Abstract
The whooping cough agent Bordetella pertussis is closely related to Bordetella bronchiseptica, which is responsible for chronic respiratory infections in various mammals and is occasionally found in humans, and to Bordetella parapertussis, one lineage of which causes mild whooping cough in humans and the other ovine respiratory infections. All three species produce similar sets of virulence factors that are co-regulated by the two-component system BvgAS. We characterized the molecular diversity of BvgAS in Bordetella by sequencing the two genes from a large number of diverse isolates. The response regulator BvgA is virtually invariant, indicating strong functional constraints. In contrast, the multi-domain sensor kinase BvgS has evolved into two different types. The pertussis type is found in B. pertussis and in a lineage of essentially human-associated B. bronchiseptica, while the bronchiseptica type is associated with the majority of B. bronchiseptica and both ovine and human B. parapertussis. BvgS is monomorphic in B. pertussis, suggesting optimal adaptation or a recent population bottleneck. The degree of diversity of the bronchiseptica type BvgS is markedly different between domains, indicating distinct evolutionary pressures. Thus, absolute conservation of the putative solute-binding cavities of the two periplasmic Venus Fly Trap (VFT) domains suggests that common signals are perceived in all three species, while the external surfaces of these domains vary more extensively. Co-evolution of the surfaces of the two VFT domains in each type and domain swapping experiments indicate that signal transduction in the periplasmic region may be type-specific. The two distinct evolutionary solutions for BvgS confirm that B. pertussis has emerged from a specific B. bronchiseptica lineage. The invariant regions of BvgS point to essential parts for its molecular mechanism, while the variable regions may indicate adaptations to different lifestyles. The repertoire of BvgS sequences will pave the way for functional analyses of this prototypic system.
Introduction
Bordetella pertussis, the whooping cough agent, is an extremely contagious pathogen that infects the upper respiratory tract of humans and causes an acute infection [1]. The expression of most virulence factors of this Gram-negative bacterium, including adhesins and toxins, is controlled by the BvgAS two-component system (TCS) [2]–[5]. TCSs regulate major physiological responses in bacteria [6]–[9]. They are composed of two proteins, a sensor kinase that perceives (a) signal(s) and autophosphorylates a conserved histidine residue, and a response regulator that becomes activated upon phosphorylation by its cognate sensor kinase and often serves as a transcriptional activator [7], [10], [11].
BvgS is an “unorthodox” sensor kinase composed of three domains potentially involved in signal perception: two tandem periplasmic Venus Fly Trap (VFT) domains, proteins composed of two lobes with a solute-binding cavity between them [12] and a cytoplasmic PAS (Per-ARNT-Sim) domain [13]. They are followed by several domains participating to a phosphorylation cascade: a histidine kinase (His-kinase) domain, an Asp-containing receiver domain and a His phosphotransfer domain (Hpt) [14]. When phosphorylated, the response regulator BvgA activates the transcription of the virulence-activated genes (vags) [15].
By controlling the intracellular concentration of phosphorylated BvgA, this system mediates a progressive transition between three phenotypic phases, Bvg− (avirulent), Bvgi (intermediate) and Bvg+ (virulent) [16]–[20]. The Bvg+ phase occurs at 37°C and is necessary for B. pertussis to cause respiratory infections in animal models [16], [21]. The Bvgi phase, in which some adhesins are produced, has been proposed to play a role in transmission and in the initial stages of infection [22]. Under laboratory conditions, negative signals such as nicotinate, MgSO4 or low temperature trigger modulation to the Bvg− phase [23]. A set of virulence-repressed genes (vrgs) is expressed in that phase [24].
Bordetella parapertussis and Bordetella bronchiseptica are close relatives of B. pertussis [25]–[28]. Two distinct lineages of B. parapertussis cause either a generally milder form of whooping cough in humans or ovine respiratory infections [29]–[33], and B. bronchiseptica causes chronic respiratory infections in various mammals and has also been occasionally isolated from humans [34], [35]. B. pertussis and B. parapertussis are thought to have derived from distinct B. bronchiseptica clones [28], [33], [36]. The virulence regulons of the three species are similar. They are also controlled by BvgAS and respond to the same negative modulators. B. bronchiseptica can survive outside its hosts in the Bvg− phase, which promotes motility and survival under nutrient-limiting conditions [37]–[39]. In contrast, B. pertussis and B. parapertussishu have no known reservoir other than humans, and the function of their Bvg− phase is unclear. While B. bronchiseptica is fairly responsive to negative modulators, the sensitivity of B. pertussis to modulators appears to vary between isolates, which might suggest that the ability to down-modulate virulence has become dispensable for this species [16].
Because of the diversity of hosts and the different types of infection caused by these three Bordetella species, we investigated here the molecular diversity and evolution of BvgAS among the three Bordetella species. Sequencing of bvgA and bvgS from a number of isolates revealed that BvgA is almost invariant, while BvgS shows marked divergences between phylogenetic groups. Two evolutionary solutions for BvgS clearly appear from the analysis. The VFT2 domain is totally conserved within each of the two major BvgS types, indicating its pivotal role for the function of the protein.
Results
Genotyping of bvgA and bvgS in Bordetella
Full genomic sequences are available thus far for B. pertussis Tohama I, the B. bronchiseptica rabbit isolate RB50, and the human B. parapertussis isolate Bpp12822 [26]. The three genomes are closely related, with orthologous genes displaying little diversity between the three species. However, the bvgS genes markedly differ between them, while in contrast the bvgA genes are highly similar, harbouring small numbers of substitutions all of which are synonymous. To characterize the diversity of bvgA and bvgS, these genes were sequenced from a number of isolates selected from a collection described earlier [28]. In that previous phylogenetic study based on the sequences of housekeeping gene fragments, 4 major complexes of Bordetella were identified [28]. Complex I includes B. bronchiseptica isolates mainly of animal origin and the B. parapertussis ovine isolates (Bppov), complex II all B. pertussis isolates, complex III all B. parapertussis human isolates (Bpphu) and complex IV B. bronchiseptica isolates mainly of human origin. We thus selected isolates to represent these 4 complexes and all the sequence types identified in that study. We attempted to maximize the diversity of hosts, geographic origins and times of isolation.
In the 82 bvgS sequences, 247 distinct single nucleotide polymorphisms (SNPs) on a total of 3,717 bp were found (6.6%). Among these, 147 SNPs are synonymous substitutions (3.8%) and 100 non-synonymous substitutions (2.6%). This apparent diversity masks major differences between the Bordetella complexes (Table 1), as well as between domains of the protein (see below). Thus, bvgS is remarkably conserved in B. pertussis (complex II) with only two -non silent- SNPs among all 29 isolates. This is illustrated by a very low diversity index (0.15), which is yet lower than that calculated from the set of housekeeping genes sequenced in [28] (0.65). In contrast, the genetic diversity indices of both bvgS and the housekeeping genes calculated for complex IV and for complex I are significantly higher than those of B. pertussis (Table 1). Although B. parapertussishu is thought to have emerged more recently than B. pertussis [28], bvgS is more diverse in complex III than in complex II, while the contrary is true for the housekeeping genes (Table 1).
Table 1. Numerical analyses of the sequence data.
| bvgA (630 bp) | bvgS (3,717 bp) | HKG (2,912 bp) | |||||||||||||
| Complexes | I | II | III | IV | Total | I | II | III | IV | Total | I | II | III | IV | Total |
| Number of isolates | 38 | 28 | 6 | 13 | 85 | 35 | 29 | 6 | 12 | 82 | 35 | 22 | 6 | 12 | 75 |
| Number of alleles | 7 | 2 | 2 | 6 | 16 | 12 | 3 | 3 | 11 | 28 | 13 | 3 | 1 | 11 | 28 |
| Total SNPs | 6 | 1 | 2 | 6 | 15 | 68 | 2 | 20 | 41 | 247 | 51 | 2 | 0 | 20 | 83 |
| Non synonymous SNPs | 2 | 0 | 0 | 1 | 3 | 38 | 2 | 6 | 14 | 100 | 26 | 0 | 0 | 12 | 45 |
| Synonymous SNPs | 4 | 1 | 2 | 5 | 12 | 30 | 0 | 14 | 27 | 147 | 25 | 2 | 0 | 8 | 38 |
| Shannon-Wiener index | 1.18 | 0.15 | 0.63 | 1.52 | 2 | 1.72 | 0.29 | 0.86 | 2.37 | 2.42 | 2.34 | 0.65 | 0 | 2.37 | 2.87 |
The sequences of bvgA and bvgS originate from this study, while those of the 7 housekeeping gene fragments (HKG) were obtained from a previous study [28] and concatenated for the analyses. All three sets of sequences were obtained from similar sets of isolates. The Shannon-Wiener index represents the genetic diversity. Complex I includes B. bronchiseptica isolates mainly of animal origin and the B. parapertussis ovine isolates (Bppov), complex II all B. pertussis isolates, complex III all B. parapertussis human isolates (Bpphu) and complex IV B. bronchiseptica isolates mainly of human origin.
bvgA was found to be almost invariant, with 15 SNPs in total (Table 1). Only three SNPs are non-synonymous, but each is found in only one isolate. This indicates an extremely strong pressure on bvgA in Bordetellae. Accordingly, genetic diversity in all 4 complexes is lower for bvgA than for housekeeping genes or bvgS. Both bvgA and bvgS are strikingly conserved in B. pertussis (complex II).
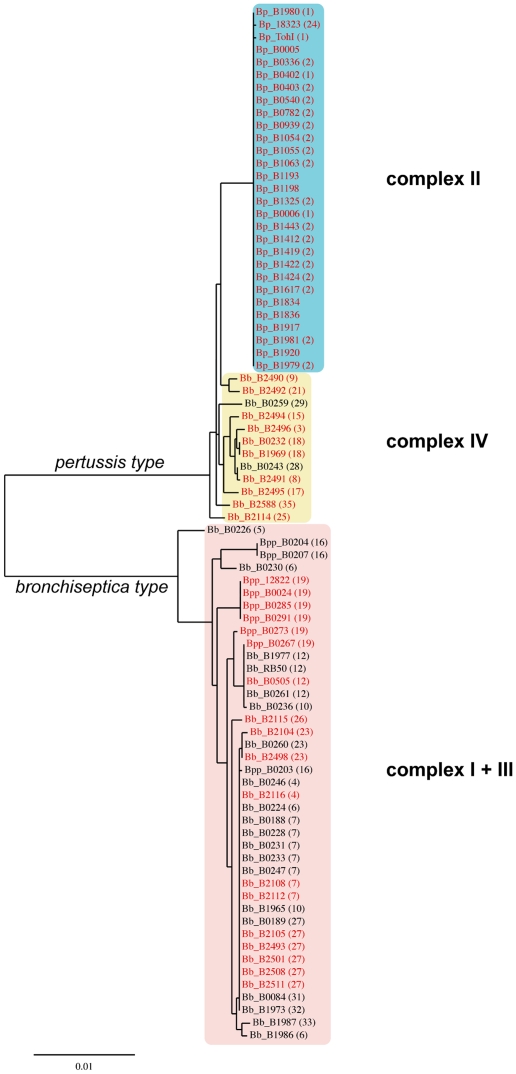
A phylogenetic tree was constructed based on the bvgS sequences (Fig. 1). This analysis clearly distinguishes two lineages of bvgS. The “pertussis type” (BvgSBp) encompasses bvgS from the 29 B. pertussis isolates as well as from the 12 B. bronchiseptica complex IV isolates. The other type, referred to hereafter as the “bronchiseptica type” (BvgSBb) encompasses bvgS from all 32 isolates that belong to the complex I of B. bronchiseptica and from the 9 isolates of B. parapertussis. Interestingly, the ovine and human B. parapertussis isolates do not form separate clusters but are included in the B. bronchiseptica complex I group, in contrast with previous studies [28], [40]. Similarly, the human isolates of B. bronchiseptica complex I are found interspersed among the animal isolates and do not form a separate group.
Figure 1. Phylogenetic tree based on the DNA sequences of bvgS.
The lengths of the branches represent the phylogenetic distances. All human B. pertussis (Bp), B. bronchiseptica (Bb) and B. parapertussis (Bpp) isolates are shown in red. The major complexes identified in an earlier study [28] are boxed in colors, and the numbers in parentheses correspond to the sequence types defined in that study.
A comparison of the bvgS-based phylogeny with that based on concatenated fragments from 7 different housekeeping genes described earlier [28] showed that while the overall topology of the two trees are similar, details differ. In addition to the absence of a separate complex III encompassing the B. parapertussis human isolates in the bvgS-based tree, some of the sequence types (ST) defined based on the housekeeping genes are split between several different branches of the bvgS-based tree, e.g. ST6, ST16 and ST19 (Fig. 1). These observations suggests that bvgS has evolved somewhat differentially from the housekeeping genes in the various branches.
Amino acid sequence analyses
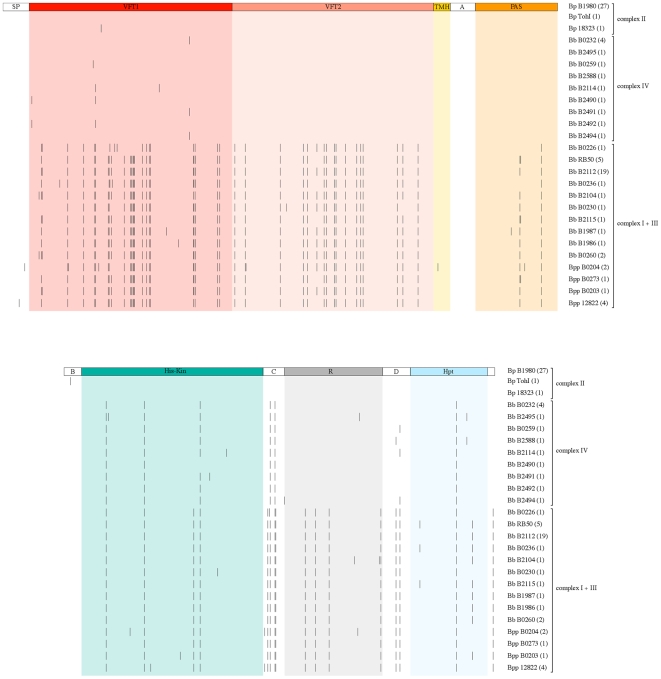
To gain insight into the evolution of the functional domains of BvgS, the predicted amino acid sequences were aligned (Fig. 2). The two types of BvgS, BvgSBb and BvgSBp, appear distinctly from this analysis. BvgS is identical among 27 B. pertussis isolates, while the two strains widely used in laboratories harbour each one substitution, E705K for Tohama I and I124T for strain 18323, respectively. BvgS of B. bronchiseptica complex IV is most similar to BvgSBp in the three domains potentially involved in signal perception, while the picture is slightly more blurred in the region corresponding to the phosphotransfer domains. BvgS from B. parapertussis resembles BvgSBb all along the sequence.
Figure 2. Schematic representation of BvgS variability at the amino acid level.
The complexes I+III, II and IV are defined as in Fig. 1. The various domains of BvgS drawn to scale are boxed in colors. The top line represents the prototype BvgS of all B. pertussis isolates but Tohama I and 18323. Each line below represents a distinct BvgS sequence, with each substitution relative to the reference sequence shown as a small vertical bar at the corresponding position in the protein. The same substitution is systematically found at any given position. Some of the BvgS variants displayed represent several isolates, the number of which is given in parentheses. SP represents the signal peptide, TMH the transmembrane helix, R the receiver domain, and A, B, C and D the four linkers connecting the domains (see text).
Remarkably, no more than two different residues are present at any given sequence position of the protein in all 82 isolates (Supporting information). Thus, in spite of the divergent evolution of BvgS between B. bronchiseptica complex I on the one hand, and B. pertussis and B. bronchiseptica complex IV on the other hand, selective pressure appears to limit the possible substitutions at the variable positions.
Our analyses revealed significant differences with respect to domain conservation. The first periplasmic domain, VFT1 totals the largest number of variable positions, although it is relatively well conserved within each type. Among the 82 isolates, 37 positions are variable in VFT1 over a total of 255 residues (14.5%). The overwhelming majority of the substitutions between the BvgSBb and the BvgSBp VFT1 are conservative in nature. In very rare instances, more drastic changes were observed, such as a Glu to Lys substitution at position 83 for two B. parapertussis ovine isolates and a Val to Glu substitution at position 206 for a single complex I isolate (Supporting information). Unlike for B. pertussis, some degree of variation is found within the complex IV VFT1s. Intriguingly, 4 of the substitutions identified in complex IV are not at the positions that differentiate BvgSBp from BvgSBb. They are thus specific of complex IV.
One isolate, B0226, stands out from the B. bronchiseptica complex I by a small portion of VFT1. At 7 positions between residues 133 and 166 it shares B. pertussis sequences, although it also harbours unique substitutions in that region.
For VFT2 essentially two types of sequence have evolved. VFT2 of BvgSBp is invariant for all 41 B. pertussis and B. bronchiseptica complex IV isolates, suggesting that this domain is under strong stabilizing selection. VFT2 of BvgSBb also displays a very limited degree of variation. Furthermore, only conservative substitutions are found between the two types except for one at position 404, where Ala in BvgSBp is replaced by Asp in BvgSBb. In total, 19 positions are different between the pertussis type and the bronchiseptica type VFT2s over a total of 254 residues (7.5%). There are no intermediates between the two VFT2 types. All these observations argue that VFT2 is pivotal to the function of BvgS and is under strong selective pressure in each lineage.
The PAS domain is well conserved among all isolates, suggesting that it is also under stabilizing pressure. In total only 5 positions are variable, and the few substitutions found in this domain are all conservative. Similar to VFT1 and VFT2, the PAS domain distinguishes B. bronchiseptica complex I from B. pertussis and B. bronchiseptica complex IV. Thus, for all three putative perception domains, B. bronchiseptica complex IV clearly sides with B. pertussis.
For the His-kinase domain, in contrast, B. bronchiseptica complex IV sequences appear to be more similar to BvgSBb than to BvgSBp. Altogether the His-kinase domain is also much more conserved than the two VFT domains, with only 11 variable positions harbouring essentially conservative substitutions over a total of 223 residues (4.9%). This probably points to strong functional constraints on this domain.
The receiver domain is more diverse, with 9 variable positions over a total of 122 residues (7.3%). Intriguingly, the receiver domain of B. bronchiseptica complex IV is of the BvgSBp type, unlike the His-kinase domain.
The Hpt domain is well conserved, with only 4 variable positions for a total of 98 residues (4.1%). Intriguingly, at position 1190, the conservative Val to Ala substitution differentiates B. pertussis from B. bronchiseptica of both complexes I and IV. Altogether, thus, complex IV appears to be intermediate between the BvgSBb and the BvgSBp types for the His-kinase and Hpt domains, in contrast with the VFT1, VFT2, PAS and receiver domains. This suggests that the distinct domains have evolved at different rates.
Distinct functional constraints appear to operate also on the various linkers that join the domains. The transmembrane helix is invariant, except for a unique conservative Gly to Ala change. Similarly, the linkers A and B between the membrane and the PAS domain and between the PAS and His-kinase domains respectively are invariant, indicating that they are under strong selective pressure (Fig. 2). One striking exception to this rule is that a Glu residue is replaced by a Lys residue in Tohama I at position 705 (see below). In marked contrast with the strict conservation of linkers A and B, the linker C between the His-kinase and receiver domains seems to accumulate a disproportionate number of mutations, with 20% positions that are variable. Linker D between the receiver and Hpt domains harbours 2 variable positions over 36 residues.
Mapping of the substitutions
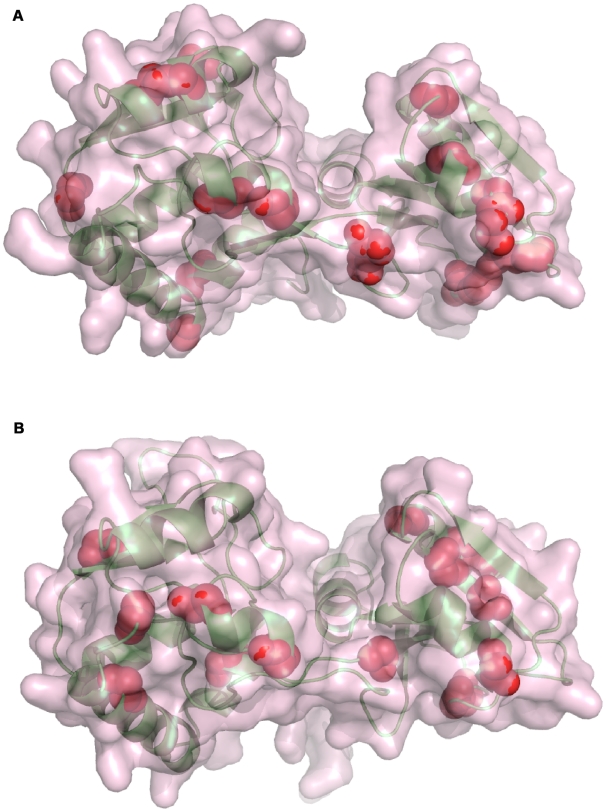
In order to map the positions of the substitutions, structural models were constructed for several domains of BvgS based on available X-ray structures of homologous proteins, since the crystal structure of BvgS is not available. VFT1 and VFT2 were modelled on the glutamine-binding protein, a bacterial periplasmic binding protein of an ABC transporter. The overwhelming majority of the substitutions between the two broad phylogenetic groups map to the external surfaces of both VFT1 and VFT2, leaving the potential solute-binding cavities conserved (Fig. 3). In addition, a few substitutions appear to be in the hydrophobic core of VFTs, but they are highly conservative and thus unlikely to affect the structure or stability of the proteins. Thus, the tertiary structures of the two VFT domains and their potential ligand-binding pockets are likely to be totally conserved, while the external surfaces of the proteins are quite different between the two BvgS types. The more drastic substitution between BvgSBp (Asp404) and BvgSBb (Ala404) is located in an external loop in the second lobe of VFT2. Unlike non-synonymous substitutions, synonymous substitutions are found both in the ligand binding cavities and on the surfaces of the VFTs, indicating differential selective pressure between cavities and surfaces.
Figure 3. Structural models of VFT1 and VFT2.
The models of VFT1 (A) and VFT2 (B) are based on the structure of the glutamine binding protein in an unliganded, open conformation (PDB 1GGG). The elements of secondary structure are shown in ribbon representation in lime green, and the surfaces of the proteins are shown in semi-transparency. The cavity of each of the two domains corresponds to the groove between the two lobes. The positions of the substitutions that distinguish the BvgSBp and BvgSBb types are depicted as red balls. Only the main chain atoms are shown. The rare substitutions have not been represented.
A model of the His-kinase domain of BvgS was also constructed. The ATP-binding pocket appeared to be conserved among all isolates, as expected for the activity of the protein (not shown). A drastic substitution located in the linker B that precedes the His-kinase domain distinguishes B. pertussis Tohama I from all other B. pertussis, B. bronchiseptica and B. parapertussis isolates. Tohama I harbours a Lys at position 705 instead of a Glu in the 81 others. This linker is predicted to form an α helix immediately preceding the dimerisation helix of the His-kinase domain. Conceivably, this substitution might affect the conformation of this region most likely crucial for signal transduction (see below).
The structure of an Hpt domain in complex with its cognate phosphorylation substrate has been reported [41], [42], showing the interface between the two proteins. The BvgS Hpt domain has two interaction partners, the receiver domain and BvgA. The surfaces of Hpt predicted to form interfaces with its partners appear to be conserved as well (not shown). Similarly, neither of the three frequent substitutions between the receiver domains of the two BvgS types map to the predicted interface with Hpt (not shown).
Regarding BvgA, only 3 unique substitutions are found each in one isolate, namely Glu to Gly, Asp to Gly and Val to Glu at positions 64, 137 and 197, respectively. Sequence alignments of response regulators homologous to BvgA as well as the X-ray structure of one such protein indicate that these three substitutions are at non-conserved positions in the superfamily (not shown). The rarity and the positions of non-synonymous replacements in BvgA strongly indicate that the protein is under stringent selection.
Functional implications of sequence variations
Thus the vast majority of the predicted amino acid differences between pertussis type and bronchiseptica type BvgS are located in the periplasmic domains, essentially at the surface of the two VFT domains that appear to have co-evolved within each lineage. This suggests that the two domains operate together through inter-domain interactions, most likely for proper signal transduction. To test this hypothesis, we determined whether the sequence variations identified between the VFT1 and VFT2 of the two types of BvgS affect its activity or its level of sensitivity to the known negative virulence modulators. BvgS harbouring the BvgSBp periplasmic domain of our laboratory strain B. pertussis BPSM, a Tohama I derivative, was substituted with BvgS harbouring the BvgSBb periplasmic domain from B. bronchiseptica RB50, a typical complex I isolate (Fig. 4A). We also performed single domain exchanges. The activities of chimeric BvgS proteins and their responses to modulation were determined using the reporter gene lacZ placed under the control of the Bvg-regulated promoter of the pertussis toxin operon.
Figure 4. ß-galactosidase activities of the recombinant BPSM(ptx::lacZ) strains harbouring BvgS variants.
(A) Schematic representation of the chimeric BvgS, with the white boxes representing BvgSBp domains and the shaded boxes BvgSBb domains. BPWT corresponds to BPSM carrying the reporter gene. HK represents the His-kinase domain. (B) and (C) The ß-gal activities were measured as a function of increasing concentrations of nicotinate or MgSO4. The measurements were performed in triplicate using different clones for each recombinant strain. The level of ß-gal activity in non-modulated medium was taken as 100%.
The replacement of both VFT1 and VFT2 of the BPSM BvgS by those of RB50 (BPBb1+2) did not affect the level of BvgS activity in the absence of modulation, but it made the recombinant bacteria much more sensitive to nicotinic acid than their parent (Fig. 4B,C). Thus, the BvgSBb and BvgSBp periplasmic domains of BvgS influence the response to this negative modulator.
Similarly, the replacement of the BPSM VFT2 by that of RB50 (BPBb2), yielding a VFT1Bp-VFT2Bb chimera strongly enhanced the sensitivity of BvgS to both nicotinic acid and MgSO4 (Fig. 4B,C). In contrast, introduction of the RB50 VFT1 into the BPSM BvgS (BPBb1) did not modify the response of the recombinant bacteria to either modulator (not shown). Thus, VFT2 is essential in determining sensitivity to modulation.
Because of its drastic character in an otherwise totally invariant linker segment, the substitution identified in the Tohama I BvgS was also investigated. Tohama I has been described as relatively insensitive to modulation compared with other B. pertussis isolates [16], and thus we tested whether Lys705 is the cause for this lack of sensitivity (Fig. 5A). Glu at position 705 in the BPSM BvgS made the recombinant strain more responsive to nicotinate than its parent, with 4 mM nicotinic acid causing the complete loss of BvgS activity (Fig. 5B). The BPE705 substitution also enhanced the responsiveness to MgSO4 (Fig. 5C). Thus, the presence of a Lys at position 705 is responsible for the lack of sensitivity to modulation of Tohama I. This substitution is quite rare in Bordetella, suggesting that the mutation acquired in Tohama I is not particularly advantageous. The other unique substitution identified in Bp18323, I124T, was also introduced into BPSM (BPT124). The level of ß-gal activity and the responsiveness to modulators of BPT124 was identical to that of BPSM, indicating that this is likely a neutral substitution (not shown).
Figure 5. ß-galactosidase activities of the recombinant BPSM(ptx::lacZ) strains harbouring BvgS variants.
(A) Schematic representation of the chimeric BvgS. The BvgS domains are represented as in Fig. 4. The star indicates that the Lys705 residue found in Tohama I was replaced by Glu as in all other isolates. (B) and (C) The ß-gal activities were measured as a function of increasing concentrations of nicotinate or MgSO4. The measurements were performed as in Fig. 4.
Since both VFT2 and the linker preceding the HK domain determine the sensitivity to modulators, we tested whether their effects are cumulative. Therefore, a new chimeric strain, BPBb2+E705 was constructed that harbours both the VFT2Bb domain and Glu at position 705 (Fig. 5A). BPBb2+E705 was hypersensitive to modulators, with as low as 0.25 mM nicotinic acid (Fig. 5B) or 5 mM MgSO4 (Fig. 5C) being sufficient for complete modulation.
Hypermodulator B. pertussis in an animal model of infection
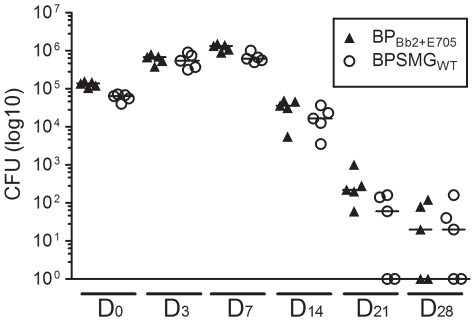
No such hypersensitive variant has been characterized in natural isolates so far [16]. Thus, it was interesting to determine whether this phenotype might affect B. pertussis's ability to colonize the respiratory tract in an animal model of infection. BPSM or BPBb2+E705 were instilled intranasally to two groups of mice, and the numbers of bacteria recovered from the lungs were counted after various periods of time. No significant difference was observed between the two strains (not shown). To determine whether BPBb2+E705 might be disadvantaged in a mixed infection, mice were also inoculated intranasally with equal numbers of the two bacterial strains together. The two strains colonized the lungs of mice in the same manner (Fig. 6). Thus, in this model of experimental infection, B. pertussis hypersensitive to modulation does not appear to be defective for colonization.
Figure 6. Colonization of mice lungs by two strains in a mixed infection.
Balb/c mice were infected intranasally by 1.6×105 CFU of each BPSMGWT and BPBb2+E705. At the indicated time points, mice were killed, and the viable bacteria present in the lungs were counted. 5 mice were analyzed per time point. The two bacterial strains were distinguished by an antibiotic marker. Medians for each group are shown.
Discussion
In this work, we investigated the molecular evolution of the BvgAS two-component system, which is central to the regulation of Bordetella virulence. BvgA is remarkably conserved. In contrast, the sequences of BvgS differ markedly between species, and in particular can be separated into two lineages of Bordetella. Interestingly, one of the two lineages includes all B. pertussis isolates and a subset of B. bronchiseptica isolates previously identified as forming a distinct B. bronchiseptica complex (called complex IV), mainly isolated from humans [28]. Our results confirm the hypothesis that complex IV strains are an evolutionary intermediate between animal and human adapted Bordetellae from which B. pertussis evolved [28]. The degree of divergence of BvgS varies significantly between lineages, as shown by the genetic diversity indices. In B. pertussis, BvgS displays an extremely low level of diversity, indicating that it has reached equilibrium in this highly successful human colonizer or has undergone a recent selective sweep. With a broad range of hosts, complex IV may be using a different evolutionary strategy, and its BvgS may still be in a process of diversification.
Our analyses do not provide evidence of an absolute correlation between the type of bvgS allele found in a given Bordetella lineage and the hosts of its members. Thus, two bona fide human pathogens, B. pertussis and B. parapertussishu, harbour BvgS of the BvgSBp type and of the BvgSBb type, respectively, although the latter type otherwise corresponds to B. bronchiseptica complex I, with isolates of essentially animal origin. Similarly, the ovine and human isolates of B. parapertussis have the same BvgS type although they infect distinct hosts. It is interesting that BvgS is monomorphic in B. pertussis but not in B. parapertussishu, as B. pertussis is assumed to be older [28]. This may reflect that B. parapertussishu is not yet optimally adapted to its host, unlike B. pertussis [33].
Remarkably, the various domains of BvgS display widely different degrees of diversity, which shows that they are under different functional constraints, and/or parts of the protein are subject to diversifying selection in some lineages. The periplasmic domains of BvgS appear to have evolved most extensively. However, their evolution has been restricted to external surfaces, strongly arguing that the binding paradigm for Venus Fly Trap domains applies to BvgS. Purifying selective pressure on the cavities argues that they bind specific ligands. The prevalent model proposes that BvgS is in a permanently activated state in the absence of ligand, unlike classical sensors of TCS. If this is correct, then the conservation of the cavities is important essentially for the perception of negative signals. However, we have obtained preliminary data that specific substitutions in the VFTs cavities markedly decrease the activity of BvgS (our unpublished data). This supports the idea that the cavities of the VFT domains also perceive positive signals, at odds with the generally accepted model.
The external surfaces of the VFT domains have evolved considerably, arguing that they undergo a lesser selective pressure or are even diversifying. However, because there are no intermediate types between the BvgSBb and BvgSBp types, the evolutionary pressure must have selected for functional combinations of the two VFT surfaces. In other words, the surfaces may have co-evolved in each of the two major lineages because interactions between the two domains must be fine-tuned for optimal function. The combination of VFT1Bp and VFT2Bb in particular appears to significantly increase the sensitivity of BvgS to modulators. We thus propose that the sensitivity to modulators depend on the efficiency of signal transduction between the two VFT domains and between VFT2 and the cytoplasmic portion through the membrane segment. The membrane-proximal position of VFT2 most likely makes it especially important in this respect. Similarly, the two types of BvgS harbour distinct sets of cytoplasmic domains, although the situation is more blurred in the case of complex IV. This suggests that there may be small functional differences in the phosphorylation cascade between the two types.
The region of the protein with the lowest degree of diversity encompasses the transmembrane segment, the following cytoplasmic linker and the PAS and His-kinase domains, emphasizing the functional constraints on that region. The transmembrane segment must transmit signals perceived by the periplasmic domain to the cytoplasmic domain in a purely mechanical manner, such as a rotational or a piston-like motion. This signal must then be conveyed mechanically by the linker to the PAS domain, and then from the PAS domain to the His-kinase domain. PAS is thus central to transduction between periplasmic and His-kinase domains. In agreement with this model, several mutations that make BvgS insensitive to negative modulating signals map to the linker and PAS domains [43], [44]. Following phosphorylation of the His-kinase, signal transmission to BvgA is enzymatic, and thus functional constraints are exerted on the interfaces between domains, for interactions in trans and/or in cis, rather than on linkers [45]. In agreement with this idea, the linkers between the domains involved in the phosphorylation cascade have diverged more rapidly, most likely because they do not undergo strong selective pressures.
Only 3 rare substitutions were identified in BvgA. BvgA forms many interactions that are all important for its function. It forms homodimers, interacts with the Hpt domain for phosphotranfer and with its target DNA sequences and the RNA polymerase. This most likely explains why BvgA undergoes such a strong stabilizing pressure in pathogenic Bordetellae.
The importance of responsiveness to modulating agents for the function of BvgS in vivo, and the relevance of nicotinate and MgSO4 are unclear. The Bvg− phase is most likely more important for B. bronchiseptica to survive outside the host and cause chronic infections [46], [47] than it is for the lifestyle of B. pertussis. Nevertheless, the fact that both types of BvgS have retained similar sensitivities to negative modulators argues that this phenotype is nevertheless relevant for both species. Thus, no natural B. pertussis isolates appear to be “locked” in the Bvg+ phase, even though such strains can arise by simple point mutations in the laboratory [43], [44], [48]. Therefore, the ability to down-modulate virulence factor expression must confer some advantage upon the bacterium, which is supported by the fact that Tohama I, with its low sensitivity to modulators, is the exception among B. pertussis. Conversely, we wondered whether B. pertussis with a BvgS variant hypersensitive to modulation would be impaired for colonisation. Surprisingly, this strain appeared to be functional in the mouse model of infection. The observation that no naturally occurring Bordetella isolate has been described to harbour such a hyper-responsive BvgS nevertheless suggests that this phenotype might be detrimental. The animal model used in this study does not reproduce all the features of a human infection, and in particular bacterial transmission. Better models may be needed to detect subtle regulation defects.
Materials and Methods
Ethics statement
All animal experiments were performed at the animal facility of the Institut Pasteur de Lille (number A59-35-064, Lille, France) according to the rules of the European Community Council guidelines (86/609/EEC) for laboratory animal experimentation. The animal protocol was approved by the local institutional review board (Comité d'Ethique en Expérimentation Animale Nord-Pas-De-Calais, CEEA 03/2009).
Sequencing of the circulating infectious Bordetella isolates
The B. pertussis, B. bronchiseptica and B. parapertussis isolates were selected from the collection described in [28]. bvgS was sequenced for 28 isolates of B. pertussis, 8 of B. parapertussis (5 Bpphu and 3 Bppov), 31 of B. bronchiseptica from complex I and 12 of B. bronchiseptica from complex IV [28] (Table S2). bvgA was sequenced from 27 isolates of B. pertussis, 8 B. parapertussis isolates (5 Bpphu and 3 Bppov), 34 B. bronchiseptica isolates from complex I and 13 from complex IV (Table S3). The strains were cultivated on BG blood agar medium during 16 hours for B. bronchiseptica and 48 hours for B. pertussis and B. parapertussis. Chromosomal DNA was extracted using the Illustra™ bacterial genomic Prep Mini Spin Kit (GE Healthcare) according to the manufacturer's instructions. For PCR amplification, several partially overlapping PCRs were performed by using several pairs of primers, bvgA′-Up and bvgA′-Lo, VFT1′-Up and VFT1′-Lo, VFT2′-Up and VFT2′-Lo, PAS/HisKin-Up and PAS/HisKin-Lo, R-Up and R-Lo and Hpt-Up and Hpt-Lo (Table S1). The following conditions for the mixes were used: HotStar Taq DNA polymerase in the presence of Q buffer and MgCl2 (Qiagen), 30 cycles of 1 min at 95°C, 1 min at 57°C and 1 min at 72°C. The PCR products were purified by using the PCR Purification Kit or the Gel Extraction Kit (Qiagen), depending on the degree of purity of the amplicons. The DNA fragments were sequenced by Genoscreen using an ABI 377 sequencer (Lille, France). The sequences were reassembled and compared by using the DNAstar software. A number of targets were amplified and sequenced twice to determine the level of error introduced by the PCR and sequencing steps. In particular, all fragments with a unique sequence type were checked in this manner. No discrepancy was obtained in any case.
Sequence data analysis
The genetic diversity for each complex was calculated using the Shannon-Wiener index of diversity using the following formula in which pi is the frequency of the ith type:
Single nucleotide polymorphisms (SNP) and their synonymous or non-synonymous characteristics were evaluated by using the DnaSP software. Neighbour-joining trees were constructed by using the www.phylogeny.fr software.
Models of the BvgS domains
The PDB codes of the X-ray structures used to build models for the various domains of BvgS are 1GGG (VFTs), 2C2A (His-kinase), 2AYX (receiver domain) and 2AOB (Hpt domain), and 3C3W for BvgA. The Modeller software was used on the following portions of BvgS: residues 33–287 for VFT1, 288–541 for VFT2, 726–946 for the His-kinase domain, 974–1095 for the receiver domain and 1133–1228 for the Hpt domain.
Construction of chimeric B. pertussis strains
B. pertussis BPSM is a Tohama I-derivative that is resistant to streptomycin [49]. The bvgAS deletion strain BPSMΔbvgAS was constructed as follows. The 5′ and 3′ extremities of the bvgAS locus were amplified by PCR using the BPSM chromosome as template and the oligonucleotides bvgA-Up and bvgA″-Lo, and bvgS-Up and bvgS-Lo as primers, respectively (Table S1). The amplicons were inserted directly into pCR®II-TOPO (Invitrogen) and sequenced. They were then successively introduced as EcoRI-KpnI and XbaI-HindIII fragments into pUC19, yielding pUC19ΔbvgAS. The resulting 1.0 kb EcoRI-HindIII fragment was introduced into the EcoRI-HindIII sites of pSORTP1, a mobilizable plasmid used for conjugation [50]. Conjugation was performed on BG-blood agar plates containing 10 mg/ml MgCl2 for 6–7 hours, and co-integrates were selected on BG-blood agar plates containing 10 µg/ml gentamycin and 30 µg/ml nalidixic acid to prevent growth of the E. coli donor. Allelic exchange was selected by two successive steps as described [50]. After 4 to 5 days growth on selective media, isolated non-haemolytic streptomycin-resistant colonies, characteristic of the Bvg− phase were analysed by PCR to confirm the deletion.
Recombinant BPSM strains containing chimeric bvgS genes were constructed as follows. Successive portions of the bvgAS locus of BPSM were amplified by PCR in order to introduce restriction sites using silent mutations at specific sites corresponding to junctions between structural domains. The sequences of the oligonucleotides used as primers are given in Table S1. In addition, an EcoRI restriction site naturally occurring in the region of the gene coding for the PAS/His-kinase portion of bvgS was removed by overlapping PCR using the pairs of primers ΔEcoRI-Up and PAS/HisKin-Lo, and ΔEcoRI-Lo and PAS/HisKin-Up. The various amplicons were sequenced and introduced successively into pUC19. The resulting “mosaic” bvgAS locus includes the 5′ and 3′ extremities of the operon remaining in BPSMΔbvgAS and necessary for allelic replacement in that strain. The mosaic bvgAS locus allowed us to replace selectively the genetic cassettes encoding each of the two VFT-like domains and the PAS/His-kinase domains of the BPSM bvgS by the corresponding cassettes of B. pertussis 18323 (to introduce Glu705) or B. bronchiseptica RB50. This procedure generated a number of bvgS variants that were each excised from pUC19 by restriction with EcoRI and HindIII and introduced into the EcoRI and HindIII sites of pSORTP1. Each bvgS variant was then introduced into BPSMΔbvgAS by allelic exchange, using conjugation as described above. Our criterion for the selection of recombinant clones was the restoration of hemolysis, which can be easily detected on BG-blood agar.
Construction of ptx-lacZ transcriptional fusions and measurement of ß-galactosidase activity
A recombinant pFUS plasmid, harbouring lacZ in transcriptional fusion with the sequence of the first gene of the pertussis toxin operon [51], was introduced into the different strains by conjugation, and the integrants were selected on BG blood agar containing 100 µg/ml streptomycin and 10 µg/ml gentamycin. The recombinant strains were grown in modified Stainer-Scholte medium (SS) [52] containing the relevant antibiotics. After overnight growth at 37°C under rotating agitation, the bacterial suspension was used to initiate cultures in 10 ml of SS medium containing increasing concentrations of nicotinate or MgSO4. The inoculation volume was adapted to compensate for slower growth in the presence of high concentrations of nicotinate. The bacteria were grown until the cultures reached an OD600 of 1.5. They were harvested by centrifugation and broken by using a Hybaid Ribolyser apparatus (35 s at speed 6 in tubes containing 0.1 mm silica spheres as the lysing matrix). ß-galactosidase activities were measured as described [51]. The experiments were performed in triplicate.
Murine respiratory tract infections
Experimental infections were performed at biosafety level 2 facilities with 8 week-old female BALB/c mice purchased from Charles River Laboratories.
Two groups of mice slightly sedated by intraperitoneal pentobarbital injection were inoculated with 1.6×106 bacteria from BPSM and BPBb2+E705 suspensions, by depositing 20 µl droplets into the nostrils. Groups of four animals per strain were sacrificed at days 0, 7, 14, 21 and 28 post-inoculation, and lungs were removed. Lung colonization was quantified by homogenizing the entire lungs in 2 to 5 ml phosphate-buffered saline (PBS), plating 100 µl aliquots of serial dilutions of the lung suspensions onto BG blood agar with 100 µg/ml of streptomycin, and counting the colonies after 4 days of incubation at 37°C.
Mixed infection essays were performed with BPSM carrying a gentamycin resistance marker at the 3′ end of the bctCBA operon [53] to distinguish between this strain (called hereafter BPSMGWT) and BPBb2+E705. In this case, the inoculum consisted of 1.6×105 CFU of each bacterial strain suspended in a final volume of 20 µl. Five animals per time point were sacrificed at days 0, 3, 7, 14, 21 and 28 post-inoculation and lung colonization was quantified as described above. Serial dilutions of each suspension were plated onto BG blood agar with 100 µg/ml of streptomycin or with 10 µg/ml of gentamycin. Colonies grown on gentamycin (corresponding to BPSMGWT) were subtracted from the number of colonies present on the streptomycin-containing medium (corresponding to both BPSMGWT and BPBb2+E705) to determine the number of BPBb2+E705 bacteria present in the mice lungs.
Supporting Information
Oligonucleotides used in this study
(0.07 MB PDF)
bvgS sequences
(0.35 MB RTF)
bvgA sequences
(0.04 MB RTF)
Acknowledgments
We thank Marjolein Van Gent for her help with the bacterial isolates and Philip Supply for his advise on data analyses. FJD is a researcher of the CNRS.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Financial support for this work was provided by Sanofi-Pasteur, the Institute Pasteur of Lille and INSERM.
References
- 1.Hewlett EL. Pertussis: current concepts of pathogenesis and prevention. Pediatr Infect Dis J. 1997;16(4 Suppl):S78–84. doi: 10.1097/00006454-199704001-00002. [DOI] [PubMed] [Google Scholar]
- 2.Uhl MA, Miller JF. Autophosphorylation and phosphotransfer in the Bordetella pertussis BvgAS signal transduction cascade. Proc Natl Acad Sci U S A. 1994;91(3):1163–1167. doi: 10.1073/pnas.91.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhl MA, Miller JF. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 1996;15(5):1028–1036. [PMC free article] [PubMed] [Google Scholar]
- 4.Beier D, Gross R. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol. 2006;9(2):143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Stibitz S. Locht C, editor. The bvg regulon. 2007. pp. 47–67. Bordetella Molecular Microbiology. Horizon bioscience.
- 6.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26(6):369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 7.Bekker M, Teixeira de Mattos MJ, Hellingwerf KJ. The role of two-component regulation systems in the physiology of the bacterial cell. Sci Prog. 2006;89(Pt 3–4):213–242. doi: 10.3184/003685006783238308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70(4):910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szurmant H, White RA, Hoch JA. Sensor complexes regulating two-component signal transduction. Curr Opin Struct Biol. 2007;17(6):706–715. doi: 10.1016/j.sbi.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egger LA, Park H, Inouye M. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells. 1997;2(3):167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 11.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 12.Felder CB, Graul RC, Lee AY, Merkle HP, Sadee W. The Venus Fly Trap of periplasmic binding proteins: an ancient protein module present in multiple drug receptors. AAPS PharmSci. 1999;1(2):E2. doi: 10.1208/ps010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63(2):479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhl MA, Miller JF. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J Biol Chem. 1996;271(52):33176–33180. doi: 10.1074/jbc.271.52.33176. [DOI] [PubMed] [Google Scholar]
- 15.Bock A, Gross R. The unorthodox histidine kinases BvgS and EvgS are responsive to the oxidation status of a quinone electron carrier. Eur J Biochem. 2002;269(14):3479–3484. doi: 10.1046/j.1432-1033.2002.03029.x. [DOI] [PubMed] [Google Scholar]
- 16.Martinez de Tejada GM, Miller JF, Cotter PA. Comparative analysis of the virulence control systems of Bordetella pertussis and Bordetella bronchiseptica. Mol Microbiol. 1996;22(5):895–908. doi: 10.1046/j.1365-2958.1996.01538.x. [DOI] [PubMed] [Google Scholar]
- 17.Coote JG. Environmental sensing mechanisms in Bordetella. Adv Microb Physiol. 2001;44:141–181. doi: 10.1016/s0065-2911(01)44013-6. [DOI] [PubMed] [Google Scholar]
- 18.Deora R. Differential regulation of the Bordetella bipA gene: distinct roles for different BvgA binding sites. J Bacteriol. 2002;184(24):6942–6951. doi: 10.1128/JB.184.24.6942-6951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deora R, Bootsma HJ, Miller JF, Cotter PA. Diversity in the Bordetella virulence regulon: transcriptional control of a Bvg-intermediate phase gene. Mol Microbiol. 2001;40(3):669–683. doi: 10.1046/j.1365-2958.2001.02415.x. [DOI] [PubMed] [Google Scholar]
- 20.Jones AM, Boucher PE, Williams CL, Stibitz S, Cotter PA. Role of BvgA phosphorylation and DNA binding affinity in control of Bvg-mediated phenotypic phase transition in Bordetella pertussis. Mol Microbiol. 2005;58(3):700–713. doi: 10.1111/j.1365-2958.2005.04875.x. [DOI] [PubMed] [Google Scholar]
- 21.Merkel TJ, Barros C, Stibitz S. Characterization of the bvgR locus of Bordetella pertussis. J Bacteriol. 1998;180(7):1682–1690. doi: 10.1128/jb.180.7.1682-1690.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cotter PA, Miller JF. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol Microbiol. 1997;24(4):671–685. doi: 10.1046/j.1365-2958.1997.3821741.x. [DOI] [PubMed] [Google Scholar]
- 23.Prugnola A, Arico B, Manetti R, Rappuoli R, Scarlato V. Response of the bvg regulon of Bordetella pertussis to different temperatures and short-term temperature shifts. Microbiology. 1995;141(Pt 10):2529–2534. doi: 10.1099/13500872-141-10-2529. [DOI] [PubMed] [Google Scholar]
- 24.Cummings CA, Bootsma HJ, Relman DA, Miller JF. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J Bacteriol. 2006;188(5):1775–1785. doi: 10.1128/JB.188.5.1775-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arico B, Gross R, Smida J, Rappuoli R. Evolutionary relationships in the genus Bordetella. Mol Microbiol. 1987;1(3):301–308. doi: 10.1111/j.1365-2958.1987.tb01936.x. [DOI] [PubMed] [Google Scholar]
- 26.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35(1):32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 27.Preston A, Parkhill J, Maskell DJ. The Bordetellae: lessons from genomics. Nat Rev Microbiol. 2004;2(5):379–390. doi: 10.1038/nrmicro886. [DOI] [PubMed] [Google Scholar]
- 28.Diavatopoulos DA, Cummings CA, Schouls LM, Brinig MM, Relman DA, et al. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 2005;1(4):e45. doi: 10.1371/journal.ppat.0010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linnemann CC, Perry EB. Bordetella parapertussis. Recent experience and a review of the literature. Am J Dis Child. 1977;131(5):560–563. doi: 10.1001/archpedi.1977.02120180074014. [DOI] [PubMed] [Google Scholar]
- 30.Cullinane LC, Alley MR, Marshall RB, Manktelow BW. Bordetella parapertussis from lambs. N Z Vet J. 1987;35(10):175. doi: 10.1080/00480169.1987.35433. [DOI] [PubMed] [Google Scholar]
- 31.Porter JF, Connor K, Donachie W. Differentiation between human and ovine isolates of Bordetella parapertussis using pulsed-field gel electrophoresis. FEMS Microbiol Lett. 1996;135(1):131–135. doi: 10.1111/j.1574-6968.1996.tb07977.x. [DOI] [PubMed] [Google Scholar]
- 32.Porter JF, Connor K, Donachie W. Isolation and characterization of Bordetella parapertussis-like bacteria from ovine lungs. Microbiology. 1994;140(2):255–261. doi: 10.1099/13500872-140-2-255. [DOI] [PubMed] [Google Scholar]
- 33.van der Zee A, Mooi F, Van Embden J, Musser J. Molecular evolution and host adaptation of Bordetella spp.: Phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J Bacteriol. 1997;179(21):6609–6617. doi: 10.1128/jb.179.21.6609-6617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodnow RA. Biology of Bordetella bronchiseptica. Microbiol Rev. 1980;44:722–738. doi: 10.1128/mr.44.4.722-738.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woolfrey BF, Moody JA. Human infections associated with Bordetella bronchiseptica. Clin Microbiol Rev. 1991;4(3):243–255. doi: 10.1128/cmr.4.3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musser JM, Hewlett EL, Peppler MS, Selander RK. Genetic diversity and relationships in populations of Bordetella spp. J Bacteriol. 1986;166(1):230–237. doi: 10.1128/jb.166.1.230-237.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porter JF, Parton R, Wardlaw AC. Growth and survival of Bordetella bronchiseptica in natural waters and in buffered saline without added nutrients. Appl Environ Microbiol. 1991;57(4):1202–1206. doi: 10.1128/aem.57.4.1202-1206.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter JF, Wardlaw AC. Long-term survival of Bordetella bronchiseptica in lakewater and in buffered saline without added nutrients. FEMS Microbiol Lett. 1993;110(1):33–36. doi: 10.1111/j.1574-6968.1993.tb06291.x. [DOI] [PubMed] [Google Scholar]
- 39.Cotter PA, Miller JF. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect Immun. 1994;62(8):3381–3390. doi: 10.1128/iai.62.8.3381-3390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cummings CA, Brinig MM, Lepp PW, van de Pas S, Relman DA. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J Bacteriol. 2004;186(5):1484–1492. doi: 10.1128/JB.186.5.1484-1492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Q, Porter SW, West AH. The yeast YPD1/SLN1 complex: insights into molecular recognition in two-component signaling systems. Structure. 2003;11(12):1569–1581. doi: 10.1016/j.str.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 42.Zhao X, Copeland DM, Soares AS, West AH. Crystal structure of a complex between the phosphorelay protein YPD1 and the response regulator domain of SLN1 bound to a phosphoryl analog. J Mol Biol. 2008;375(4):1141–1151. doi: 10.1016/j.jmb.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller JF, Johnson SA, Black WJ, Beattie DT, Mekalanos JJ, et al. Constitutive sensory transduction mutations in the Bordetella pertussis bvgS gene. J Bacteriol. 1992;174(3):970–979. doi: 10.1128/jb.174.3.970-979.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goyard S, Mireau H, Ullmann A. Mutations which result in constitutive expression of the Bordetella pertussis filamentous haemagglutinin gene. Res Microbiol. 1995;146(5):363–370. doi: 10.1016/0923-2508(96)80282-8. [DOI] [PubMed] [Google Scholar]
- 45.Beier D, Schwarz B, Fuchs TM, Gross R. In vivo characterization of the unorthodox BvgS two-component sensor protein of Bordetella pertussis. J Mol Biol. 1995;248(3):596–610. doi: 10.1006/jmbi.1995.0245. [DOI] [PubMed] [Google Scholar]
- 46.Lacey BW. Antigenic modulation of Bordetella pertussis. J Hyg. 1960;31:423–434. doi: 10.1017/s0022172400038134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coote JG. Antigenic switching and pathogenicity: environmental effects on virulence gene expression in Bordetella pertussis. J Gen Microbiol. 1991;137(11):2493–2503. doi: 10.1099/00221287-137-11-2493. [DOI] [PubMed] [Google Scholar]
- 48.Manetti R, Arico B, Rappuoli R, Scarlato V. Mutations in the linker region of BvgS abolish response to environmental signals for the regulation of the virulence factors in Bordetella pertussis. Gene. 1994;150(1):123–127. doi: 10.1016/0378-1119(94)90870-2. [DOI] [PubMed] [Google Scholar]
- 49.Menozzi FD, Mutombo R, Renauld G, Gantiez C, Hannah JH, et al. Heparin-inhibitable lectin activity of the filamentous hemagglutinin adhesin of Bordetella pertussis. Infect Immun. 1994;62(3):769–778. doi: 10.1128/iai.62.3.769-778.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stibitz S, Black W, Falkow S. The construction of a cloning vector designed for gene replacement in Bordetella pertussis. Gene. 1986;50(1–3):133–140. doi: 10.1016/0378-1119(86)90318-5. [DOI] [PubMed] [Google Scholar]
- 51.Antoine R, Alonso S, Raze D, Coutte L, Lesjean S, et al. New virulence-activated and virulence-repressed genes identified by systematic gene inactivation and generation of transcriptional fusions in Bordetella pertussis. J Bacteriol. 2000;182(20):5902–5905. doi: 10.1128/jb.182.20.5902-5905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imaizumi A, Suzuki Y, Ono S, Sato Y, Sato H. Heptakis (2,6-O-dimethyl)beta-cyclodextrin: a novel growth stimulant for Bordetella pertussis phase I. J Clin Microbiol. 1983;17(5):781–786. doi: 10.1128/jcm.17.5.781-786.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antoine R, Huvent I, Chemlal K, Deray I, Raze D, et al. The periplasmic binding protein of a tripartite tricarboxylate transporter is involved in signal transduction. J Mol Biol. 2005;351(4):799–809. doi: 10.1016/j.jmb.2005.05.071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotides used in this study
(0.07 MB PDF)
bvgS sequences
(0.35 MB RTF)
bvgA sequences
(0.04 MB RTF)