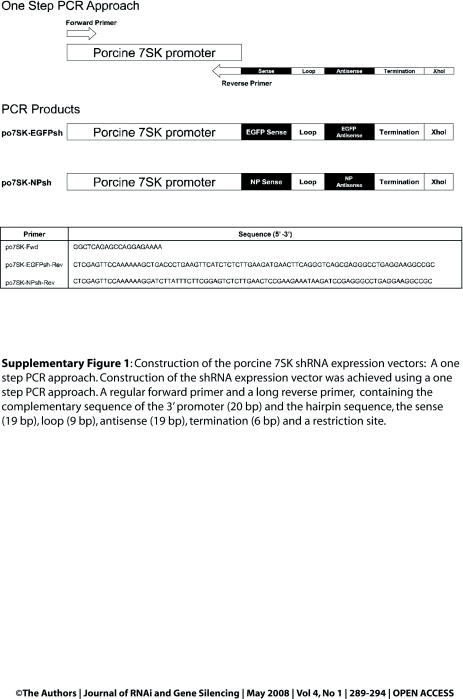
Expression vectors containing an EGFP shRNA (po7SK-EGFPsh) and an irrelevant shRNA control targeting the influenza virus nucleocapsid protein (NP; po7SK-NPsh) under the control of the po7SK promoter were constructed. The po7SK-EGFPsh and po7SK-NPsh expression vectors were constructed using the one-step PCR approach described by Lambeth et al, 2005 (Supplementary Figure 1). Briefly, the pGEM-po7SK construct was used as template to amplify the po7SK promoter using a short forward oligonucleotide and a long reverse oligonucleotide containing complementary sequence to the po7SK promoter along with the sense, loop, antisense and stop sequence of the shRNA. The amplified PCR product containing the promoter and hairpin sequence was cloned into pGEM-T Easy. The EGFP shRNA expression vectors under the control of the bovine 7SK (pEZ-b7SKshEGFP), chicken 7SK (pch7SK-shEGFP) and mouse U6 (pmU6-shEGFP) promoter have previously been described (Lambeth et al, 2006; Bannister et al, 2007). The bovine, chicken and mouse EGFP shRNA expression constructs were renamed b7SK-EGFPsh, c7SK-EGFPsh and mU6-EGFPsh for this investigation.
Detection of expressed shRNA molecules
Small enriched RNAs were isolated from transfected ST cells at 72 hr post-transfection using the mirVana miRNA isolation kit (Ambion). Detection of expressed EGFP shRNA molecules was achieved using an RNAse protection assay (RPA) using the RNA probe LL91 (named shEGFP probe in this investigation) described previously (Lambeth et al, 2006). RNA samples were prepared and hybridised with the radiolabelled (γ-32P) shEGFP probe in solution overnight at 42°C. The shEGFP hybridised samples were RNase A/T1 treated as recommended. Samples were separated on a 15% (w/v) polyacrylamide (8M urea) gel and then exposed to Hyperfilm™ ECL (Amersham Biosciences) within an EC-AWU cassette (Fuji) and placed at -80°C overnight. The film was developed using an X-ray processor FPM-100A (Fuji).
RNAi assay
EGFP expression was analysed at 48 hr post-transfection. Fluorescence microscopy images were captured using a Leica DM LB Fluorescence Microscope coupled to a Leica DC200F digital camera (Leica Microsystems) 100× magnification. Flow cytometry was used to measure the EGFP Mean Fluorescence Intensity (MFI) of each sample. Transfected cells were harvested using 0.25% (w/v) trypsin-EDTA, pelleted at 2000 rpm for 5 min, washed in phosphate buffered saline-A (PBSA) (Oxoid) and re-suspended in FACS-solution (1% fetal calf serum (v/v) in PBSA). Analysis was performed using a FACScalibur (Becton Dickinson) fluorescent activated cell sorter and CELLQuest software (Becton Dickinson). The MFI was calculated as a percentage of the non-silencing control (po7SK-NPsh) for each cell line.
RESULTS
Identification and isolation of the porcine 7SK promoter
To identify the po7SK promoter, a bioinformatics search of the porcine genome using the 331 nucleotide human 7SK snRNA (GenBank Accession Number NR001445) was conducted. Several matches were identified with significant similarity observed. The porcine sequence (CU62466.2) showing the greatest similarity to the human 7SK snRNA was selected for further analysis and an alignment between the human, chicken and bovine 7SK snRNA sequences carried out to identify the presence of the porcine full-length 7SK snRNA and delineate sequence similarities (Figure 1).
Figure 1:
Alignment of the 7SK snRNA. An alignment of the porcine (po7SK snRNA; CU62466.2), human (h7SK snRNA; NR001445), bovine (b7SK snRNA; CR810253) and chicken (c7SK snRNA; AJ890101) 7SK snRNA sequences using ClustalW (Chenna et al, 2003). Dot (.) represent identical base. Dash (-) represents gap/no base. Base changes are indicated with the appropriate base.
PCR was used to amplify 436 bp upstream of the porcine 7SK snRNA. Analysis of the cloned po7SK sequence by alignment predicted the following; TATA box at -25 to -32 bp, PSE at -47 to -67 bp, SPH domain at -194 to -212 and an OCT motif at -226 to -234. An alignment of the po7SK promoter sequence motifs are shown in Figure 2.
Figure 2:
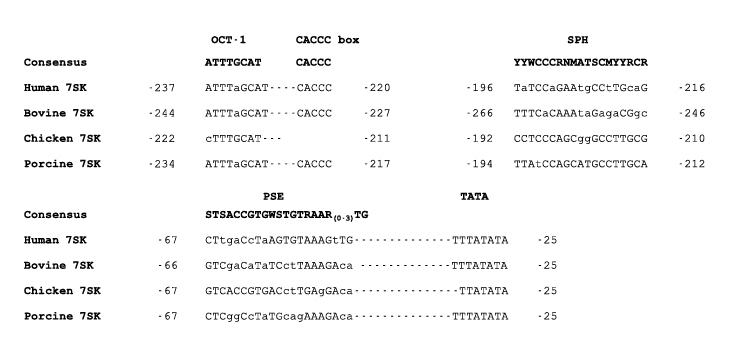
Alignment of the 7SK promoter motifs. The sequence elements for the enhancer region (OCT and SPH) and core region (PSE and TATA) are shown for the human 7SK (h7SK; X04992), bovine 7SK (b7SK; CR810253), chicken 7SK (c7SK; EF488955) and porcine 7SK (po7SK; EU39883) promoters. The consensus sequence for OCT, SPH, PSE and TATA are indicated (OCT consensus - Strum et al, 1988; SPH consensus - Kleinert et al, 1990; PSE consensus - Dahlberg and Lund, 1988). Dashes (-) represent a single nucleotide
Detection of expressed EGFP shRNA
The activity of the putative po7SK promoter was verified by the detection of expressed EGFP shRNAs. An RNAse protection assay was performed on ST cells transfected with the plasmids po7SK-EGFPsh, b7SK-EGFPsh, c7SK-EGFPsh, mU6-EGFPsh and negative controls pEGFP-N1 and po7SK-NSsh (Figure 3). A 19 nucleotide band was detected in the lanes representing the po7SK-EGFPsh, b7SK-EGFPsh, c7SK-EGFP and mU6-EGFPsh transfected ST cells. No bands were identified in the pEGFP-N1 or po7SK-NPsh control samples. These results demonstrated that the cloned po7SK promoter was functional as indicated by the detection of the expressed shRNAs.
Figure 3:

Detection of RNA pol III expressed EGFP shRNAs. Swine testis cells were transfected with the constructs indicated. Small RNA's were isolated at 72 h post-transfection and radiolabelled with an EGFP shRNA probe (shEGFP) to detect the expressed shRNA's.
Activity of the porcine 7SK promoter measured by EGFP knockdown
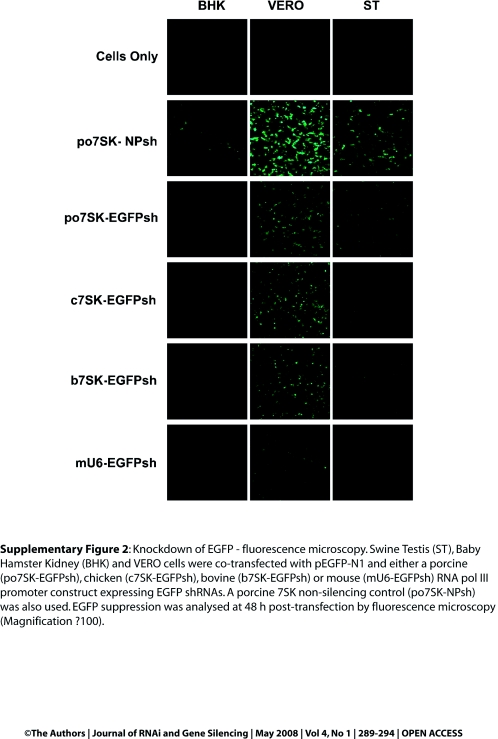
To determine the activity of the po7SK promoter, EGFP silencing experiments in ST, BHK and VERO cells were conducted. The pEGFP-N1 construct was co-transfected with either po7SK-EGFPsh or the non-silencing control po7SK-NPsh. Given co-transfection of the shRNA expression plasmid and the reporter construct are considered to be 100% efficient for RNAi validation (Cullen, 2006), a reduction in EGFP fluorescence was considered a result of RNAi-mediated EGFP knockdown. In addition, the constructs b7SK-EGFPsh, c7SK-EGFPsh and mU6-EGFPsh were also co-transfected along with pEGFP-N1 and fluorescence microscopy used to observe EGFP knockdown (Supplementary Figure 2). The level of gene silencing was compared relative to the level of fluorescence observed in the pEGFP-N1/po7SK-NPsh control cells. Results indicated that the po7SK promoter induced specific knockdown of EGFP in all three cell lines. To quantify the level of EGFP knockdown mediated by po7SK compared with that of the other RNA pol III promoters, those constructs were co-transfected into the three cell lines as described above. At 48 hr post-transfection, cells were harvested and analysed for their MFI by flow cytometry (Figure 4). The MFI was calculated as a percentage of the pEGFP-N1/po7SK-NPsh co-transfected cells. The po7SK promoter was capable of silencing EGFP by 30% in VERO cells and by 80% in both BHK and ST cells. These results demonstrated that the po7SK promoter was functional in a range of cell types. However, it appeared that in each of the cell types the bovine, chicken and mouse RNA pol III promoters induced greater EGFP knockdown than the po7SK promoter.
Figure 4:
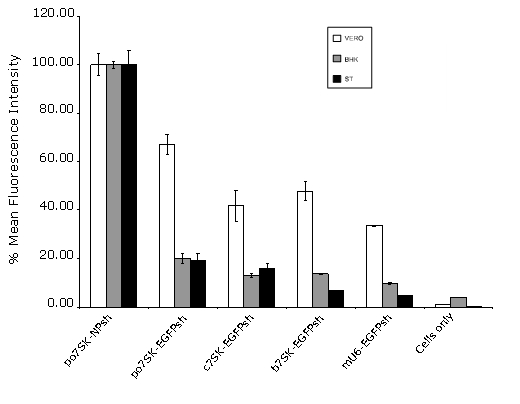
Knockdown of EGFP - flow cytometry. Swine Testis (ST), Baby Hamster Kidney (BHK) and VERO cells were co-transfected with pEGFP-N1 and either a porcine (po7SK-EGFPsh), chicken (c7SK-EGFPsh), bovine (b7SK-EGFPsh) or mouse (mU6-EGFPsh) RNA pol III promoter construct expressing EGFP shRNAs. Flow cytometry was used to determine EGFP fluorescence at 48 h post-transfection. Values are shown as a percentage of the porcine 7SK non-silencing control (po7SK-NPsh) as the mean of three replicates ± standard error.
DISCUSSION
The identification and characterisation of promoters for the expression of shRNA molecules has been limited to species with known genome sequence. The recent release of the porcine genome sequence will enable the identification of porcine promoters for shRNA expression. Porcine related RNAi research has focused on silencing porcine specific viruses such as porcine reproductive and respiratory syndrome virus, porcine circovirus and porcine transmissible gastroenteritis virus using commercially available constructs and promoters (He et al, 2007; Sun et al, 2007; Zhou et al, 2007). In this investigation, we are the first to identify and characterise a porcine RNA polymerase III promoter and demonstrate its functionality for shRNA expression.
The conserved nature of the 7SK snRNA enabled the porcine 7SK snRNA to be identified by comparison with the human 7SK snRNA sequence (Humphries et al, 1987). Although many 7SK pseudogenes are known to exist, we were able to identify a full-length porcine snRNA sequence with high similarity to the human, chicken and bovine 7SK snRNA (Figure 1). Given that only a single functional 7SK promoter has been identified upstream of the 7SK snRNA in the human, bovine, chicken and mouse genomes (Murphy et al, 1987; Lambeth et al, 2006; Moon and Krause, 1991), it was likely that a functional promoter would be located upstream of the porcine 7SK snRNA. Typical pol III motifs TATA, PSE, OCT and SPH were identified in the po7SK and showed significant sequence identity to motifs in the h7SK, b7SK and c7SK promoters.
Activity of the po7SK promoter was determined by its ability to express EGFP shRNAs and silence plasmid expressed EGFP. A shRNA expression vector was constructed and tested in homologous and heterologous cell lines to test functionality, efficiency and specificity. Our results clearly demonstrated the promoter was functional through the detection of shRNAs (Figure 3) and silencing of the reporter gene EGFP (Figure 4). Although the po7SK was capable of inducing gene silencing at varying levels in different cell types, the po7SK promoter was not as efficient as the bovine 7SK, chicken 7SK or mouse U6 promoters in either porcine, hamster or monkey derived cells. This difference in efficiency is similar to a study by Bannister et al, (2007), who demonstrated that the chicken 7SK promoter was not as efficient as the b7SK or mU6 promoters in chicken DF1 cells. Previous studies (Lambeth et al, 2007; Koper-Emde et al, 2004) have shown 7SK promoters perform more efficiently in cell lines that are of the same species as the origin of the promoter. Greater silencing efficiencies for the po7SK promoter were not observed in this study or the study by Bannister et al, (2007), suggesting that promoter sequence is the most important factor for promoter activity rather than the species of promoter origin.
Comparison of the bovine and porcine 7SK promoter elements at the nucleotide level revealed significant similarities, although the activity of the bovine promoter was as much as 20% more effective at gene silencing than the porcine promoter. The TATA and PSE elements of the bovine and porcine promoters are similarly spaced and match the consensus sequence at the same locations. The porcine promoter has greater sequence similarity to the SPH consensus than the bovine, although the bovine SPH is located upstream of the OCT domain. Perfect similarity and spacing is observed between the OCT and CACCC box for both the porcine and bovine promoters. Although the bovine elements are located an additional 10 bp upstream of the start site, relative to the porcine promoter. Co-dependence has been demonstrated for the OCT and SPH elements in Xenopus laevis tRNAsec promoter and human U6 promoters for efficient enhancer activity (Myslinski et al, 1992; Kunkel et al, 1996). Whereas for the human 7SK, optimal efficiency is not dependent on the presence of an SPH region (Boyd et al, 2000). If this feature applies to both bovine and porcine 7SK promoter elements and given the SPH motif of the bovine promoter has been suggested to be located 5′ to OCT motif, then the downstream spacing and sequence could be responsible for the increased promoter efficiency observed in the bovine promoter. Additionally, differences between the chicken and porcine sequence motifs were also observed. The largest difference is the absence of a CACCC box and a C/A substitution at position 1 (bp -222) in the chicken 7SK OCT motif. The CACCC box is a notable feature of 7SK promoters and has been reported to function in enhancing the transcriptional activity of the human 7SK promoter (Kleinert et al, 1990). Mutational studies within the OCT sequence of the human 7SK promoter have been shown to have the greatest effect on transcription (Boyd et al, 2000). Given there is greater sequence variation between the po7SK and c7SK, it would be hypothesised that promoter efficiency would be greatest for the porcine 7SK because of the more conserved nature of the sequence motifs. However, in this study we observed that the chicken 7SK performed marginally better in both BHK and VERO cell lines and was substantially more efficient in ST cells. This data further suggests that promoter sequence variation and not the species of promoter origin plays the major role in directing promoter activity.
CONCLUSIONS
In this report, we identified and characterised the po7SK promoter for the delivery of shRNAs. Analysis revealed that the po7SK promoter efficiently expressed shRNAs and induced gene silencing. The identification of additional porcine pol III promoters including the U6 family of promoters should provide greater insight into the mechanisms directing transcriptional efficiency between different promoter types within the same species. Furthermore, the identification and characterisation of new promoters will enable the development of tools for shRNA expression not only for porcine specific applications, but also for the field of RNAi.
Acknowledgments
We would like to thank Anthony Keyburn and Pauline Cottee for critically reviewing this manuscript. A special thanks also to Terry Wise and Stephanie Bannister for their helpful discussions.
LIST OF ABBREVIATIONS
- BHK
Baby hamster kidney
- BME
Basal Medium Eagle
- b7SK
bovine 7SK
- c7SK
chicken 7SK
- EMEM
Eagles Minimal Essential Media
- FCS
fetal calf serum
- HEPES
N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid
- LAH
lactalbumin hydrolysate
- mU6
mouse U6
- OCT
octamer motif
- pol III
polymerase III
- po7SK
porcine 7SK
- PSE
proximal sequence element
- snRNA
small nuclear RNA
- SPH
post-octamer Homology domain
- ST
Swine testis
- VERO
African green monkey kidney cells
COMPETING INTERESTS
None declared.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amarzguioui M, Rossi JJ, Kim D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett. 2005;579:5974–5981. doi: 10.1016/j.febslet.2005.08.070. [DOI] [PubMed] [Google Scholar]
- Bannister SC, Wise TG, Cahill DM, Doran TJ. Comparison of chicken 7SK and U6 RNA polymerase III promoters for short hairpin RNA expression. BMC Biotech. 2007;7:79. doi: 10.1186/1472-6750-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd DC, Greger IH, Murphy S. In vivo footprinting studies suggest a role for chromatin in transcription of the human 7SK gene. Gene. 2000;247:33–44. doi: 10.1016/s0378-1119(00)00134-7. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acid Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Induction of stable RNA interference in mammalian cells. Gene Ther. 2006;13:503–508. doi: 10.1038/sj.gt.3302656. [DOI] [PubMed] [Google Scholar]
- Dahlberg JE, Lund E. The genes and transcription of the major small nuclear RNAs. In: Birnstiel ML, editor. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Berlin, Germany: Springer-Verlag; 1988. pp. 38–70. [Google Scholar]
- Domitrovich AM, Kunkel GR. Multiple, dispersed human U6 small nuclear RNA genes with varied transcriptional efficiencies. Nucleic Acids Research. 2003;31:2344–52. doi: 10.1093/nar/gkg331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. Journal of Molecular Biology. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- He YX, Hua RH, Zhou YJ, Qiu HJ, Tong GZ. Interference of porcine reproductive and respiratory syndrome virus replication on MARC-145 cells using DNA-based short interfering RNAs. Antiviral Res. 2007;74:83–91. doi: 10.1016/j.antiviral.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Humphries P, Russell SE, McWilliam P, McQuaid S, Pearson C, Humphries MM. Observations on the structure of two human 7SK pseudogenes and on homologous transcripts in vertebrate species. Biochem J. 1987;245:281–284. doi: 10.1042/bj2450281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinert H, Bredow S, Benecke BJ. Expression of a human 7SK RNA gene in vivo requires a novel pol III upstream element. EMBO J. 1990;9:711–718. doi: 10.1002/j.1460-2075.1990.tb08164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koper-Emde D, Herrmann L, Sandrock B, Benecke BJ. RNA interference by small hairpin RNAs synthesised under control of the human 7S K RNA promoter. Biol Chem. 2004;385:791–794. doi: 10.1515/BC.2004.103. [DOI] [PubMed] [Google Scholar]
- Kunkel GR, Cheung TC, Miyake JH, Urso O, McNamara-Schroeder KJ, Stumph WE. Identification of a SPH element in the distal region of a human U6 small nuclear RNA gene promoter and characterization of the SPH binding factor in HeLa cell extracts. Gene Expr. 1996;6:59–72. [PMC free article] [PubMed] [Google Scholar]
- Lambeth LS, Wise TG, Moore RJ, Muralitharan MS, Doran TJ. Comparison of bovine RNA polymerase III promoters for short hairpin RNA expression. Anim Genet. 2006;37:369–372. doi: 10.1111/j.1365-2052.2006.01468.x. [DOI] [PubMed] [Google Scholar]
- Lambeth LS, Moore RJ, Muralitharan M, Dalrymple BP, McWilliam S, Doran TJ. Characterisation and application of a bovine U6 promoter for expression of short hairpin RNAs. BMC Biotechnol. 2005;5:13. doi: 10.1186/1472-6750-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon IS, Krause MO. Common RNA polymerase I, II, and III upstream elements in mouse 7SK gene locus revealed by the inverse polymerase chain reaction. DNA Cell Biol. 1991;10:23–32. doi: 10.1089/dna.1991.10.23. [DOI] [PubMed] [Google Scholar]
- Murphy S, Di Liegro C, Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987;51:81–87. doi: 10.1016/0092-8674(87)90012-2. [DOI] [PubMed] [Google Scholar]
- Myslinski E, Krol A, Carbon P. Optimal tRNA ((Ser)Sec) gene activity requires an upstream SPH motif. Nucleic Acid Res. 1992;20:203–209. doi: 10.1093/nar/20.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Strum RA, Das G, Herr W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988;2:1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- Sun M, Liu X, Cao S, He Q, Zhou R, Ye J, et al. Inhibition of porcine circovirus type 1 and type 2 production in PK-15 cells by small interfering RNAs targeting the Rep gene. Vet Microbiol. 2007;123:203–209. doi: 10.1016/j.vetmic.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JF, Hua XG, Cui L, Zhu JG, Miao DN, Zou Y, et al. Effective inhibition of porcine transmissible gastroenteritis virus replication in ST cells by shRNAs targeting RNA-dependent RNA polymerase gene. Antiviral Res. 2007;74:36–42. doi: 10.1016/j.antiviral.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]