Summary
As terminally differentiated vital cells, neurons may be specialized to fight viral infections without undergoing cellular self-destruction. The cellular lysosomal degradation pathway, autophagy, is emerging as one such mechanism of neuronal antiviral defence. Autophagy has diverse physiological functions, such as cellular adaptation to stress, routine organelle and protein turnover, and innate immunity against intracellular pathogens, including viruses. Most of the in vivo evidence for an antiviral role of autophagy is related to viruses that specifically target neurons, including the prototype alphavirus, Sindbis virus, and the α-herpesvirus, herpes simplex virus type 1 (HSV-1). In the case of HSV-1, viral evasion of autophagy is essential for lethal encephalitis. As basal autophagy is important in preventing neurodegeneration, and induced autophagy is important in promoting cellular survival during stress, viral antagonism of autophagy in neurons may lead to neuronal dysfunction and/or neuronal cell death. This review provides background information on the roles of autophagy in immunity and neuroprotection, and then discusses the relationships between autophagy and viral neurovirulence.
Introduction
Autophagy is a highly conserved mechanism for recycling cellular contents by delivering cytoplasmic material to the lysosome for degradation (Yorimitsu and Klionsky, 2005; Xie and Klionsky, 2007). To date, yeast genetic analyses have identified at least 31 genes (ATG genes) that are required for autophagy (e.g. ATG1–10, 12–14, 16–18, 29 and 31) and related pathways; many of these are conserved in mammals (e.g. atg1, 4–10, 12, 16 and 18) (see Fig. 1 for overview of autophagy pathway and machinery). Through genetic deletion or knockdown of these autophagy genes in model eukaryotic organisms, we have learned much about the biological functions of autophagy in the past decade (reviewed in Levine and Klionsky, 2004; Klionsky, 2007). The major known biological roles of autophagy relate to its ability to recycle nutrients and energy, and to its ability to degrade unwanted cytoplasmic constituents. With respect to recycling, the autophagy pathway plays an evolutionary conserved role in nutrient stress adaptation and cell survival. With respect to the degradation of unwanted cytoplasmic constituents, autophagy is the primary pathway for ridding cells of damaged organelles and toxic aggregate-prone proteins, and thereby, functions in lifespan extension and protection against neurodegeneration.
Fig. 1.
The autophagy pathway and machinery.
A. Schematic illustration of events in autophagosome initiation, formation and maturation. Autophagy functions at basal levels in many cell types, but can be upregulated in response to stresses, such as nutrient deprivation and infection.
B. Upstream signalling events converge on a class III PI3-K complex containing Beclin 1, as well as positive (UVRAG, Bif1, Ambra-1) and negative (Bcl-2/XL) regulators of initiation.
C. The initiation complex is thought to generate PIP-3, which recruits additional Atg proteins involved in two ubiquitin-like conjugation systems that are required for autophagosome elongation and completion. Autophagosomes then fuse with lysosomes containing hydrolytic enzymes that degrade the contents, which are recycled for use in protein synthesis or energy production.
As part of its integral role in maintaining cellular homeostasis, autophagy also defends cells from assaults by diverse microorganisms. Increasing evidence suggests that autophagy functions in innate and adaptive immunity, and these studies have recently been reviewed elsewhere in detail (Levine and Deretic, 2007; Schmid and Munz, 2007). The ability of autophagy to serve as a ‘topological conduit’, delivering cytoplasmic material to cellular compartments that are continuous with the extracellular space, helps explain some previous paradoxes in viral immunology and also provides insight into the role of autophagy in the activation of innate and adaptive immune responses. First, many peptides presented by MHC class II molecules were found to be of cytoplasmic origin (Chicz et al., 1993; Rammensee et al., 1999; Dongre et al., 2001), but it was unknown how these peptides were delivered to MHC class II loading compartments. It is now clear that autophagy can deliver cytosolic antigens for class II presentation in antigen presenting cells (Nimmerjahn et al., 2003; Dengjel et al., 2005; Paludan et al., 2005; Schmid et al., 2007), providing one potential explanation for this paradox. Second, some viruses were found to require an intermediate cytoplasmic replication step to activate type I interferon (IFN) production via endosomal toll-like receptor (TLR) stimulation, but it was unknown why viral cytoplasmic replication was required. A recent study indicates that autophagy may deliver viral nucleic acids to TLR7-containing endosomes to stimulate innate immune activation (Lee et al., 2007), providing an explanation for this second paradox.
By sequestering large portions of the cytosol in a double membrane autophagosome that is delivered to the lysosome (Fig. 1A), autophagy also has the unique capacity to target for degradation intact intracellular pathogens that reside in vesicles or in the cytoplasm. The targeting of cytoplasmic microorganisms to autophagosomes for degradation may be an important clearance mechanism, and has been termed xenophagy (or ‘to eat what is foreign’) (Levine, 2005). Two landmark studies were published nearly simultaneously demonstrating a role for autophagy genes in bactericidal activity. Nakagawa et al. (2004) demonstrated that group A Streptococcus localizes to autophagosomes and is degraded by autophagy in wild-type cells, but can invade into and survive in the cytoplasm of cells deleted of an essential autophagy gene, atg5. Gutierrez et al. (2004) first demonstrated that mycobacterial-containing phagosomes were targeted by autophagy, and that autophagic restriction of mycobacterial growth could be pharmacologically and immunologically enhanced. Subsequent studies have confirmed these findings and extended the list of intracellular bacteria and parasites targeted by autophagy to include Listeria monocytogenes, Salmonella enterica, Francisella tularensis and Toxoplasma gondii, which have been reviewed elsewhere (Swanson, 2006; Huang and Klionsky, 2007; Levine and Deretic, 2007).
Autophagy also targets RNA and DNA viruses, including Sindbis virus and herpes simplex virus, for sequestration and elimination (Seay et al., 2005; Levine, 2006; Talloczy et al., 2006), although other viruses such as poliovirus may subvert the pathway, utilizing components of the autophagic machinery to generate double-membrane compartments that serve as sites of assembly of viral RNA replication complexes (Jackson et al., 2005; Taylor and Kirkegaard, 2007; Lee et al., 2008). The first paper published on a mammalian autophagy protein, Beclin 1, described an antiviral role for this protein in Sindbis virus encephalitis in mice (Liang et al., 1998). A more recent study has indicated that fatal encephalitis caused by herpes simplex virus type 1 (HSV-1) requires the inactivation of Beclin 1 by an HSV-1-encoded neurovirulence factor (Orvedahl et al., 2007). These findings, coupled with independent lines of research demonstrating that autophagy is important in protection against neurodegeneration, underscore a potential critical role for the host autophagic machinery in defence against viral infections of the central nervous system (CNS). This review will provide background on the role of autophagy in neurons, highlight the role of autophagy in CNS viral diseases, and speculate on potential implications of viral inhibition of autophagy in acute and chronic CNS viral pathogenesis.
Autophagy and neurons
In post-mitotic long-lived cell types, such as neurons, where protein quality control and organelle turnover are more important than in self-renewing cell types, basal and stress-induced autophagy may be especially important in maintaining cellular health (Boland and Nixon, 2006) (Fig. 2, left). Basal levels of neuronal autophagy are neuroprotective, as mice with neuronal-specific deletion of atg5 or atg7 accumulate ubiquitinated protein aggregates and develop neurodegenerative disease (Hara et al., 2006; Komatsu et al., 2006). Further, mice with Purkinje cell-specific atg7 deletion develop dystrophic Purkinje cell axons, suggesting that autophagy may protect against axonal pathology associated with neurodegeneration (Komatsu et al., 2007).
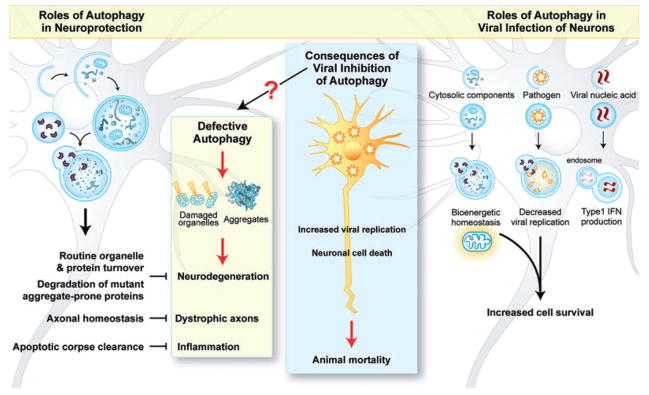
Fig. 2.
Autophagy in neurons and infection. Basal levels of autophagy in neurons are important for preventing the accumulation of damaged organelles or protein aggregates in neurons, promoting axonal homeostasis, and for efficient clearance of cell corpses (left). Autophagy is also important to restrict viral replication and promote survival of infected neurons (right). Speculatively, autophagy may also promote survival through maintaining bioenergetic stores within infected neurons, and may contribute to immune activation through IFN production (right). Viral inhibition of autophagy results in increased viral replication and neuronal death, ultimately leading to animal mortality (middle). It is also possible that viral inhibition of autophagy may underlie chronic neuronal diseases through interruption of homeostatic neuronal autophagy (arrow with question mark).
In addition to routine organelle and protein quality control functions, neuronal autophagy is important in preventing the accumulation of aggregate-prone disease-causing proteins (Rubinsztein et al., 2005). In Caenorhabditis elegans, autophagy prevents mutant polyQ-containing protein aggregate accumulation and neurodegeneration (Jia et al., 2007; Khan et al., 2008). Mammalian cells may target ubiquitinated proteins, including the polyQ-containing mutant huntingtin disease protein (htt), for autophagy by the adaptor protein, p62/SQSTM1 (Bjorkoy et al., 2005), which contains a ubiquitin-binding motif and also directly interacts with the autophagosomal protein, LC3 (Pankiv et al., 2007). Importantly, upregulation of autophagy enhances the clearance of mutant htt and tau proteins in Drosophila models of neurodegeneration (Ravikumar et al., 2002; Berger et al., 2006). There are also data suggesting that autophagy may play a role in preventing Alzheimer’s disease (AD) (Nixon, 2007). In worms, autophagy promotes the clearance of AD-related protein (Florez-McClure et al., 2007). Furthermore, a recent study indicates that Beclin 1 expression is decreased in brain tissue from human patients with AD, and β-amyloid accumulation and AD pathology are exacerbated by heterozygous deletion of beclin 1 in mice (Pickford et al., 2008). Together, these findings indicate that autophagy is likely essential for maintaining normal neuronal function and for the prevention of neurodegenerative diseases.
Autophagy and alphavirus encephalitis
Although functions of autophagy in innate and adaptive immunity have been described in in vitro studies for diverse pathogens, much of our understanding of autophagy as a bona fide in vivo host defence pathway is based on studies with two different neurotropic viruses, Sindbis virus and HSV-1. Sindbis virus is a positive-stranded RNA virus in the alphavirus genus. It is transmitted by mosquitoes and causes mild rheumatological diseases in humans, but serves as a useful mouse model for studying human alphavirus encephalitides, such as those caused by Eastern and Western equine encephalitis viruses (Strauss and Strauss, 1994). In mice, Sindbis virus produces an age-dependent fatal encephalitis that can be prevented by inhibitors of apoptotic cell death, including cellular Bcl-2, cowpox virus-encoded CrmA, and cellular regulators of the mitochondrial membrane permeability transition, such as the peripheral benzodiazepine receptor (Levine et al., 1996; Nava et al., 1998; Johnston et al., 2001). In a search to understand the mechanism by which Bcl-2 regulates Sindbis virus pathogenesis, a yeast two-hybrid screen was performed of a mouse brain library using Bcl-2 as a bait, leading to the identification of a novel Bcl-2-interacting coiled-coil protein, Beclin 1.
Beclin 1 is the mammalian homologue of yeast Atg6 and the first identified mammalian autophagy protein. Enforced neuronal expression of Beclin 1 was found to protect mice against fatal Sindbis virus encephalitis, reduce CNS Sindbis virus replication and reduce Sindbis virus-induced neuronal apoptosis (Liang et al., 1998). Although nearly two decades earlier cytomegalovirus (CMV) and herpes simplex virus virions were noted within autophagosomes (Smith and de Harven, 1978), the observations by Liang et al. provided the first direct evidence that an autophagy gene played an important role in the host antiviral response.
Interestingly, the expression of Beclin 1 lacking the Bcl-2-binding domain not only failed to protect against fatal Sindbis virus encephalitis, but also resulted in increased neuronal death (Liang et al., 1998). Subsequent studies have shown that this domain of Beclin 1 is dispensable for its autophagy activity, but is essential for physiological regulation of its levels of autophagy activity by Bcl-2-like family members. The expression of Beclin 1 mutants that cannot bind to Bcl-2 in mammalian cells results in non-physiological levels of autophagy that are associated with cell death that is inhibitable by siRNA targeted against downstream autophagy genes (Pattingre et al., 2005). The studies in mice infected with Sindbis virus expressing wild-type Beclin 1 or a Bcl-2-binding defective mutant of Beclin 1 suggest that, similar to in vitro, physiological regulation of Beclin 1-dependent autophagy by Bcl-2 may also be critical to ensure that autophagy is an adaptive, pro-survival process, rather than a maladaptive, pro-death response during CNS viral infection.
The ability of enforced neuronal expression of Beclin 1 to protect against lethal Sindbis virus encephalitis suggests a protective role for autophagy in host defence against a neurotropic infection. However, several unanswered questions remain. First, as the yeast orthologue of Beclin 1, Atg6, functions in both autophagy and other class III PI3-K-dependent intracellular trafficking pathways (Kihara et al., 2001), it is formally possible that autophagy-independent functions of Beclin 1 are responsible for its neuroprotective effects. Arguing against this, autophagy-defective mutants of Beclin 1 fail to protect against lethal Sindbis virus encephalitis (X.H. Liang and B. Levine, unpubl. data), and CNS-specific deletion of other autophagy genes increases Sindbis virus neurovirulence (S. MacPherson et al., unpubl. data). Second, it is unclear whether the effect of Beclin 1 on animal mortality is related to reduced Sindbis virus replication, reduced Sindbis virus-induced apoptosis, other protective effects of autophagy on neurons during viral infection or other actions of autophagy in the innate immune response in neurons. Third, it is unclear whether Sindbis virus replication is reduced as a result of xenophagic degradation, increased innate immune signalling or other as-of-yet-undefined mechanisms. Fourth, it is unclear whether Sindbis virus-induced neuronal apoptosis is decreased merely as a consequence of reduced replication, or whether such a reduction reflects a more direct antiapoptotic role of autophagy in virally infected neurons. Future studies examining the pathogenesis of Sindbis virus and other neurotropic viruses in mice lacking functional autophagy genes in their neurons should help address these questions.
Autophagy and HSV-1 encephalitis
HSV-1, a double-stranded DNA α-herpesvirus, infects the majority of humans. It causes a variety of clinical syndromes of differing severity, ranging from mild mucocutaneous disease to the most common cause of sporadic viral encephalitis. HSV-1 encephalitis is a devastating disease, with a 70% fatality rate in the absence of treatment and a 20–30% fatality rate with presently available antiviral therapy (Whitley and Roizman, 2001). A major breakthrough in understanding HSV-1 neurovirulence was the identification of the HSV-1 neurovirulence factor ICP34.5, which is essential for fatal encephalitis both in mice (Chou et al., 1990) and in humans (Harrow et al., 2004). The C-terminal domain of ICP34.5 (the GADD34 domain) recruits a host phosphatase, PP1α, to reverse PKR-mediated eIF2α phosphorylation and host cell translational shutoff (He et al., 1997). However, mutant strains that retain the ability to reverse host cell shutoff remain avirulent in vivo, suggesting additional functions of ICP34.5 are required for neurovirulence (He et al., 1996; Markovitz et al., 1997). In the past few years, the ability to inhibit host autophagy has been identified as an additional function of ICP34.5, and a recent study has provided strong evidence that this function is critical for HSV-1 neurovirulence (Orvedahl et al., 2007).
PKR, a double-stranded RNA-dependent serine-threonine kinase, stimulates autophagy induction through eIF2α phosphorylation, and ICP34.5 antagonizes this response (Talloczy et al., 2002). Furthermore, PKR-mediated autophagy functions to degrade virions and viral proteins in neurons infected with an ICP34.5-deleted virus (Talloczy et al., 2006). As PKR functions dually in host cell shutoff and autophagy, a critical question has been whether PKR-mediated autophagy, independent of PKR-dependent host cell shutoff, is important in host defence against HSV-1 encephalitis. This question has recently been addressed by identifying a second mechanism that ICP34.5 uses to antagonize host autophagy, and genetically dissecting the autophagy-inhibitory activity of ICP34.5 from its antagonism of PKR function. The N-terminal domain of ICP34.5, which is not required for PP1α binding or reversal of PKR-mediated eIF2α phosphorylation, binds to Beclin 1 and inhibits its autophagy function. Studies with a mutant HSV-1 virus that expresses an ICP34.5 deletion protein (lacking amino acids 68–87) incapable of binding to Beclin 1 underscore the importance of ICP34.5 antagonism of autophagy in HSV-1 neurovirulence (Orvedahl et al., 2007).
The ICP34.5 mutant HSV-1 virus (lacking amino acids 68–87) behaves similarly to wild-type HSV-1 with respect to replication in cultured cell lines, antagonism of eIF2α phosphorylation and blockade of host cell shutoff. However, unlike the wild-type virus, this mutant virus fails to inhibit virus-induced autophagy in primary neurons, and mice infected with this mutant virus exhibit decreased CNS viral replication, fewer numbers of dead neurons and increased survival compared with mice infected with wild-type virus (Orvedahl et al., 2007). Furthermore, the neurovirulence of this ICP34.5 mutant HSV-1 strain incapable of antagonizing Beclin 1 function is fully restored in PKR knockout mice, suggesting that PKR lies genetically upstream of Beclin 1 in the autophagy pathway in vivo (Orvedahl et al., 2007). Together, these HSV-1 and Sindbis virus mouse pathogenesis studies suggest that autophagy is important for protection against CNS disease caused by both DNA and RNA neurotropic viruses.
Multiple protective mechanisms of autophagy in CNS viral infections?
As discussed, autophagy promotes normal cellular functioning through constitutive turnover of potentially harmful cytosolic contents, and can be upregulated under stress conditions to promote survival. In addition, the routine turnover of cellular proteins by autophagy may prevent their accumulation and aggregation. As long-lived, non-regenerating cells, neurons rely extensively on autophagy to recycle organelles and proteins; without such basal autophagy, neurodegeneration can ensue (Boland and Nixon, 2006). Similarly, like cytosolic contents, neurons may also rely extensively on autophagy to remove cytoplasmic viral proteins or particles. In contrast to most cell types in the organism where viral clearance occurs largely through cytotoxic T lymphocyte killing of virally infected cells (Wong and Pamer, 2003), the destruction of virally infected neurons would be detrimental to the host, perhaps creating a unique reliance on non-cytolytic mechanisms of viral clearance. Along these lines, previous studies have demonstrated different non-cytolytic mechanisms for clearing Sindbis and other viruses from neurons, including antibody-mediated and cytokine-mediated restriction of viral gene expression (Levine et al., 1991; Kimura and Griffin, 2000; Binder and Griffin, 2001). Recent data with Sindbis virus and HSV-1 infections in mice raise the possibility that autophagy may represent a newly described non-cytolytic mechanism for clearing viruses from neurons (Fig. 2, right).
Beyond its role in viral clearance, it is possible that autophagy may exert other protective functions during infection with neurotropic viruses (Fig. 2, right). For example, another mechanism contributing to the protective effects of autophagy against viral CNS disease may be its role in promoting the survival of infected neurons. As noted above, decreased apoptosis is observed in the brains of mice infected with a Sindbis virus construct that expresses Beclin 1 and decreased neuronal death is observed in the brains of mice infected with a mutant HSV-1 virus that cannot inhibit Beclin 1-mediated autophagy. Besides direct cytoprotective effects in virally infected neurons, it is also possible that autophagy indirectly reduces neuronal death in virally infected brains by decreasing the total CNS viral burden and number of infected cells. Further, the homeostatic function of autophagy may not only help keep neurons alive, but also minimize neuronal dysfunction, by protecting neurons against endoplasmic reticulum (ER) and oxidative stress that occurs during infection and/or by facilitating the removal of protein aggregates and damaged organelles.
Two lines of evidence support the hypothesis that the autophagic machinery exerts antiviral activity directly in virally infected neurons. First, Beclin 1 overexpressed in neurons from a double-subgenomic Sindbis virus promoter is sufficient to protect against fatal encephalitis (Liang et al., 1998). Second, HSV-1 ICP34.5 exerts its autophagy inhibitory activity in HSV-1-infected neurons (Orvedahl et al., 2007). However, these observations do not preclude non-cell autonomous effects of neuronal autophagy. In addition to cell autonomous roles in infected cells of the CNS, autophagy may function to protect against viral infection at a tissue and organismal level by activating innate and adaptive immunity.
As noted above, it has been demonstrated that autophagy may deliver both cytosolic and exogenous antigens to MHC class II molecules (Schmid et al., 2007; A. Iwasaki et al., unpubl. data), and it is known that the CNS cell types, microglia and astrocytes, express class II molecules (Collawn and Benveniste, 1999). Therefore, it is possible that autophagy may contribute to class II presentation of viral antigens by microglia or astrocytes during CNS infection. Likewise, it is possible that the recently demonstrated role for autophagy in IFN production in peripheral dendritic cells (Lee et al., 2007) may be conserved in neurons, a cell type that is known to produce high amounts of IFN in response to Sindbis virus and other neurotropic viral infections (Delhaye et al., 2006) (Fig. 2, right). Thus, autophagy may play an integral role as both a primary barrier to productive viral infection in neurons and in activating immune responses in neurons and other cells in the CNS such as microglia and astrocytes.
While the precise mechanisms by which neuronal autophagy protects against CNS viral infection are unclear, it seems likely that autophagy functions to restrict viral replication. Ultrastructural analysis reveals the presence of Sindbis virions and ICP34.5 deletion mutant HSV-1 virions within autophagosomes (Seay, 2005; Levine, 2006; Talloczy et al., 2006). Moreover, metabolic labelling of viral proteins demonstrates increased rates of degradation in the ICP34.5 deletion mutant virus-infected cells compared with wild-type HSV-1 virus-infected cells (Talloczy et al., 2006). These results suggest that virions are targeted for xenophagic degradation in vitro, and studies in mice provide correlative evidence that xenophagic degradation also restricts viral replication of neurotropic viruses in vivo (Liang et al., 1998; Orvedahl et al., 2007).
Although Beclin 1 binding is important for promoting HSV-1 replication and neurovirulence in vivo, studies in atg5−/− MEFs suggest that autophagy may play a lesser role in restricting HSV-1 replication in vitro (Alexander et al., 2007). While PKR deletion or mutation of the eIF2α phosphorylation site is sufficient to restore wild-type levels of replication in MEFs infected with HSV-1 lacking the ICP34.5 gene (Talloczy et al., 2006), atg5 deletion does not significantly increase the replication of this mutant virus in MEFs. These findings suggest that, while ICP34.5 does inhibit autophagy in vitro, the primary determinant of efficient replication of HSV-1 in vitro is ICP34.5-mediated regulation of translational arrest rather than autophagy. Alexander and Leib (2008) speculate that the differences observed between the apparent effects of autophagy in restricting HSV-1 replication in vivo and in atg5−/− MEFs may be due to cell type-specific factors or differences between cell culture and in vivo environments. These differences highlight the potential unique importance of autophagy in restricting viral replication in neurons, which may explain the requirement for some neurovirulent viruses (e.g. HSV-1) to evade the autophagy pathway.
Viruses outsmart autophagy in the CNS
The central role of autophagy in innate and adaptive immunity may have provided the selective pressure for the evolution of viral escape mechanisms. As discussed above, the HSV-1-encoded neurovirulence factor ICP34.5 possesses at least two distinct mechanisms for blocking host autophagy: it blocks the PKR signalling pathway which is required for virus-induced autophagy and it also directly antagonizes Beclin 1-mediated autophagy (Orvedahl et al., 2007). The latter activity is required for HSV-1 acute fatal encephalitis, suggesting that viral evasion of autophagy may be essential for HSV-1 neurovirulence. Interestingly, other viruses that cause CNS disease may also block host autophagy. The β-herpesvirus CMV, which causes severe CNS infections in neonates and immunocompromised adults (Gandhi and Khanna, 2004; Griffiths, 2004), antagonizes host autophagy through an as-of-yet-undefined mechanism (Chaumorcel et al., 2008). Preliminary indications suggest that HIV may also negatively regulate host autophagy during macrophage infection (He and Orvedahl, 2007; Zhou and Spector, 2008), and it is intriguing to speculate that HIV disruption of autophagy may play a role in HIV-related encephalopathies. It is worth noting that in addition to the neurotropic α-herpesvirus, HSV-1, other viruses, such as the oncogenic γ-herpesviruses, murine γHV68 and Kaposi’s sarcoma-associated herpesvirus, that target extraneural sites also encode inhibitors of Beclin 1 and autophagy (Pattingre et al., 2005; Ku et al., 2008). While the significance of this is not yet known for γ-herpesvirus pathogenesis, these observations suggest that viral evasion of autophagy is not likely to be restricted to viruses that specifically infect the CNS.
Does viral inhibition of autophagy contribute to chronic CNS diseases?
In addition to contributing to neurovirulence during acute viral infection, one interesting speculation is that viral antagonism of host autophagy may also underlie sequelae of CNS viral infections (Fig. 2, middle). These long-term effects may include permanent neurological impairment that follows ‘recovery’ from acute encephalitis, post-infectious alterations in immunity and inflammation, and potential relationships between neurotropic viruses and neurodegenerative disorders. As basal autophagy is important in maintaining normal neuronal function, the transient interruption of autophagy by virally encoded inhibitors during acute encephalitis could contribute to the irreversible neurological dysfunction that is common following HSV-1 and other CNS infections. In addition, it is possible that this transient interruption of neuronal autophagy contributes to post-infectious autoimmune and inflammatory disorders. Recently, it has been demonstrated the autophagy promotes the clearance of dying cell corpses during mammalian development, through the generation of ‘eat me’ and ‘come get me’ signals (Qu et al., 2007). As efficient removal of apoptotic corpses is important in preventing autoimmunity, viral inhibition of autophagy may underlie autoimmune disorders following CNS infection. Viruses that have been associated with post-infectious autoimmune encephalitis (Tenembaum et al., 2007) have also been demonstrated to inhibit autophagy, including HSV-1 (Orvedahl et al., 2007) and CMV (Chaumorcel et al., 2008), so it is possible that viral inhibition of autophagy may contribute to development of these disorders after the acute infection has resolved.
Another open question is whether viral inhibition of autophagy may contribute to ‘non-infectious’ neurodegenerative diseases, such as AD. As discussed above, increasing evidence suggests that autophagy functions in preventing AD and other neurodegenerative diseases, and previous clinical epidemiological studies have linked HSV-1 infection and AD, in association with APOE4 alleles (Itzhaki et al., 1997; Hill et al., 2005). In vitro studies have also implicated a role for HSV-1 in the generation of the main components of amyloid plaques in AD brains (e.g. β-amyloid and abnormally phosphorylated tau) (Shipley et al., 2005; Wozniak et al., 2007; Itzhaki et al., 2008). Therefore, it is possible that HSV-1 inhibition of autophagy may contribute to development of AD, as has been postulated recently (Orvedahl et al., 2007; Itzhaki et al., 2008). The findings that ICP34.5 directly antagonizes host autophagy through its interaction with Beclin 1 (Orvedahl et al., 2007), coupled with the evidence that impaired Beclin 1 function increases β-amyloid accumulation and confers susceptibility to AD (Pickford et al., 2008), provide a potential molecular mechanistic link between HSV-1, AD and the protective role of autophagy in AD. Future studies using animal models will be required to determine the role of viral inhibition of autophagy in the development of neurodegenerative and autoimmune CNS diseases.
Conclusion
Autophagy plays an integral role in neuronal homeostasis and in response to stress, including viral infections. Genetic deletion of autophagy genes results in neurodegeneration, and the host autophagic machinery plays a role in protection against encephalitis caused by viruses from two distinct classes, alphaviruses and α-herpesviruses. Further, α-herpesviruses, and potentially other viruses that cause CNS disease, have evolved mechanisms to evade this pathway. While much remains to be learned about the precise mechanisms by which autophagy functions in CNS antiviral defence, these findings suggest novel approaches to treating CNS viral infections. Pharmacological agents that augment host autophagy or that block interactions between viral neurovirulence proteins and their host autophagy targets may prove to be beneficial in reducing the acute mortality and/or chronic morbidity associated with CNS viral infections. Future progress in this field will undoubtedly reveal important insight into the functions of autophagy in the CNS and into innate antiviral host defences in general.
Acknowledgments
We apologize to authors’ whose work could not be included due to space restrictions. We thank Renee Talley for administrative support, and Angela Diehl for expert medical illustration. The work in the authors’ laboratory was supported by NIH Grants R01 A10151367 (B.L.) and T32 A1007520 (A.O.), and an Ellison Medical Foundation Senior Scholars Award in Infectious Diseases.
References
- Alexander DE, Leib DA. Xenophagy in herpes simplex virus replication and pathogenesis. Autophagy. 2008;4:101–103. doi: 10.4161/auto.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J Virol. 2007;81:12128–12134. doi: 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- Binder GK, Griffin DE. Interferon-gamma-mediated site-specific clearance of alphavirus from CNS neurons. Science. 2001;293:303–306. doi: 10.1126/science.1059742. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Nixon RA. Neuronal macroautophagy: from development to degeneration. Mol Aspects Med. 2006;27:503–519. doi: 10.1016/j.mam.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Chaumorcel M, Souquère S, Pierron G, Codogno P, Esclatine A. Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy. 2008;4:46–53. doi: 10.4161/auto.5184. [DOI] [PubMed] [Google Scholar]
- Chicz RM, Urban RG, Gorga JC, Vignali DA, Lane WS, Strominger JL. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J, Kern ER, Whitley RJ, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- Collawn JF, Benveniste EN. Regulation of MHC class II expression in the central nervous system. Microbes Infect. 1999;1:893–902. doi: 10.1016/s1286-4579(99)00228-2. [DOI] [PubMed] [Google Scholar]
- Delhaye S, Paul S, Blakqori G, Minet M, Weber F, Staeheli P, Michiels T. Neurons produce type I interferon during viral encephalitis. Proc Natl Acad Sci USA. 2006;103:7835–7840. doi: 10.1073/pnas.0602460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongre AR, Kovats S, deRoos P, McCormack AL, Nakagawa T, Paharkova-Vatchkova V, et al. In vivo MHC class II presentation of cytosolic proteins revealed by rapid automated tandem mass spectrometry and functional analyses. Eur J Immunol. 2001;31:1485–1494. doi: 10.1002/1521-4141(200105)31:5<1485::AID-IMMU1485>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD. Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in C. elegans. Autophagy. 2007;3:569–580. doi: 10.4161/auto.4776. [DOI] [PubMed] [Google Scholar]
- Gandhi MK, Khanna R. Human cytomegalovirus: clinical aspects, immune regulation, and emerging treatments. Lancet Infect Dis. 2004;4:725–738. doi: 10.1016/S1473-3099(04)01202-2. [DOI] [PubMed] [Google Scholar]
- Griffiths P. Cytomegalovirus infection of the central nervous system. Herpes. 2004;11 (Suppl 2):95A–104A. [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Harrow S, Papanastassiou V, Harland J, Mabbs R, Petty R, Fraser M, et al. HSV1716 injection into the brain adjacent to tumour following surgical resection of high-grade glioma: safety data and long-term survival. Gene Ther. 2004;11:1648–1658. doi: 10.1038/sj.gt.3302289. [DOI] [PubMed] [Google Scholar]
- He B, Chou J, Liebermann DA, Hoffman B, Roizman B. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the gamma(1)34.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J Virol. 1996;70:84–90. doi: 10.1128/jvi.70.1.84-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Gross M, Roizman B. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Orvedahl A. 2007 keystone symposium on autophagy in health and disease. Autophagy. 2007;3:527–536. doi: 10.4161/auto.4595. [DOI] [PubMed] [Google Scholar]
- Hill JM, Gebhardt BM, Azcuy AM, Matthews KE, Lukiw WJ, Steiner I, et al. Can a herpes simplex virus type 1 neuroinvasive score be correlated to other risk factors in Alzheimer’s disease? Med Hypotheses. 2005;64:320–327. doi: 10.1016/j.mehy.2003.11.045. [DOI] [PubMed] [Google Scholar]
- Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- Itzhaki RF, Lin WR, Shang D, Wilcock GK, Faragher B, Jamieson GA. Herpes simplex virus type 1 in brain and risk of Alzheimer’s disease. Lancet. 1997;349:241–244. doi: 10.1016/S0140-6736(96)10149-5. [DOI] [PubMed] [Google Scholar]
- Itzhaki RF, Cosby SL, Wozniak MA. Herpes simplex virus type 1 and Alzheimer’s disease: the autophagy connection. J Neurovirol. 2008;14:1–4. doi: 10.1080/13550280701802543. [DOI] [PubMed] [Google Scholar]
- Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Hart AC, Levine B. Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy. 2007;3:21–25. doi: 10.4161/auto.3528. [DOI] [PubMed] [Google Scholar]
- Johnston C, Jiang W, Chu T, Levine B. Identification of genes involved in the host response to neurovirulent alphavirus infection. J Virol. 2001;75:10431–10445. doi: 10.1128/JVI.75.21.10431-10445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan LA, Yamanaka T, Nukina N. Genetic impairment of autophagy intensifies expanded polyglutamine toxicity in Caenorhabditis elegans. Biochem Biophys Res Commun. 2008;368:729–735. doi: 10.1016/j.bbrc.2008.01.150. [DOI] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Griffin DE. The role of CD8(+) T cells and major histocompatibility complex class I expression in the central nervous system of mice infected with neurovirulent Sindbis virus. J Virol. 2000;74:6117–6125. doi: 10.1128/jvi.74.13.6117-6125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X, et al. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Lee YR, Lei HY, Liu MT, Wang JR, Chen SH, Jiang-Shieh YF, et al. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374:240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Levine B. Autophagy in Antiviral Host Defense. Weinheim: Wiley-VCH; 2006. [Google Scholar]
- Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Levine B, Hardwick JM, Trapp BD, Crawford TO, Bollinger RC, Griffin DE. Antibody-mediated clearance of alphavirus infection from neurons. Science. 1991;254:856–860. doi: 10.1126/science.1658936. [DOI] [PubMed] [Google Scholar]
- Levine B, Goldman JE, Jiang HH, Griffin DE, Hardwick JM. Bc1-2 protects mice against fatal alphavirus encephalitis. Proc Natl Acad Sci USA. 1996;93:4810–4815. doi: 10.1073/pnas.93.10.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovitz NS, Baunoch D, Roizman B. The range and distribution of murine central nervous system cells infected with the gamma(1)34.5- mutant of herpes simplex virus 1. J Virol. 1997;71:5560–5569. doi: 10.1128/jvi.71.7.5560-5569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nava VE, Rosen A, Veliuona MA, Clem RJ, Levine B, Hardwick JM. Sindbis virus induces apoptosis through a caspase-dependent, CrmA-sensitive pathway. J Virol. 1998;72:452–459. doi: 10.1128/jvi.72.1.452-459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, Mautner J. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003;33:1250–1259. doi: 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120:4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, et al. The autophagy protein Beclin 1 is reduced in early Alzheimer’s disease and regulates Aβ accumulation in vivo. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, DiFiglia M, Heintz N, Nixon RA, Qin ZH, Ravikumar B, et al. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seay M, Dinesh-Kumar S, Levine B. Digesting oneself and digesting microbes: autophagy as a host response to viral infection. In: Palese P, editor. Modulation of Host Gene Expression and Innate Immunity by Viruses. Dordrecht: Springer; 2005. pp. 245–279. [Google Scholar]
- Shipley SJ, Parkin ET, Itzhaki RF, Dobson CB. Herpes simplex virus interferes with amyloid precursor protein processing. BMC Microbiol. 2005;5:48. doi: 10.1186/1471-2180-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, de Harven E. Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. III. Cytochemical localization of lysosomal enzymes in infected cells. J Virol. 1978;26:102–109. doi: 10.1128/jvi.26.1.102-109.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS. Autophagy: eating for good health. J Immunol. 2006;177:4945–4951. doi: 10.4049/jimmunol.177.8.4945. [DOI] [PubMed] [Google Scholar]
- Talloczy Z, Jiang W, Virgin HWT, Leib DA, Scheuner D, Kaufman RJ, et al. Regulation of starvation-and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talloczy Z, Virgin HWT, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J Virol. 2007;81:12543–12553. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenembaum S, Chitnis T, Ness J, Hahn JS. Acute disseminated encephalomyelitis. Neurology. 2007;68:S23–S36. doi: 10.1212/01.wnl.0000259404.51352.7f. [DOI] [PubMed] [Google Scholar]
- Whitley RJ, Roizman B. Herpes simplex virus infections. Lancet. 2001;357:1513–1518. doi: 10.1016/S0140-6736(00)04638-9. [DOI] [PubMed] [Google Scholar]
- Wong P, Pamer EG. CD8 T cell responses to infectious pathogens. Annu Rev Immunol. 2003;21:29–70. doi: 10.1146/annurev.immunol.21.120601.141114. [DOI] [PubMed] [Google Scholar]
- Wozniak MA, Itzhaki RF, Shipley SJ, Dobson CB. Herpes simplex virus infection causes cellular beta-amyloid accumulation and secretase upregulation. Neurosci Lett. 2007;429:95–100. doi: 10.1016/j.neulet.2007.09.077. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12 (Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. Aids. 2008;22:695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]