Abstract
We recently reported a β3-decapeptide, βWWI-1, that binds a validated gp41 model in vitro and inhibits gp41-mediated fusion in cell culture. Here we report six analogs of βWWI-1 containing a variety of non-natural side chains in place of the central tryptophan of the WWI-epitope. These analogs were compared on the basis of both gp41 affinity in vitro and fusion inibition in live, HIV-infected cells. One new β3-peptide, βWXI-a, offers a significantly improved CC50/EC50 ratio in the live cell assay.
Linear peptides derived from the C-terminus of HIV-1 gp41 (C-peptides) are potent HIV fusion inhibitors1. These molecules bind to the gp41 N-peptide region and inhibit an intramolecular protein-protein interaction that drives fusion of viral and host cell membranes2–4. Previous work has shown that the protein-protein interface consists of a highly conserved pocket on the N-peptide surface that is occupied by three C-peptide side chains: Trp-628, Trp-631 and Ile-6353–5. These three residues comprise the WWI epitope3–5. Simple6–9 and constrained10–13 α-peptides, aromatic foldamers14, peptide-small molecule conjugates15, and small molecules16, 17 that bind this N-peptide surface pocket inhibit gp41-mediated cell fusion with IC50 values ranging from 250 pM for α-peptides to 5 μM for small molecules. We previously reported a set of β3-decapeptides that present a WWI epitope on one face of a salt bridge18–21 and macrodipole-stabilized22 14-helix23, 24. One of these molecules, βWWI-1, binds a validated gp41 model in vitro and inhibits gp41-mediated fusion in cell culture25. Past work by Chan and co-workers6 demonstrated the importance of the three epitope residues, particularly the central tryptophan, in both gp41 affinity and viral infectivity. Here we report six analogs of βWWI-1 containing a variety of nonnatural side chains in place of the central tryptophan of the WWI-epitope. These analogs were compared on the basis of both gp41 affinity in vitro and fusion inibition in live, HIV-infected cells. One new β3-peptide, βWXI-a, offers a significantly improved CC50/EC50 ratio in the live cell assay.
We synthesized a small collection of β3-decapeptides (βWXI-a—f) containing a variety of nonnatural side chains in place of the central tryptophan of the WWI-epitope (Figure 1). These nonnatural residues included those with both entended or alternative π-systems (βWXI-b,d) and halogen-substituted aromatic rings (βWXI-a,c,e,f) to probe the steric and electronic requirements of the N-peptide surface pocket in the context of a β3-peptide. βWWI-1, a previously described β-peptide HIV fusion inhibitor25, was synthesized as a positive control.
Figure 1.
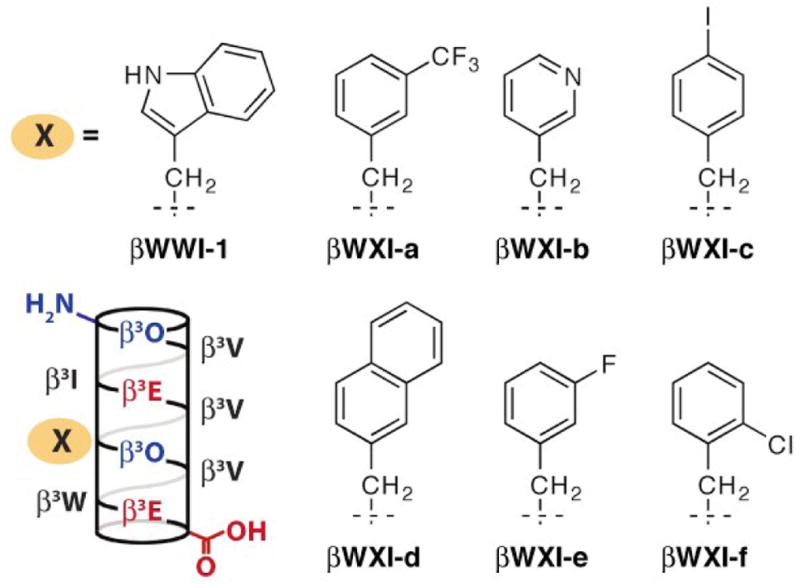
Helical net representations of βWWI-125 and βWXI-a—f. β3-homoamino acids are identified by the single letter code used for the corresponding α-amino acid. O represents ornithine.
Each β-peptide was labeled at the N-terminus with 6-(fluorescein-5(6)-carboxamido) hexanoic acid N-hydroxy-succinimidyl ester (Flu) and employed in a direct fluorescence polarization (FP) assay to determine its affinity for IQN17, a fusion protein containing 17-residues from the gp41 N-terminus joined to a 29 residue isoleucine zipper10. IQN17 exists as a stable trimer in solution10 and effectively recreates the N-peptide surface pocket for C-peptide-like ligands. β-peptides βWXI-a-fFlu bound IQN17 with equilibrium dissociation constants between 12.1 μM (βWXI-d) and 105.4 mM (βWXI-b) (Table 1, Figure 1A). With the exception of pyridyl-containing βWXI-b, all new β-peptides bound IQN17 about as well as βWWI-1 (KD = 16.5 ± 0.6 μM). These results are significant if not surprising, given the loss off affinity that typically results from altering the central tryptophan residue6, 25.
Table 1.
Binding affinity and MTT assay results for peptides βWWI-1 and βWXI-a-f.
| Peptide | KDa (μM) | EC50b (μM) | CC50c (μM) | Selectivity (CC50/EC50) |
|---|---|---|---|---|
| βWWI-1 | 16.5 ± 0.6 | 56 ± 5.9 | 100 ± 19.6 | 1.8 |
| βWXI-a | 10.2 ± 0.3 | 19 ± 1.7 | 150 ± 3.3 | 7.9 |
| βWXI-b | 104.5 ± 8.2 | > 250 | > 250 | N/Ad |
| βWXI-c | 14.1 ± 2.3 | 8.9 ± 1.3 | 23 ± 4.6 | 2.6 |
| βWXI-d | 12.2 ± 0.9 | 8.2 ± 5.0 | 23 ± 5.9 | 2.8 |
| βWXI-e | 15.7 ± 1.3 | 18 ± 3.7 | 50 ± 4.5 | 2.8 |
| βWXI-f | 13.3 ± 1.4 | 8.8 ± 7.4 | 31 ± 9.1 | 3.5 |
For 50% binding of IQN17; binding curves were measured in triplicate.
For 50% protection in MT-2 cells; antiviral curves used triplicate samples at each concentration.
For 50% inhibition of MT-2 cell growth; toxicity curves also used triplicate samples.
Not active.
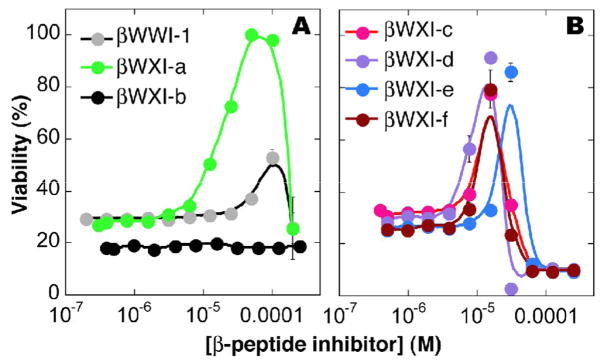
All seven β-peptides were evaluated for the ability to promote cell survival in an MTT colorimetric assay26, 27. In this method, MT-2 human T-cells are plated with varying concentrations of β-peptide inhibitor and cultured with wild-type HIV-1 IIIB28–30. After 5 days incubation, the number of live cells that remain is determined by addition of (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT). MTT is reduced in the mitochondria of live cells to formazan (λmax = 595 nm) and quantified by UV. The EC50 values reported represent the β-peptide concentration required to achieve 50% survival of infected cells (Figure 2; Table 1).
Figure 2.
Plots illustrating survival of HIV-infected MT-2 cells in the presence of the indicated β-peptide. EC50 values reported represent the β-peptide concentration required to achieve 50% survival of infected cells; CC50 values represent the concentration required to achieve 50% survival of uninfected cells. Viability was measured with an MTT colorimetric assay26, 27 as described in the text.
The EC50 values of β-peptides βWXI-a through f vary between 8.2 μM (βWXI-d) and > 250 μM (βWXI-b). With the exception of βWXI-b, which is inactive (EC50> 250 μM), all of the new β-peptides (8.2 μM ≤ EC50 ≤ 19 μM) are more potent than βWWI-1 (EC50 = 56 μM) at promoting the survival of HIV-infected cells. Interestingly, two of the most potent new β-peptides (βWXI-c and f) share little structural similarity, with halogen substituents at para- and ortho- positions, respectively. βWXI-a and e, with EC50 = 18–19 μM, share a fluorine-containing substituent at the meta position of the phenyl side chain.
We also compared the new β-peptides in terms of cytotoxicity, determined as the viability of uninfected cells in the presence of inhibitor alone (Figure S1, Table 1). The CC50 values reported represent the β-peptide concentration required to inhibit MT-2 cell growth by 50%. CC50 values range from 31 μM (βWXI-f) to > 250 μM (βWXI-b), with a value of 100 μM for βWWI-1. Interestingly, although βWXI-d and f are characterized by the lowest EC50 values, each was cytotoxic at concentrations close to this value, with CC50/EC50 ratios less than 4. Importantly, one new β-peptide, βWXI-a, exhibits an CC50/EC50 ratio of 8, representing a significant improvement relative to βWXI-1 as well as βWXI-c-f.
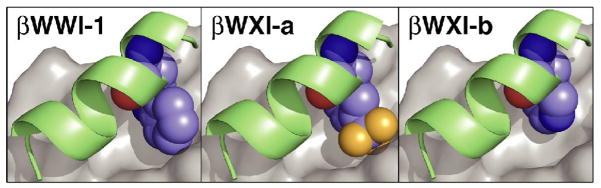
The ability of βWXI-a to bind IQN17 and inhibit fusion in the MTT assay may be partially rationalized by a simple model in which the indole side chain of the central tryptophan is replaced by the central aromatic side chains of our β-peptides (Figure 3). A crystal structure of the gp41 fusion peptide solved by Sia et. al.11 depicts the epitope-containing β-peptide C14linkmid bound to IQN17 and clearly shows association between the indole side chain and the N-peptide surface pocket. Substitution of the Trp indole ring of C14linkmid with the m-trifluoromethylphenyl side chain in βWXI-a suggests that the trifluoro-methylbenzene side chain is a reasonable structural mimic of the indole ring, whereas the 3-pyridyl side chain is not. Although βWXI-a is not as potent as Fuzeon in the MTT assay (EC50 = 37.5 nM), it has a significantly lower mass (1457 Da vs. 4492 for Fuzeon), and higher metabolic and proteolytic stablity31–35. Furthermore, due to the ability of the 14-helical scaffold to tolerate changes to the epitope face, it may be possible to identify β3-peptides with further improved activity and decreased toxicity through combinatorial optimization36, 37.
Figure 3.
Models representing the interface between the N-peptide surface pocket (grey) and the central epitope residue of βWWI-1, βWXI-a and βWXI-b. Models were constructed using the programs Spartan (Wavefunction, Inc.) and PyMOL (DeLano Scientific, LLC) and the high-resolution structure10 1gzl of the α-peptide C14linkmid bound to IQN17.
Supplementary Material
Acknowledgments
This work was partially supported by NIH grant GM49551 (KSA) and a research award from Bristol-Myers Squibb (ADB). ADB thanks EJP and JSA for helpful discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Proc Natl Acad Sci USA. 1994;91:9770. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu M, Blacklow SC, Kim PS. Nat Struct Mol Biol. 1995;2:1075. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 3.Chan DC, Fass D, Berger JM, Kim PS. Cell. 1997;89:263. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 4.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Nature. 1997;387:426. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 5.Tan K, Liu JH, Wang JH, Shen S, Lu M. Proc Natl Acad Sci USA. 1997;94:12303. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan DC, Chutkowski CT, Kim PS. Proc Natl Acad Sci USA. 1998;95:15613. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin BS, Ryu JR, Ahn K, Yu YG. AIDS Res Hum Retroviruses. 2000;16:1797. doi: 10.1089/08892220050195757. [DOI] [PubMed] [Google Scholar]
- 8.Sia SK, Kim PS. Proc Natl Acad Sci. 2003;100:9756. doi: 10.1073/pnas.1733910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welch BD, VanDemark AP, Heroux A, Hill CP, Kay MS. Proc Natl Acad Sci USA. 2007;104:16828. doi: 10.1073/pnas.0708109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Cell. 1999;99:103. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 11.Sia SK, Carr PA, Cochran AG, Malashkevich VN, Kim PS. Proc Natl Acad Sci USA. 2002;99:14664. doi: 10.1073/pnas.232566599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dwyer JJ, Wilson KL, Davison DK, Freel SA, Seedorff JE, Wring SA, Tvermoes NA, Matthews TJ, Greenberg ML, Delmedico MK. Proc Natl Acad Sci USA. 2007;104:12772. doi: 10.1073/pnas.0701478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Xiao Y, Song H, Liang Q, Ju D, Chen X, Lu H, Jing W, Jiang S, Zhang L. J Biol Chem. 2008;283:11126. doi: 10.1074/jbc.M800200200. [DOI] [PubMed] [Google Scholar]
- 14.Ernst JT, Kutzki O, Debnath AK, Jiang S, Lu H, Hamilton AD. Angew Chem Int Ed. 2002;41:278. doi: 10.1002/1521-3773(20020118)41:2<278::aid-anie278>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 15.Ferrer M, Kapoor TM, Strassmaier T, Weissenhorn W, Skehel JJ, Oprian D, Schreiber SL, Wiley DC, Harrison SC. Nat Struct Mol Biol. 1999;6:953. doi: 10.1038/13324. [DOI] [PubMed] [Google Scholar]
- 16.Debnath AK, Radigan L, Jiang S. J Med Chem. 1999;42:3203. doi: 10.1021/jm990154t. [DOI] [PubMed] [Google Scholar]
- 17.Frey G, Rits-Volloch S, Zhang XQ, Schooley RT, Chen B, Harrison SC. Proc Natl Acad Sci USA. 2006;103:13938. doi: 10.1073/pnas.0601036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arvidsson PI, Rueping M, Seebach D. Chem Commun. 2001:649. [Google Scholar]
- 19.Cheng RP, DeGrado WF. J Am Chem Soc. 2001;123:5162. doi: 10.1021/ja010438e. [DOI] [PubMed] [Google Scholar]
- 20.Rueping M, Mahajan YR, Jaun B, Seebach D. Chem-Eur J. 2004;10:1607. doi: 10.1002/chem.200305571. [DOI] [PubMed] [Google Scholar]
- 21.Kritzer JA, Hodsdon ME, Schepartz A. J Am Chem Soc. 2005;127:4118. doi: 10.1021/ja042933r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart SA, Bahadoor ABF, Matthews EE, Qiu XJ, Schepartz A. J Am Chem Soc. 2003;125:4022. doi: 10.1021/ja029868a. [DOI] [PubMed] [Google Scholar]
- 23.Bautista AD, Craig CJ, Harker EA, Schepartz A. Curr Opin Chem Biol. 2007;11:685. doi: 10.1016/j.cbpa.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kritzer JA, Stephens OM, Guarracino DA, Reznik SK, Schepartz A. Bioorg Med Chem. 2005;13:11. doi: 10.1016/j.bmc.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens OM, Kim S, Welch BD, Hodsdon ME, Kay MS, Schepartz A. J Am Chem Soc. 2005;127:13126. doi: 10.1021/ja053444+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tai-Shun L, Mei-Zhen L, Mao-Chin L, Pai SB, Dutschman GE, Yung-Chi C. Biochem Pharmacol. 1994;47:171. doi: 10.1016/0006-2952(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 27.Ray AS, Yang Z, Chu CK, Anderson KS. Antimicrob Agents Chemother. 2002;46:887. doi: 10.1128/AAC.46.3.887-891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popovic M, Read-Connole E, Gallo R. Lancet. 1984;324:1472. doi: 10.1016/s0140-6736(84)91666-0. [DOI] [PubMed] [Google Scholar]
- 29.Popovic M, Sarngadharan MG, Read E, Gallo RC. Science. 1984;224:497. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 30.Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich B, Josephs SF, Doran ER, Rafalski JA, Whitehorn EA, Baumeister K, Ivanoff L, Petteway SR, Pearson ML, Lautenberger JA, Papas TS, Ghrayeb J, Chang NT, Gallo RC, Wong-Staal F. Nature. 1985;313:277. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 31.Seebach D, Overhand M, Kuhnle FNM, Martinoni B, Oberer L, Hommel U, Widmer H. Helv Chim Acta. 1996;79:913. [Google Scholar]
- 32.Seebach D, Abele S, Schreiber JV, Martinoni B, Nussbaum AK, Schild H, Schulz H, Hennecke H, Woessner R, Bitsch F. Chimia. 1998;52:734. [Google Scholar]
- 33.Frackenpohl J, Arvidsson PI, Schreiber JV, Seebach D. Chem BioChem. 2001;2:445. doi: 10.1002/1439-7633(20010601)2:6<445::aid-cbic445>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 34.Wiegand H, Wirz B, Schweitzer A, Camenisch GP, Perez MIR, Gross G, Woessner R, Voges R, Arvidsson PI, Frackenpohl J, Seebach D. Biopharm Drug Dispos. 2002;23:251. doi: 10.1002/bdd.334. [DOI] [PubMed] [Google Scholar]
- 35.Wiegand H, Wirz B, Schweitzer A, Gross G, Perez MIR, Andres H, Kimmerlin T, Rueping M, Seebach D. Chem Biodiversity. 2004;1:1812. doi: 10.1002/cbdv.200490136. [DOI] [PubMed] [Google Scholar]
- 36.Kritzer JA, Luedtke NW, Harker EA, Schepartz A. J Am Chem Soc. 2005;127:14584. doi: 10.1021/ja055050o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray JK, Farooqi B, Sadowsky JD, Scalf M, Freund WA, Smith LM, Chen J, Gellman SH. J Am Chem Soc. 2005;127:13271. doi: 10.1021/ja052733v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.