Abstract
Lupus nephritis (LN) is among the main determinants of poor prognosis in systemic lupus erythematosus (SLE). The objective of this study was to 1) isolate and identify proteins contained in the LN urinary protein signature (PS) of children with SLE; 2) assess the usefulness of the PS-proteins for detecting activity of LN over time. Using surface-enhanced or matrix assisted laser desorption/ionization time of flight mass spectrometry, the proteins contained in the LN urinary PS were identified. They were transferrin (Tf), ceruloplasmin (Cp), α1-acid-glycoprotein (AGP), lipocalin-type prostaglandin-D synthetase (L-PGDS), albumin and albumin-related fragments. Serial plasma and urine samples were analyzed using immunonephelometry or ELISA in 98 children with SLE (78% African-American) and 30 controls with juvenile idiopathic arthritis. All urinary PS-proteins were significantly higher with active versus inactive LN or in patients without LN (all p<0.005), and their combined area under the receiver operating characteristic curve was 0.85. As early as 3 months before a clinical diagnosis of worsening LN, significant increases of urinary Tf, AGP (both p < 0.0001) and L-PGDS (p < 0.01) occurred, indicating that these PS-proteins are biomarkers of LN activity and may help anticipate the future course of LN.
Systemic Lupus Erythematosus (SLE) is an inflammatory autoimmune disease and lupus nephritis (LN) is one of the main determinants of poor prognosis (1). Currently, LN is gauged by measuring circulating and excreted indicators of renal dysfunction, with supporting information from kidney biopsies. The latter constitute the current standard for diagnosing LN, providing a direct assessment of the presence, severity and activity of LN, and the degree of renal damage (2). Due to the invasive nature of kidney biopsies, clinicians base LN activity and its therapy on the results of urinary protein excretion, urinary sediment, creatinine clearance and serum albumin. These traditional markers are not accurate in assessing whether active LN is present or not, and none of them is predictive, i.e. can anticipate the course of LN.
Using Surface-Enhanced Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (SELDI-TOF MS) technology, we previously identified a LN urinary protein signature (PS), consisting of eight candidate biomarkers at the mass-to-charge ratios (m/z) of 2.763, 22, 23, 44, 56, 79, 100, and 133 kDa (3).
In this study, we present the identification of the specific proteins contained in this PS of children with LN. We further assayed plasma and urine samples of SLE patients and controls with juvenile idiopathic arthritis (JIA) to investigate the concurrent and predictive validity of the PS-proteins to serve as biomarkers of LN activity.
MATERIALS AND METHODS
SLE Patients
Children diagnosed with SLE (4) prior to the age of 16 years (n=98) were studied every 3 months for up to 18 months. At each study visit, blood and random spot urine samples for research were obtained, and information on the following laboratory measures was collected: BUN (urea), serum creatinine, serum complement levels C3 and C4, presence of anti-dsDNA antibodies, urine protein:creatinine ratio (normal < 0.2), and creatinine clearance approximated according to the Schwartz formula. At the participating centers, kidney biopsies are obtained in SLE patients when abnormal urinalyses cannot be explained by mechanisms other than SLE. Thus all children without kidney biopsies were considered to have SLE without LN. The study was approved by the Institutional Review Board (IRB) of the Cincinnati Children’s Hospital Medical Center, and the IRBs of all other participating centers, with informed consent obtained prior to any study-related procedures.
SLE Disease Measures
At each study visit two widely accepted measures of disease activity were completed: the 2k-version of the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) (5), and the British Isles Lupus Activity Group (BILAG) Index (6), an index that has been developed specially to assess organ-specific disease activity. SLEDAI or BILAG scores of 0 indicate inactive disease, and higher scores represent higher disease activity. Renal disease activity corresponds to SLEDAI or BILAG renal scores of > 0 or >1, respectively. At study entry, the Systemic Lupus International Collaborating Clinics/ACR Damage Index (SDI; 0 = no damage) was completed (7).
Controls with Juvenile Idiopathic Arthritis (JIA)
Like SLE, JIA is an inflammatory autoimmune disease. Although it rarely involves the kidney primarily, theoretically nephrotoxic medications are the mainstay of JIA therapy. Thirty children with JIA [26 Caucasians, 4 African-American; F:M = 27:3] served as controls, none of them had current or preceding laboratory abnormalities suggestive of a chronic renal disease. Only cross-sectional data of 20 patients with active and 10 with inactive JIA (as rated by their pediatric rheumatologist) were available for analysis.
Peptide Mapping and Protein Identification
Details on the approach to develop the LN protein signature (PS) with its eight candidate biomarkers have been published (3). Briefly, these biomarkers were detected on at least two different ProteinChips, and displayed a >100-fold increase in peak intensity between groups. Subsequently, three urine samples from each WHO-class of patients with LN and controls were lyophilized and re-dissolved with Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA, USA) for SDS-PAGE, using 8% or 12% Tris-Glycine gels with molecular weight standard markers (Invitrogen, Carlsbad, CA, USA). Bands that showed the same molecular weights as the candidate biomarker proteins (SELDI-TOF MS) were excised, then digested with trypsin, and recovered for mass spectrometry (8, 9). One third of the individual band was treated with elution solution [50% formic acid, 25% acetonitrile, 15% isopropranolol, 10% water] to extract the proteins contained in each band. These proteins were analyzed on a normal binding ProteinChip to confirm the aimed mass spectrum. Peptides recovered from the in-gel digest were identified either via peptide mass fingerprints (PMFs) on the SELDI-TOF platform, or MALDI-TOF/TOF MS via MS/MS fragmentation with sequencing individual peptides. The use of both methods was necessary as albumin or albumin fractions often were present, and none of the various albumin removal approaches [albumin depletion kit (QIAGEN, Qproteome albumin/IgG depletion kit, Valencia, CA, USA), immunoprecipitation (Dynabeads Protein G, Invitrogen, Carlsbad, CA, USA), urea treatment, and anion exchange spin column (ProteinChip Q spin column, Bio-Rad laboratories Inc., Hercules, CA, USA)] succeeded in removing the albumin fraction effectively.
For protein identification by SELDI-TOF MS, samples were dried on a surface chip target plate followed by matrix application. In this format, the SELDI system may be comparable to a conventional MALDI-TOF instrument and can be used to collect PMFs spectra directly. Alternatively, samples were applied onto the ProteinChip SEND-ID Array. The peptide mapping data were standardized using the All-in-One-Peptide Software (Bio-Rad Laboratories Hercules, CA, USA).
For MALDI-TOF/TOF MS, the excised peptides were desalted and concentrated on C18-micro-ZipTips as recommended by the vendor (Millipore, Billerica MA) and then spotted on the target plate in 2.5 mg/mL CHCA containing 10 mM monobasic ammonium phosphate dissolved in 50% acetonitrile. The monobasic ammonium phosphate suppresses ionization of matrix clusters and enhances low mass range detection of peptides (10). PMFs and MS/MS-fragmentation data were collected for each sample. Both MALDI-TOF and TOF/TOF approaches were used, since the extreme abundance of albumin fragments interfered with the PMF identification for many of the bands. The acquired peptide data from SELDI-TOF MS were searched via Mascot (Matrix Science, Boston, MA, USA) database search engine and the International Protein Index (IPI) human protein database. For the MALDI-TOF MS/MS spectra, data were processed using an integrated GPSExplorer interface from Applied Biosystems coupled to a local Mascot Server (Matrix Science) with database searches against the entire NCBInr database. In either case, standard Mascot statistical criteria were used to indicate positive protein identification.
Quantitative testing of the identified proteins
We measured plasma and urinary transferrin (Tf), plasma ceruloplasmin (Cp), plasma α-1-acid-glycoprotein (AGP, also: orosomucoid), as well as plasma and urine lipocalin-type prostaglandin-D synthetase (L-PGDS) by immunonephelometry (Dade Behring BNII Prospect, Marburg, Germany). Urinary Cp was quantified by ELISA (Human Ceruloplasmin ELISA Quantitation Kit; Genway Biotech, Inc., San Diego, CA, USA); and urinary AGP by ELISA (Human Orosomucoid ELISA Quantitation Kit; Genway Biotech, Inc., San Diego, CA, USA).
Statistical analysis
We inspected the central tendency, dispersion and skewness of PS-protein levels and found them to all fit well into normal distributions after log transformation. Hence, log transformed PS-protein levels were used in the formal statistical analyses, and results related to the PS-proteins are presented using geometric means after their log transformed means were converted back to original values by taking exponentials.
Using data from the first study visit only, PS-protein concentrations in three groups of SLE patients (children with active LN; those with inactive LN and those without LN) and among two control groups with active JIA or inactive JIA, respectively, were tested for statistically important differences under a multivariate fixed effect model (or ANOVA model) framework, after adjusting for patients’ characteristics such as age, gender and race. Active LN was defined as a renal SLEDAI score > 0 or a renal BILAG score > 1 (11), respectively. For analysis of longitudinal data with repeated observations on each patient, a random effect (i.e. the patient) was added to the previous fixed effect models to account for within-patient correlation. Receiver operating characteristic (ROC) curves were constructed, and the area under each ROC curve (AUCROC: range 0 – 1) was calculated (12) to assess performance of the PS-proteins in discriminating between the presence versus absence of LN activity. An AUCROC of 1.0 represents a perfect biomarker whereas a value of 0.5 is no better than expected by chance. Statistical computations were conducted using SAS version 9.1 (SAS, Cary, NC, USA) software. P-values < 0.05 were considered statistically significant.
RESULTS
Characteristics of Patients with SLE
Characteristics of the children with SLE are summarized in Table 1. At study entry, there were 26 patients with active LN (renal SLEDAI > 0), 36 with inactive LN, and 36 who never had LN. For SLE patients a total of 347 visits (249 follow-up visits) were available for analysis with all children having had a minimum of two study visits.
Table 1.
Demographics and disease features of children with SLE at baseline
| Parameter | n | Mean (SE) | Number of patients with score = 0 | |
|---|---|---|---|---|
| Gender (female: male) | 81:17 | 98 | ||
| Race | American-Indian | 2 | ||
| Asian | 2 | |||
| African-American | 76 | |||
| Pacific Islander | 1 | |||
| Caucasian | 17 | |||
| Ethnicity | Hispanic | 12 | ||
| Non-Hispanic | 86 | |||
| Age (in years) | 98 | 15.4 (0.49) | ||
| Disease duration (in years) | 98 | 4.3 (1.11) | ||
| Current medications | Prednisone (mg/day) | 67 | 17.2 (2.0) | |
| Azathioprine, mycophenolate mofetil, methotrexate | 52 | |||
| Cyclophosphamide§ | 23 | |||
| Angiotensin blocking agents | 29 | |||
| No lupus nephritis | 36 | |||
| Lupus nephritis* | WHO Class 2 | 5 | ||
| WHO Class 3 | 17 | |||
| WHO Class 4 | 22 | |||
| WHO Class 5 | 18 | |||
| Disease activity | SLEDAI** | |||
| renal | 2.0 (0.35) | 68 | ||
| extrarenal | 98 | 3.5 (0.31) | 25 | |
| BILAG¶ | ||||
| renal | 2.0 (0.34) | 61 | ||
| extrarenal | 3.4 (0.31) | 13 | ||
| Disease damage | SDI|| | |||
| renal | 98 | 0.07 (0.03) | 91 | |
| extrarenal | 0.45 (0.11) | 72 | ||
Six patients were treated with cyclophosphamide at enrollment.
Classified as per Churg J, Bernstein J, Glassock RJ. Renal disease: classification and atlas of glomerular diseases. 2nd ed. New York: Igaku-Shoin; 1995.
SLEDAI: Systemic Lupus Erythematosus Disease Activity Index, version 2k; 0 = inactive disease. Renal disease activity equal the sum of the items addressing renal disease. Extrarenal disease activity considers scores of all but the renal domain items.
BILAG: British Isles Lupus Activity Group index. Renal disease activity corresponds to the renal domain score of the BILAG. Extrarenal disease activity considers all other BILAG domain scores. Alphabetical BILAG score were converted into numericals as follows: A=9; B=3; C=1; D or E = 0; 0 = inactive disease.
SDI: Systemic Lupus International Collaborating Clinics/ACR Damage Index; 0 = no damage.
Identification LN Protein Signature Proteins
The LN urinary PS consisted of eight proteins with MS peaks-to-charge ratios (m/z) of 2.763, 22, 23, 44, 56, 79, 100, and 133 kDa (3). We identified the 23 kDa band as L-PGDS; the 56 kDa as AGP or orosomucoid; the 79 kDa as Tf; and the 133 kDa as Cp, respectively. The remaining four bands of the LN urinary PS represented albumin or albumin fragments, which were not further, examine for their relationship to the features of LN because we were unable to extract any specific proteins that might have been contained in these bands by our methods.
In the following, the plasma concentrations of Tf, Cp, AGP and L-PGDS are reported in mg/dL; urinary concentrations of the PS-proteins are reported as 1) absolute concentrations in the urine: Tf and L-PGDS in mg/dL, Cp and AGP in ng/mL urine, respectively; 2) corrected for urinary creatinine (in mg/mL); and 3) corrected for nonselective proteinuria as estimated by the protein:creatinine ratio.
Differences between JIA and SLE
At the first study visit, the mean ± SE of the urinary concentrations (per ml urine) of Tf, Cp, AGP (all p < 0.0001), and L-PGDS (p < 0.0025) were markedly higher in children with SLE than those with JIA. Plasma levels of all PS-proteins were comparable between children with SLE vs. JIA, with the exception of plasma Tf, where levels were higher with JIA than SLE (JIA: 304 ± 9.6 vs. SLE: 250 ± 5.8; p < 0.002).
PS-proteins in SLE patients
PS-proteins were unrelated to SLE patients’ weight, gender, race, ethnicity (Hispanic/Non-Hispanic), the use of angiotensin blocking medications, or disease duration.
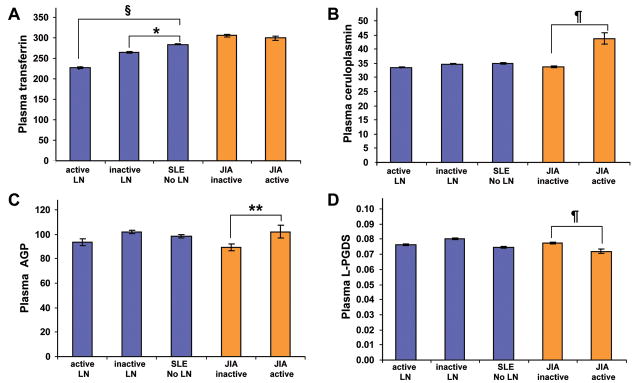
Comparison of PS-protein plasma concentrations in the three groups of SLE patients (no LN, active LN, inactive LN) and the two groups of controls (active JIA, inactive JIA) is shown in Figure 1, supporting only statistically significant differences of Tf plasma levels among the groups of SLE patients, while the plasma levels of the other PS-proteins appear not to be relevant biomarkers of LN.
Figure 1. Plasma concentrations of the PS-proteins.
Values are means and SE in mg/dL. Significant differences are based on Tukey post-hoc testing. The histograms show the level of the PS-proteins, Tf (A), Cp (B), AGP (C) and L-PGDS (D). SLE patients with active lupus nephritis (LN), inactive LN or without LN are compared with groups were defined by the SLEDAI. Twenty children with active and 10 with inactive JIA served as controls. Significant differences between groups are indicated as follows: *= P<0.02; **= P<0.01; ¶= P<0.001; §= P<0.0002.
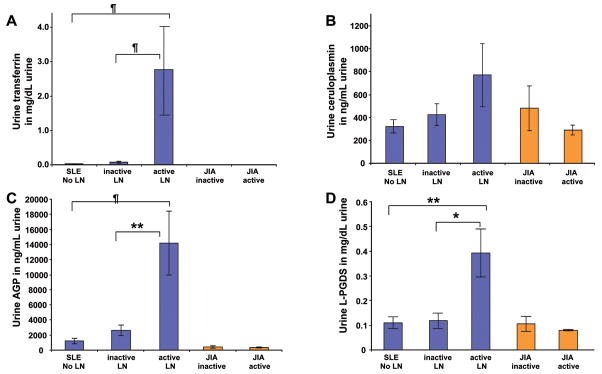
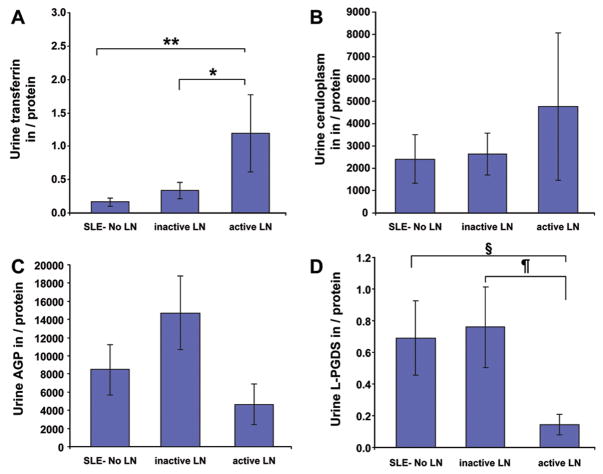
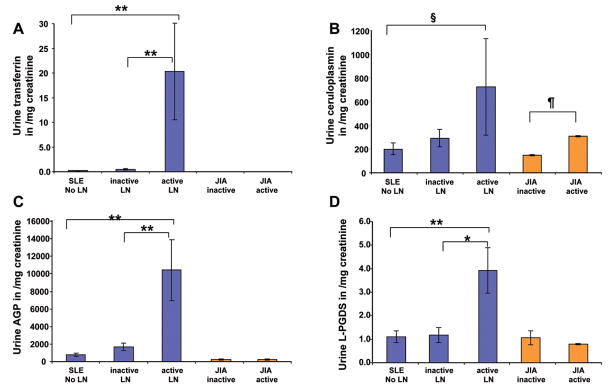
Figures 2–4 depict the comparison of urinary concentrations of the PS-proteins considering absolute levels (per dL or mL of urine; Figure 2), levels standardized by urinary creatinine (Figure 3) or nonselective proteinuria (Figure 4), respectively. SLE patients with active LN had much higher levels of all PS-proteins per mL or dL of urine (Figure 2) or standardized by urinary creatinine (Figure 3), with statistically significant differences indicated in the figures. Corrected for nonselective proteinuria, only urinary Tf and CP levels continued to be higher with active LN, suggesting that their excretion increases to a higher degree than nonselective proteinuria. Conversely, although urinary AGP and L-PGDS were significantly higher with active compared to inactive LN (see Table 3), increases were less pronounced than those of nonselective proteinuria. Significant differences of between SLE groups persisted only for Tf and L-PGDS once PS-proteins were corrected for nonselective proteinuria, as is indicated in Figure 4. Use of the BILAG instead of the SLEDAI to classify SLE groups according to LN activity yielded comparable results as shown for the SLEDAI in Figures 1 to 4.
Figure 2. Urinary concentration of the PS-proteins.
Values are means and SE. Significant differences are based on Tukey post-hoc testing. The histograms show urinary concentrations of Tf (A), Cp (B), AGP (C) and L-PGDS (D) for the groups defined as Figure 1. Uncorrected PS-protein levels (per mL or dL of urine) are depicted. Significant differences between groups are indicated as follows: * = P<0.004; **= P<0.002; ¶= P<0.00001.
Figure 4. Urinary concentration of the PS-proteins.
Values are means and SE. Significant differences are based on Tukey post-hoc testing. The histograms show urinary concentrations of Tf (A), Cp (B), AGP (C) and L-PGDS (D) for the groups defined as Figure 1. PS-protein excretion standardized by nonselective proteinuria is depicted with significant differences between groups indicated as follows: * = P<0.05; ** = P<0.005; § = P<0.02; ¶ = P<0.009.
Figure 3. Urinary concentration of the PS-proteins.
Values are means and SE. Significant differences are based on Tukey post-hoc testing. The histograms show urinary concentrations of Tf (A), Cp (B), AGP (C) and L-PGDS (D) for the groups defined as Figure 1. PS-protein excretion standardized by urine creatinine (mg/mL urine) is shown. Significant differences between groups are indicated as follows: *= P<0.0005; **= P<0.0001; §= P<0.05; ¶ = P<0.001.
Table 3.
Area under the ROC curve of protein biomarkers for LN activity and damage¶
| Measure of LN | SLEDAI-2k | BILAG | SDI |
|---|---|---|---|
|
Protein signature- proteins | |||
| Plasma Tf | 0.54 | 0.57 | 0.69 |
| Urinary Tf | 0.80 | 0.81 | 0.84 |
| Urinary Cp | 0.68 | 0.80 | 0.73 |
| Urinary AGP | 0.76 | 0.81 | 0.87 |
| Urinary L-PGDS | 0.71 | 0.73 | 0.79 |
| All LN Protein Biomarkers | 0.84 | 0.85 | 0.88 |
|
Traditional renal biomarkers | |||
| Creatinine clearance | 0.45 | 0.50 | 0.39 |
| Protein-creatinine ratio | 0.91 | 0.85 | 0.76 |
| Complement C3 | 0.58 | 0.63 | 0.75 |
| Complement C4 | 0.60 | 0.49 | 0.64 |
PS-proteins differentiate better than traditional measures with the features of LN
Table 2 provides a cross-sectional comparison of PS-proteins and traditional laboratory measures for their ability to identify active LN or renal damage. Besides the protein:creatinine ratio, the levels of none of the other traditional laboratory markers, including serum creatinine and BUN (data not shown), demonstrated important differences among patients with active vs. inactive LN. Amongst seven SLE patients with renal damage, both the levels of plasma Tf and all urinary PS proteins were significantly higher than in SLE patients without renal damage. However, six patients with renal damage had concomitantly active LN.
Table 2.
Concurrent validity in SLE - Biomarker concentrations with active and inactive lupus nephritis§
| Disease activity | Protein Signature proteins* | Traditional biomarkers** | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Urinary Tf | Urinary Cp /1000 |
Urinary AGP /1000 |
Urinary L-PGDS |
GFR | C3 | C4 | Protein creatinine ratio |
||
| SLEDAI renal | absent | 0.01 (0.01, 0.03) | 0.49 (0.36, 0.66) | 1.55 (1.15,2.08) | 0.09 (0.07,0.12) | 78 (67, 133) | 64.3 (56.1, 73.7) | 9.5 (8.1, 11.0) | 0.05 (0.04,0.08) |
| present | 0.09 (0.04, 0.22) | 0.98 (0.63, 1.50) | 2.53 (1.74,3.68) | 0.16 (0.12,0.22) | 64 (50, 128) | 57.9 (47.3, 70.9) | 11.3 (8.9, 14.3) | 0.32 (0.19,0.52) | |
| p-value | <0.0001 | 0.004 | 0.005 | <0.0001 | NS¶ | NS | NS | <0.0001 | |
| SLEDAI extrarenal | absent | 0.02 (0.01, 0.04) | 0.45 (0.29, 0.72) | 1.45 (0.98,2.15) | 0.09 (0.07,0.13) | 67 (52,86) | 72.5 (58.8, 89.3) | 13.3 (10.4,16.9) | 0.09 (0.05,0.17) |
| present | 0.03 (0.01, 0.06) | 0.66 (0.48, 0.90) | 1.91 (1.40,2.61) | 0.11 (0.09,0.15) | 76 (66,89) | 59.4 (52.0, 67.9) | 9.01 (7.8,10.4) | 0.09 (0.06,0.14) | |
| p-value | NS | NS | NS | NS | NS | NS | 0.007 | NS | |
| BILAG renal | absent | 0.01 (0.00, 0.02) | 0.44 (0.32, 0.61) | 1.33 (0.98,1.81) | 0.09 (0.07,0.12) | 80 (68, 95) | 66.5 (57.5, 77.0) | 9.7 (8.2,11.5) | 0.04 (0.03, 0.06) |
| present | 0.11 (0.05, 0.26) | 0.92 (0.62, 1.37) | 2.67 (1.88,3.79) | 0.14 (0.11,0.19) | 64 (52, 79) | 55.3 (46.2, 66.3) | 10.2 (8.3, 12.5) | 0.23 (0.16,0.35) | |
| p-value | <0.0001 | 0.003 | <0.0001 | 0.004 | NS | NS | NS | <0.0001 | |
| BILAG extrarenal | absent | 0.01 (0.00, 0.03) | 0.54 (0.31, 0.95) | 1.54 (0.97,2.44) | 0.08 (0.06,0.12) | 63 (46, 86) | 72.7 (56.6, 93.4) | 10.8 (7.9,14.6) | 0.11 (0.05, 0.23) |
| present | 0.03 (0.01, 0.06) | 0.60 (0.44, 0.81) | 1.80 (1.32,2.46) | 0.12 (0.09,0.15) | 76 (66, 88) | 59.9 (52.6, 68.3) | 9.73 (8.4,11.3) | 0.09 (0.06,0.13) | |
| p-value | 0.032 | NS | NS | NS | NS | NS | NS | NS | |
| SDI renal | absent | 0.02 (0.01, 0.04) | 0.59 (0.41, 0.85) | 1.53 (1.07,2.17) | 0.10 (0.07,0.13) | 82 (69, 98) | 62.4 (53.7,72.4) | 9.10 (7.5,11.0) | 0.09 (0.06, 0.15) |
| present | 2.48 (0.15, 42) | 2.02 (0.55, 7.44) | 9.65 (2.23, 41.65) | 0.36 (0.13,0.99) | 51 (28, 91) | 42.2 (25.0, 71.4) | 10.6 (5.5,20.6) | 0.83 (0.17,3.98) | |
| p-value | 0.001 | 0.080 | 0.018 | 0.016 | NS | NS | NS | 0.011 | |
| SDI extrarenal | absent | 0.02 (0.01, 0.04) | 0.65 (0.43, 0.98) | 1.47 (0.99,2.20) | 0.09 (0.07,0.13) | 84 (69,102) | 61.9 (52.2, 73.2) | 8.92 (7.2,11.0) | 0.10 (0.06,0.18) |
| present | 0.08 (0.02, 0.37) | 0.65 (0.32, 1.31) | 2.61 (1.28,5.31) | 0.17 (0.10,0.29) | 68 (49, 94) | 57.2 (43.2, 75.7) | 10.0 (7.1,14.3) | 0.14 (0.06,0.36) | |
| p-value | NS | NS | NS | NS | NS | NS | NS | NS | |
Values are geometric means (95% confidence intervals).
Urinary concentrations of Tf in mg/dL, Cp in ng/mL, AGP in ng/mL, and L-PGDS in mg/dL.
C3 and C4 all in mg/dL; GFR in mL/min/1.73m2; protein:creatinine ratio (no units).
NS not significant.
For additional legend, see Tables 1
The AUCROC was calculated to assess the concurrent validity of the PS-proteins and the traditional renal biomarkers to diagnose the presence of active LN as measured by the SLEDAI and the BILAG, respectively (Table 3). Individual urinary PS-Proteins in performed all in the fair to good range according to current ROC interpretation standards (12), they were all better diagnostic markers of active LN than traditional renal biomarkers (all AUCROC <0.63) with the exception was the urine protein:creatinine ratio with an AUCROC at 0.91 (SLEDAI) and 0.85 (BILAG), respectively.
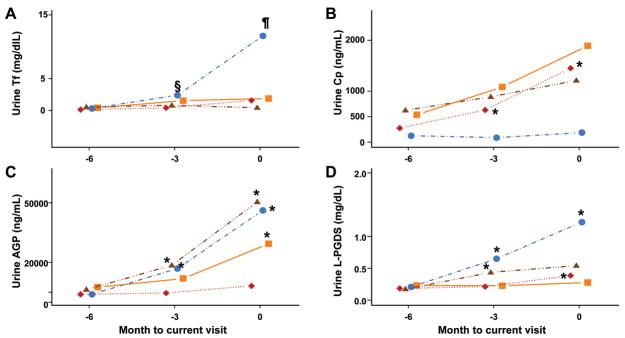
PS-Proteins may predict the future course of LN
Figure 5 depicts the absolute levels of urinary PS-proteins over time in relation to changes in LN activity as measured by the SLEDAI. Urinary levels of Tf, AGP and L-PGDS significantly increased (SLEDAI: all p < 0.009) at least 3 months prior to the clinical diagnosis of worsening LN activity (month 0) and continued to be elevated at the time of the clinically diagnosed LN flare. Cp levels did not show a consistent pattern with the course of LN. None of the traditional biomarkers, including the protein: creatinine ratio was predictive of the course of LN. Similar results were observed when the BILAG instead of the SLEDAI was used to determine the course of LN.
Figure 5. Changes of the PS-proteins in relationship to future changes in LN activity.
Values are geometric means of uncorrected urinary levels of Tf (A), Cp (B), AGP (C) and L-PGDS (D) at months -6, -3 and 0, respectively. Month 0 is the time point when the clinical diagnosis of the course of LN is made and months -3 corresponds to the timepoint of 3 months prior to the clinical diagnosis of the LN flare. ‘Improved LN’ describes the course of LN with decreasing renal SLEDAI scores; ‘worse LN’ describes the course of LN associated with an increase of the renal SLEDAI scores; ‘stable active LN’ describes patients with stable renal SLEDAI scores > 0; and ‘inactive LN’ describes the course of continuously inactive LN (renal SLEDAI = 0). Significant differences in the levels between two consecutive visits are indicated in the plots as follows. § = P< 0.009; ¶ = P<0.0001; * = P<0.001. The above defined LN courses are depicted as follows: Improved LN, squares; Worsened LN, circles; Stable active LN, triangles; Inactive LN, diamonds.
DISCUSSION
There is a need for high-quality accurate biomarkers to judge LN activity and renal damage with SLE. In this study, we chose a proteomic approach for the discovery of novel LN biomarkers and identified a set of PS-proteins (i.e. Tf, Cp, AGP and L-PGDS). In quantitative analysis, particularly urinary rather than plasma levels of the PS-proteins increased significantly with the presence of active LN. The increased urinary excretion of the PS-proteins could not simply be explained by concomitant increases of nonselective proteinuria. Different from all traditional laboratory measures of LN, we have initial evidence that Tf, AGP and L-PGDS constitute predictive biomarkers of worsening LN activity.
We found high and increasing urinary levels of Tf associated with active LN and impending worsening of LN flares. Tf is co-regulated by interferon-α, involved in iron delivery, and the innate immune system. Plasma Tf levels were correlated to global SLE disease activity in the past (13). Thus, our study confirms these earlier findings in SLE, and new evidence is provided that urinary Tf excretion may represent a predictive biomarker for LN.
Cp plays a critical physiological role in controlling the rate of iron efflux from cells with mobilizable iron (14). Like Tf, plasma Cp has been recommended as a marker of global SLE disease activity (13, 15). Conversely, our results support that urinary Cp concentrations only differ with LN activity rather than extrarenal disease activity. Possibly, because urinary Cp levels vary widely in SLE, we were unable to detect a meaningful relationship to the course of LN.
AGP is a predictive biomarker for diabetic renal disease (16), and we provide initial evidence that this is also the case for LN. More importantly, urine concentrations of AGP (similar to Tf and L-PGDS) appear useful to anticipate LN flares, i.e. these markers may allow clinicians to preemptively adjust therapy prior to the appearance of overt worsening of LN. Previous studies proposed plasma AGP to be a biomarker of SLE global disease activity (17, 18). Our results support this (data not shown) but we also provide evidence that urinary AGP constitutes a biomarker of LN rather than extrarenal disease activity.
Lipocalins play a role in many biological processes, among them immune responses and prostaglandin synthesis. L-PGDS, a lipocalin, is involved in nitric oxide regulation and the induction of apoptosis in the kidney. L-PGDS has not been previously found to be a LN biomarker. Urine and plasma L-PGDS are considered sensitive indicators of chemotherapy-induced renal damage and diabetes-associated hypertension (19,20). We found urinary L-PGDS unrelated to the creatinine clearance in both JIA and SLE; L-PGDS also did not significantly change with cyclophosphamide exposure in SLE patients. Reason for these discrepancies might be that our patients had all normal or only minimally decreased creatinine clearance, and that there was at least a 3-week time-lag from a previous intravenous cyclophosphamide dose.
We confirm the result of a recent study that Tf and AGP are part of the LN protein signature (21) but were unable to detect hepcidin, a protein recently identified via SELDI-TOF MS (22). This might have been due to differences in the experimental approach, including the study of children instead of adults with SLE. Biomarker discovery in children may have a distinct advantage as children generally lack co-morbidities, increasing the likelihood of detecting specific biomarkers.
Further research is needed to compare the usefulness of the PS-proteins to that of other recently discovered LN biomarkers, including neutrophil gelatinase associate lipocalin (11) or various urinary chemokines.
At present, there is no agreement how best to interpret protein biomarkers derived by protein profiling. Hence, it remains unclear whether absolute uncorrected concentrations in the urine, levels standardized by urine creatinine excretion, or even protein-adjusted levels are most suited for analysis. Like others (22), however, our data support that correction of absolute biomarker levels for urinary creatinine does not importantly change the principal properties of the PS-proteins for detecting LN activity. Even when we corrected the PS-proteins for the nonselective proteinuria, statistically significant differences between SLE patients without LN, inactive LN, and active LN often persisted. The results of the latter analyses will need to be confirmed under consideration of the limitations of protein:creatinine ratios, possibly correcting for microalbuminuria rather than nonselective proteinuria (23).
At present, there is no universally accepted gold standard for the measurement of LN activity. For this study, we chose to use the two widely accepted SLE disease activity indices (SLEDAI, BILAG). The relevance of our findings is strengthened by the fact that the PS-proteins performed similarly well to capture and anticipate the course of LN, irrespective of the index used. Compared to the BILAG, the SLEDAI considers only proteinuria and the urinary sediment in the calculation of the LN activity score. Thus, a close association between the protein:creatinine ratio and LN activity and an AUCROC were expected when using the SLEDAI. Given the sensitivity of moderately elevated protein:creatinine ratio to angiotensin blocking medications and its unproven ability for predicting LN flares, we consider Tf, CP, AGP and L-PGDS to be promising LN biomarkers, as their levels do not seem to change with the use of angiotensin inhibiting medications and even help discriminate patients who are at risk of a future LN flare.
A limitation of our study may be that we could not consider effectively the findings of renal biopsy specimens for their relationship to the PS-proteins. We did not have a sufficient numbers of urine samples collected at the time of kidney biopsy to present sound results on the relationship of the PS-proteins and the complex features of LN histology. Similarly, the relationship of the PS-proteins to the presence or development of renal damage will need further study, as the majority of children with renal damage also had active LN concurrently.
In summary, Tf, Cp, AGP and L-PGDS are promising LN biomarkers. Their initial validation suggests superior measurement properties compared to most traditional LN biomarkers and that Tf, AGP and L-PGDS are candidates of a novel set of predictive LN biomarkers. Additional validation studies are mandatory to evaluate the usefulness of such a LN Renal Panel to predict the course of LN, the severity of kidney pathology, and the future development of renal damage with SLE.
Acknowledgments
This study is supported by a clinical research grant NIAMS P60 AR47784 and the CCHMC Translational Research Initiative. H.I.B. and P.D. are supported by a grant from the Alliance for Lupus Research and P.D. is supported by grants from the NIH/NIDDK (RO1-DK53289) and Department of Defense (PR064328). K.W. is supported by T32 AR074594 and the NIH Loan Repayment Program.
U.C.: Ms. Thelma Kathman for her technical assistance, Dr. Susan Thompson (sample storage), Drs. Bob Colbert, Thomas Griffin, Alexei Grom, Daniel Lovell, Amber Khan, (all data collection); Shannen Nelson & Jamie Meyers-Eaton (study coordinator & data collection), Shweta Srivastava (sample processing).
Drs. James N. Jarvis and Michael Hendrickson, Yukiko Kimura, Suzanne Li and Jennifer Weiss, Cabral, Petty, Houghton, Turvey, Guzman and Malleson, Earl D. Silverman, Nora Singer, Linda Wagner-Weiner, James Nocton and Calvin Williams, Elizabeth Roth-Wojicki, PNP, Marsha Malloy (all data collection), Lisa Kempke, Linda Menifee, Mary Ellen Riordan, America Uribe MD, Tony Hong MS, Lawrence Ng, Michelle Walette, Becky Puplava, Dina Blair, Joshua Kapfhamer, & Noshaba Khan (study coordinators). Drs. Gary Ross & Nathan Harris for data analyses.
Abbreviations
- AGP
α1-acid-glycoprotein
- BILAG
British Isles Lupus Activity Group
- Cp
Ceruloplasmin
- JIA
Juvenile Idiopathic Arthritis
- L-PGDS
Lipocalin-type prostaglandin-D synthetase
- LN
Lupus nephritis
- MALDI-TOF MS
Matrix Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry
- PS
Protein signature
- NGAL
Neutrophil gelatinase associated lipocalin
- PMFs
Peptide mass fingerprints
- AUCROC
Area under the receiver operating characteristic
- SELDI-TOF MS
Surface Enhanced Laser Desorption/Ionization-Time of Flight Mass Spectrometry
- SLE
Systemic lupus erythematosus
- SDI
Systemic Lupus International Collaborating Clinics/ACR Damage Index
- Tf
Transferrin
- SLEDAI
2k-version of the Systemic Lupus Erythematosus Disease Activity Index
References
- 1.Faurschou M, Starklint H, Halberg P, Jacobsen S. Prognostic factors in lupus nephritis: diagnostic and therapeutic delay increases the risk of terminal renal failure. J Rheumatol. 2006;33:1563–1569. [PubMed] [Google Scholar]
- 2.Wu T, Xie C, Wang HW, Zhou XJ, Schwartz N, Calixto S, Mackay M, Aranow C, Putterman C, Mohan C. Elevated urinary VCAM-1, P-selectin, soluble TNF receptor-1, and CXC chemokine ligand 16 in multiple murine lupus strains and human lupus nephritis. J Immunol. 2007;179:7166–7175. doi: 10.4049/jimmunol.179.10.7166. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki M, Ross GF, Wiers K, Nelson S, Bennett M, Passo MH, Devarajan P, Brunner HI. Identification of a urinary proteomic signature for lupus nephritis in children. Pediatr Nephrol. 2007;22:2047–2057. doi: 10.1007/s00467-007-0608-x. [DOI] [PubMed] [Google Scholar]
- 4.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 5.Brunner HI, Feldman BM, Bombardier C, Silverman ED. Sensitivity of the Systemic Lupus Erythematosus Disease Activity Index, British Isles Lupus Assessment Group Index, and Systemic Lupus Activity Measure in the evaluation of clinical change in childhood-onset systemic lupus erythematosus. Arthritis Rheum. 1999;42:1354–1360. doi: 10.1002/1529-0131(199907)42:7<1354::AID-ANR8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 6.Hay EM, Bacon PA, Gordon C, Isenberg DA, Maddison P, Snaith ML, Symmons DP, Viner N, Zoma A. The BILAG index: a reliable and valid instrument for measuring clinical disease activity in systemic lupus erythematosus. Q J Med. 1993;86:447–458. [PubMed] [Google Scholar]
- 7.Brunner HI, Silverman ED, To T, Bombardier C, Feldman BM. Risk factors for damage in childhood-onset systemic lupus erythematosus: cumulative disease activity and medication use predict disease damage. Arthritis Rheum. 2002;46:436–444. doi: 10.1002/art.10072. [DOI] [PubMed] [Google Scholar]
- 8.Jarrold B, DeMuth J, Greis K, Burt T, Wang F. An effective skeletal muscle prefractionation method to remove abundant structural proteins for optimized two-dimensional gel electrophoresis. Electrophoresis. 2005;26:2269–2278. doi: 10.1002/elps.200410367. [DOI] [PubMed] [Google Scholar]
- 9.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 10.Smirnov IP, Zhu X, Taylor T, Huang Y, Ross P, Papayanopoulos IA, Martin SA, Pappin DJ. Suppression of alpha-cyano-4-hydroxycinnamic acid matrix clusters and reduction of chemical noise in MALDI-TOF mass spectrometry. Anal Chem. 2004;76:2958–2965. doi: 10.1021/ac035331j. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki M, Wiers KM, Klein-Gitelman MS, Haines KA, Olson J, Onel KB, O’Neil K, Passo MH, Singer NG, Tucker L, Ying J, Devarajan P, Brunner HI. Neutrophil gelatinase-associated lipocalin as a biomarker of disease activity in pediatric lupus nephritis. Pediatr Nephrol. 2008;23:403–412. doi: 10.1007/s00467-007-0685-x. [DOI] [PubMed] [Google Scholar]
- 12.Hanley JA, Negassa A, Edwardes MD, Forrester JE. Statistical analysis of correlated data using generalized estimating equations: an orientation. Am J Epidemiol. 2003;157:364–375. doi: 10.1093/aje/kwf215. [DOI] [PubMed] [Google Scholar]
- 13.Yilmaz A, Sari RA, Gundogdu M, Kose N, Dag E. Trace elements and some extracellular antioxidant proteins levels in serum of patients with systemic lupus erythematosus. Clin Rheumatol. 2005;24:331–335. doi: 10.1007/s10067-004-1028-y. [DOI] [PubMed] [Google Scholar]
- 14.Hellman NE, Gitlin JD. Ceruloplasmin metabolism and function. Annu Rev Nutr. 2002;22:439–458. doi: 10.1146/annurev.nutr.22.012502.114457. [DOI] [PubMed] [Google Scholar]
- 15.Hrycek A, Pochopien-Kenig G, Scieszka J. Selected acute phase proteins and interleukin-6 in systemic lupus erythematosus patients treated with low doses of quinagolide. Autoimmunity. 2007;40:217–222. doi: 10.1080/08916930701306817. [DOI] [PubMed] [Google Scholar]
- 16.Gomes MB, Nogueira VG. Acute-phase proteins and microalbuminuria among patients with type 2 diabetes. Diabetes Res Clin Pract. 2004;66:31–39. doi: 10.1016/j.diabres.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Lacki JK, Leszczynski P, Kelemen J, Muller W, Mackiewicz SH. Cytokine concentration in serum of lupus erythematosus patients: the effect on acute phase response. J Med. 1997;28:99–107. [PubMed] [Google Scholar]
- 18.Meijer C, Huysen V, Smeenk RT, Swaak AJ. Profiles of cytokines (TNF alpha and IL-6) and acute phase proteins (CRP and alpha 1AG) related to the disease course in patients with systemic lupus erythematosus. Lupus. 1993;2:359–365. doi: 10.1177/096120339300200605. [DOI] [PubMed] [Google Scholar]
- 19.Hirawa N, Uehara Y, Yamakado M, Toya Y, Gomi T, Ikeda T, Eguchi Y, Takagi M, Oda H, Seiki K, Urade Y, Umemura S. Lipocalin-type prostaglandin d synthase in essential hypertension. Hypertension. 2002;39:449–454. doi: 10.1161/hy0202.102835. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa M, Hirawa N, Tsuchida T, Eguchi N, Kawabata Y, Numabe A, Negoro H, Hakamada-Taguchi R, Seiki K, Umemura S, Urade Y, Uehara Y. Urinary excretions of lipocalin-type prostaglandin D2 synthase predict the development of proteinuria and renal injury in OLETF rats. Nephrol Dial Transplant. 2006;21:924–934. doi: 10.1093/ndt/gfk009. [DOI] [PubMed] [Google Scholar]
- 21.Varghese SA, Powell TB, Budisavljevic MN, Oates JC, Raymond JR, Almeida JS, Arthur JM. Urine biomarkers predict the cause of glomerular disease. J Am Soc Nephrol. 2007;18:913–922. doi: 10.1681/ASN.2006070767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Jin M, Wu H, Nadasdy T, Nadasdy G, Harris N, Green-Church K, Nagaraja H, Birmingham DJ, Yu CY, Hebert LA, Rovin BH. Biomarkers of lupus nephritis determined by serial urine proteomics. Kidney Int. 2008;74:799–807. doi: 10.1038/ki.2008.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birmingham DJ, Rovin BH, Shidham G, Bissell M, Nagaraja HN, Hebert LA. Relationship between albuminuria and total proteinuria in systemic lupus erythematosus nephritis: diagnostic and therapeutic implications. Clin J Am Soc Nephrol. 2008;3:1028–1033. doi: 10.2215/CJN.04761107. [DOI] [PMC free article] [PubMed] [Google Scholar]