Abstract
The transcription factor MafA regulates glucose-responsive expression of insulin. MafA-deficient mice have a normal proportion of insulin+ cells at birth but develop diabetes gradually with age, suggesting that MafA is required for maturation and not specification of pancreatic β-cells. However, several studies show that ectopic expression of MafA may have a role in specification as it induces insulin+ cells in chicken gut epithelium, reprograms adult murine acinar cells into insulin+ cells when in combination with Ngn3 and Pdx1, and triggers the lens differentiation. Hence, we examined whether MafA can induce specification of β-cells during pancreatic development. When the MafA transgene is expressed in Pdx1+ pancreatic progenitors, both pancreatic mass and proliferation of progenitors are reduced, at least partially due to induction of cyclin kinase inhibitors p27 and p57. Expression of MafA in Pdx1+ cells until E12.5 was sufficient to cause these effects and to disproportionately inhibit the formation of endocrine cells in the remnant pancreas. Thus, in mice, MafA expression in Pdx1+ pancreatic progenitors is not sufficient to specify insulin+ cells but in fact deters pancreatic development and the differentiation of endocrine cells. These findings imply that MafA should be used to enhance maturation, rather than specification, of β-cells from stem/progenitor cells.
Keywords: MafA, pancreas, β-cell, insulin gene transcription factor, endocrine differentiation, pancreatic development
INTRODUCTION
Cloning of the insulin gene transcription factor RIPE3b1 as MafA (Kataoka et al., 2002; Matsuoka et al., 2003; Olbrot et al., 2002) initiated a systematic examination of the function of Maf factors in the endocrine pancreas. Maf factors are subdivided into two classes, “large” (236-370 aa; c-Maf, MafB, NRL and MafA/L-Maf) and “small” (149-162 aa; MafF, MafG and MafK) (Blank et al., 1997; Matsushima-Hibiya et al., 1998; Ogino et al., 1998; Ring et al., 2000). All “large Maf” factors have an N-terminal serine/proline/threonine rich acidic activation domain that is absent in “small Maf” factors (Blank et al., 1997; Benkhelifa, et al. 2001). Large Maf factors are important regulators of cellular differentiation: c-Maf has been implicated in lymphopoiesis and lens development (Ho et al., 1996; Kajihara et al., 2001; Kim et al., 1999); loss of MafB function causes segmentation abnormalities in caudal hindbrain, defective inner ear development, abnormal differentiation of kidney podocytes, hematopoietic cells, and pancreatic α- and β- cells (Artner et al., 2007; Cordes and Barsh, 1994; Nishimura et al., 2008; Sadl et al., 2002; Sieweke et al., 1996); another large Maf factor MafA/L-Maf (in quail/chicken) is crucial for lens development (Benkhelifa et al., 1998; Ogino and Yasuda, 1998; Ring et al., 2000).
Redundant and discrete functions of large Maf proteins determine the lens induction and fiber differentiation program. In the lens differentiation program, ectopically expressed MafA can convert ectodermal cells into lens fibers (Benkhelifa et al., 1998; Ogino and Yasuda, 1998; Reza et al., 2007b; Reza and Yasuda, 2004). During this process, MafA is expressed earlier than cMaf, which is followed by MafB expression, and Maf proteins expressed early positively regulate expression of later-expressed one(s) while MafB negatively regulates expression of early Maf factors (Reza et al., 2007b). Misexpression of any of the large Mafs in surface ectoderm outside the lens-forming area was sufficient to induce expression of δ-crystallin, although misexpression of MafA resulted in the highest amount of δ-crystallin expression. Furthermore, over-expression of MafA and cMaf did not perturb normal development whereas ectopic expression of MafB resulted in improperly organized lens cells. These results demonstrate that MafA can trigger the lens differentiation program.
MafA, MafB and cMaf are all expressed in pancreatic endocrine cells (Matsuoka et al., 2003; Nishimura et al., 2006). In adult pancreas, expression of MafA and MafB is restricted to β- and α-cells respectively, while cMaf is expressed in both cell types. During embryonic development, MafA expression remains restricted to insulin-expressing (insulin+) cells, but MafB is expressed not only in glucagon+ but also in insulin+ cells (Nishimura et al., 2006). We showed that maturation of insulin+ cells is an ordered process wherein we first detect MafB+insulin+ cells, followed by the expression of Pdx1high and then MafA in these insulin+ cells. The ability of large Maf factors to bind Maf recognition elements (MARE) in insulin and glucagon promoters and to activate their expression in transient transfection assays (Matsuoka et al., 2004; Nishimura et al., 2006) supports that MafB can activate insulin expression during embryonic development. MafB-deficient mice exhibit reduced number of cells expressing insulin, glucagon, Pdx1 and MafA while no change was seen in the number of cells expressing Ngn3, Nkx6.1, Nkx2.2, Pax6, Isl1 and NeuroD1 (Artner et al., 2007; Nishimura et al., 2008). These results suggest that MafB is indispensable for the lineage commitment of at least a subset of insulin+ cells. In contrast, increasing evidence supports a role of MafA not in the commitment of β-cells but in their maturation. MafA-deficient mice have a normal proportion of β-cells at birth, but the ratio of β to α cells is gradually reduced, resulting in glucose intolerance and diabetes (Zhang et al., 2005). Identification of downstream targets of MafA as critical regulators of insulin synthesis and secretion, including insulin, Pdx1, Nkx6.1, Glut2, GK, GLP1 receptor, prohormone convertase 1/3, pyruvate carboxylase and granuphilin (Kato et al., 2006; Wang et al., 2007), further supports the role of MafA as a regulator of β-cell maturation. These observations suggest that the primary role of MafA in pancreatic β-cells is not to regulate their specification but to regulate maturation and possibly survival/proliferation of these cells. Consistent with this suggestion, MafA expression accompanied the acquisition of glucose responsiveness in insulin+ cells differentiated from human ES cells (Kroon et al., 2008). Even so, as shown for the lens differentiation (Benkhelifa et al., 1998; Ogino and Yasuda, 1998; Reza et al., 2007b; Reza and Yasuda, 2004), it is possible that MafA has the potential to function as a differentiation factor during pancreatic development in addition to its role as a β-cell maturation factor. This suggestion is supported by the demonstration that in ovo electroporation of MafA, but not MafB, in embryonic chick endoderm resulted in the formation of insulin+ cells (Artner et al., 2008). Similarly, expression of MafA together with Ngn3 and Pdx1 was able to reprogram adult acinar cells into insulin+ cells (Zhou et al., 2008). These findings as well as the above-mentioned ability of large Maf factors to bind to MARE elements in insulin and glucagon promoters and to activate their expression in transient transfection assays (Matsuoka et al., 2004; Nishimura et al., 2006) suggest that MafA may be able to specify β-cell development if it is expressed before MafB. This may provide a novel approach not only for initiating formation of β-cells but also triggering their maturation.
Here, we examined whether early expression of MafA in the pancreatic progenitors affected pancreatic development and differentiation of endocrine cells. We generated transgenic mice expressing MafA (tetOMafA) under the control of tetracycline activator (tTA) expressed from Pdx1tTA/+ knockin mice. We show that the expression of MafA transgene (tetOMafA) in early embryonic pancreas inhibits proliferation of pancreatic progenitors and reduces both the pancreatic size and number of endocrine cells. Furthermore, tetOMafA+ cells in the early pancreatic epithelium do not express insulin. Our results show that expression of MafA in mouse embryonic pancreatic epithelium is not sufficient to trigger the differentiation program; on the contrary, it inhibits proliferation of pancreatic progenitors. Our results suggest that MafA-based strategies to generate insulin-producing cells from stem/progenitor cells should use MafA for maturation rather than specification of insulin+ cells.
MATERIALS AND METHODS
Generation of transgenic mice
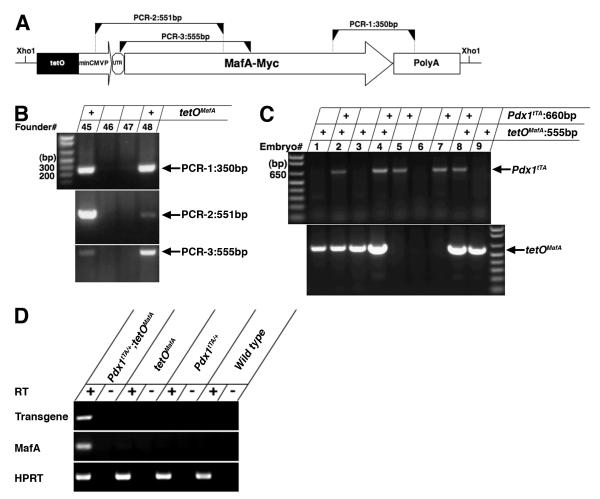
The vector for generation of transgenic mice tetOMafA contained the MafA coding region (Olbrot et al., 2002) followed by Myc tag (EQKLISEEDL) sequence cloned into pTRE-Tight (Clontech Laboratories, Mountain View, CA) containing tetracycline responsive elements (tetO) followed by the minimal cytomegalovirus promoter (miniCMVP) (Fig. 1A), and mice were generated at JDRF transgenic core at Joslin. Positive founders were identified (9 out of 39) by PCR amplifications with 3 different oligonucleotide primer sets. The copy number of integrated transgene was determined by qRT-PCR with transgene specific primers and primers that amplified a single copy gene. Of 9 founders, 6 transmitted the transgene to the F1 generation, and were then crossed with transgenic mice Pdx1tTA/+ (Holland et al., 2002) (purchased from Jackson Laboratory, Bar Harbor, ME) to generate Pdx1tTA/+;tetOMafA bigenic mice. If not specifically indicated, tetOMafA littermates were used as controls in this study.
Figure 1. Generation of Pdx1tTA/+;tetOMafA transgenic mice.
A Schematic of transgene tetOMafA shows the MafA coding region followed by Myc tag sequence (EQKLISEEDL) that was cloned into pTRE-Tight (Clontech) containing tetO followed by the minimal CMV promoter (miniCMVP). Three independent regions, detected by PCR, for the selection of positive clone are shown. B PCR of genomic DNA shows the presence of transgene. DNA from 9 founders were identified as positive to contain all three different PCR regions, image shows identification of two positive founders, #s 45 and 48 by PCR. C Genomic DNA was used in PCR to identify bigenic Pdx1tTA/+;tetOMafA embryos. Image of PCR products shows identification of three Pdx1tTA/+;tetOMafA embryos (#s 2, 4 and 8). D RNA from controls and bigenic E12.5 pancreas was used in RT-PCR analysis, in the absence (-) or presence (+) of reverse transcriptase (RT). Image shows the presence of PCR product corresponding to the transgene MafA-Myc and MafA, only in the pancreas from E12.5 bigenic embryos. Primers specific for HPRT when used in RT-PCR reaction amplified the product from control and bigenic pancreatic RNAs.
Animals
The day of vaginal-plug discovery was designated as E (embryonic day) 0.5. Treatment with doxycycline was performed with 1 mg/ml in drinking water containing 0 calorie sweetener. 21, 15 and 23 embryos of Pdx1tTA/+;tetOMafA, Pdx1tTA/+and tetOMafA respectively were examined for phenotype of reduced pancreatic size at E17.5. Blood glucose values were measured on blood from tail snip using One-Touch glucometer (Life Scan, Milpitas, CA). All animal experiments were approved by the Joslin Institutional Animal Care and Use Committee.
RNA preparation and RT-PCR analysis
Total RNA was prepared from dissected pancreases using the RNeasy Mini Kit and RNase-Free DNase Set (QIAGEN, Valencia, CA) following manufacturer’s instructions. RT-PCR was carried out using random hexamer primers for reverse transcription as per manufacturer’s instructions using the High Capacity cDNA Reverse kit (Applied Biosystems, Carlsbad, CA). Forward and reverse primers used to amplify the genes were as follows: Transgene: 5′-GTGCCAACTCCAGAGCCAGGTG-3′ and 5′-GTTTCAGGTTCAGGGGGAGGTGTG-3′; MafA: 5′-TTCAGCAAGGAGGAGGTCAT-3′ and 5′-CTCTGGAGCTGGCACTTCTC-3′; HPRT: 5′-GTTGGATACAGGCCAGACTTTGTTG-3′ and 5′-GAGGGTAGGCTGGCCTATAGGCT-3′
Immunohistochemistry and quantification
Immunostaining analyses of at least 3 embryos of each genotype were performed on 4 μm sections as described previously (Nishimura et al., 2006). The primary antibodies used were: guinea pig anti-insulin (Linco, Billerica, MA); guinea pig anti-glucagon (Linco); rabbit anti-glucagon (provided by Dr. Appel); rabbit anti-somatostatin (Chemicon International, Billerica, MA); rabbit anti-pancreatic polypeptide (Linco); rabbit anti-ghrelin (Phoenix Pharmaceuticals, Belmont, CA); mouse anti-Nkx2.2 (Developmental Studies Hybridoma Bank, Iowa City, IA); rabbit anti-Nkx6.1 (provided by Dr. Serup); mouse anti-Ngn3 (provided by Drs. Serup and Madsen, Beta Cell Biology Consortium); rabbit anti-Pdx1 (provided by Dr. Slack); mouse anti-synaptophysin (Chemicon International); rabbit anti-Myc (Cell Signaling Technology, Danvers, MA); rabbit anti-amylase (Sigma, Saint Louis, MO); rabbit anti-Sox9 (Chemicon International); rabbit anti-p21(Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti-p27 (BD Pharmingen); goat anti-p57 (Santa Cruz Biotechnology). Rabbit anti-MafA antibody was described previously (Nishimura et al., 2006). For amplification, biotinylated anti-rabbit, anti-mouse or anti-goat antibodies (Jackson ImmunoResearch, West Grove, PA) were used at a 1:400 dilution followed by streptavidin-conjugated Texas red (1:400) or streptavidin-conjugated Alexa fluor 488 (1:400) (Molecular Probes, Eugene, OR). The secondary antibodies were FITC- or Texas red-conjugated anti-rabbit, anti-mouse or anti-guinea pig IgG (Jackson ImmunoResearch). DAPI mounting medium (Vector) provided labeling of nuclei. Immunofluorescent images were obtained using Zeiss LSM410 (Zeiss, Thornwood, NY) in confocal mode; immunoperoxidase images were obtained using an Olympus BH2 bright field microscope. Acquired images were identically processed using Adobe Photoshop CS2; NIH Image J software was used for quantification. The results of quantification were derived generally from 3 to 8 embryos for each genotype, while 11 embryos were used to quantify expression of Pdx1, Myc and MafA at E12.5. For proportion of insulin+, glucagon+, and amylase+ area per total pancreatic epithelial area at E17.5, every 10th section was stained and quantified. Other results were determined by counting stained cells in at least 20 random microscope fields. In most cases, we counted 250 to 1000 cells; we counted at least 50 cells where proportion of cells expressing the gene of interest was very low (Pdx1+ or Myc+ cells at E17.5, Ngn3+ cells at E15.5, and Sox9+ or Hes1+ cells at E12.5 in bigenic embryos). The data are displayed as averages ± s.e.m.; statistical significance was determined using the two-tailed unpaired Student’s t test.
RESULTS
Early expression of MafA under the control of endogenous Pdx1 promoter inhibits pancreatic development and endocrine differentiation
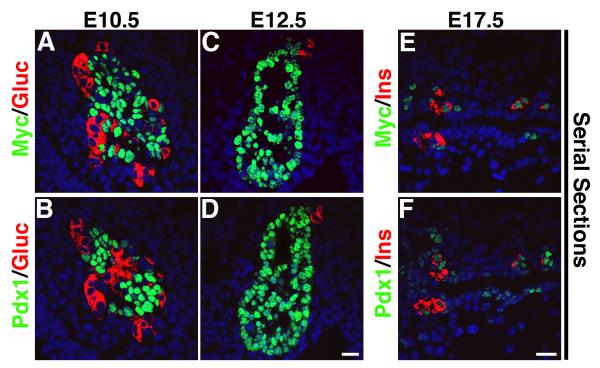
To investigate whether MafA can differentiate pancreatic precursors into insulin+ cells, transgenic mice were generated from a MafA transgenic construct containing Myc-tagged MafA transgene cloned downstream of tetracycline operator (tetO) and minimal CMV promoter (Fig. 1). Nine founders were obtained of which six (#s 12, 14, 23, 48, 50, 53) transmitted MafA transgene (tetOMafA). The six lines were crossed with Pdx1tTA/+mice (Holland et al., 2002) to generate Pdx1tTA/+;tetOMafA bigenic mice in which tetOMafA should be expressed in all Pdx1+ cells, and the administration of doxycycline should inhibit this expression. Based on the expression of Pdx1 during pancreatic development, we would expect the transgene to be expressed broadly in pancreatic epithelium at E12.5 (see supplemental Fig. S1). At this stage endogenous MafA expression is not detected in wild type pancreas, and cDNAs prepared from isolated E12.5 bigenic and control pancreas showed expression of MafA and transgene only in the bigenic pancreas (Fig. 1D). After E12.5, we expect Pdx1 and transgene expression to become restricted to beta cells and possibly some ductal cells. However, such expression pattern of the transgene will only be true if the transgene does not affect endocrine differentiation or Pdx1 expression. Different lines of bigenic mice were examined for MafA transgene protein (tetOMafA) expression by immunohistochemistry using anti-Myc antibody. High level of transgene expression was detected in three Pdx1tTA/+;tetOMafA lines (#s 23, 48 and 53), limited expression in another two lines (#s 14 and 50), and no expression in one line (#12) (data not shown). We examined the expression specificity of transgene in Pdx1+ cells in high tetOMafA expressing lines. Both anti-Pdx1 and anti-Myc antibodies used in the study were raised in rabbits and the detection of these proteins requires biotin/streptavidin-based amplification. Hence, we stained serial pancreatic sections from E10.5, E12.5, and E17.5 to examine the expression efficiency of Myc in Pdx1+ cells. At these stages the patterns of Pdx1 and Myc (tetOMafA) expression were almost identical (Fig. 2). The proportion of pancreatic epithelial cells expressing Pdx1 and Myc were quantified to determine the relative proportion of Pdx1+ cells expressing transgene. Quantification of Pdx1+ and Myc+ cells showed that 91.4±2.5% and 82.3±6.7% of Pdx1+ cells in the pancreatic epithelium at E10.5 and E12.5 in Pdx1tTA/+;tetOMafA embryos, respectively, also expressed Myc (tetOMafA) in adjacent sections. The proportion of Pdx1+ and Myc+ cells in the serial sections from E17.5 showed that at this stage 83.3±6.4% of Pdx1+ cells also expressed Myc (tetOMafA) (Fig. 2). Sections of E12.5 pancreases that were sequentially stained with rabbit anti-Myc and rabbit anti-Pdx1 antibodies with images taken between staining showed transgene expression in nearly all Pdx1+ cells (see supplemental Fig. S2). These results demonstrate that high transgene expressing bigenic lines faithfully express tetOMafA transgene in Pdx1+ cells. We analyzed three high expressing lines to determine the effect of MafA expression in pancreatic progenitors; results from all three lines were similar, hence detailed analysis from line #48 is presented in this study.
Figure 2. TetOMafA transgene is expressed in significant proportion of Pdx1+ cells at different stages of embryos.
In bigenic mice pancreases from E10.5, E12.5 and E17.5 show a significant proportion of Pdx1+ cells (BDF) expressed transgene (Myc+) (ACE). Myc (green ACE), Pdx1 (green BDF), glucagon (red A-D) and insulin (red EF). Bars: 20 μm.
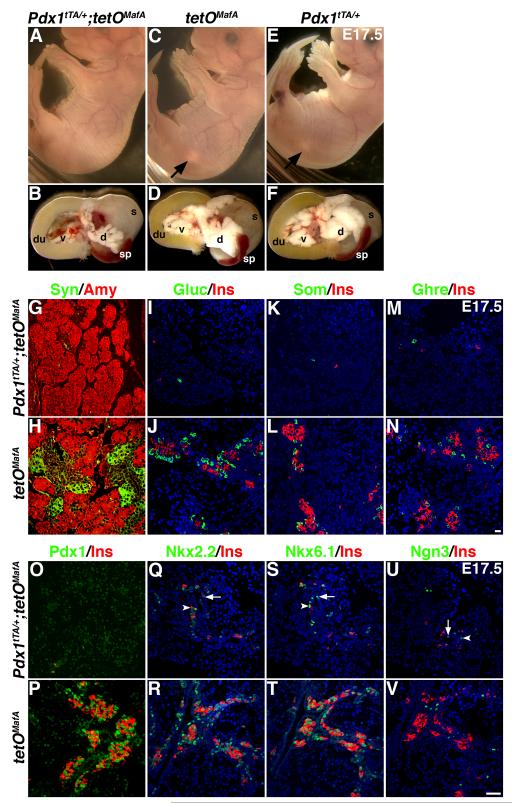
When compared with control (tetOMafA and Pdx1tTA/+) littermates (Fig. 3 C-F), E17.5 bigenic embryos showed significantly reduced pancreatic size (Fig. 3B), such that the pancreas was not visible in intact embryos (Fig. 3A). In the bigenic pancreas, the proportion of synaptophysin+ endocrine cells was severely reduced, while the organization of amylase+ acinar cells was comparable to that in control pancreases (Fig. 3G,H). Misexpression of MafA affected all endocrine cells; only a few cells expressing insulin, glucagon, somatostatin or ghrelin were seen in bigenic pancreases (Fig. 3I-N); cells expressing multiple hormones were not observed. Interestingly, cells expressing Pdx1, Nkx2.2, Nkx6.1 or Ngn3 also were dramatically reduced in E17.5 bigenic pancreas (Fig. 3O-V). Only few Pdx1high cells were observed in the bigenic pancreas, and the few Nkx2.2, Nkx6.1, Ngn3 or insulin-expressing cells were detected near or within the ductal structures. The reduced number of cells expressing these transcription factors suggests that the transgene expression does not selectively inhibits the expression of hormones and synaptophysin in endocrine precursors but rather appears to inhibit the entire endocrine differentiation program. Consistent with these observations, Pdx1tTA/+;tetOMafA bigenic mice are born with reduced pancreatic weights, they rapidly develop hyperglycemia and die with in 48-72h (data not shown).
Figure 3. MafA transgene (tetO MafA) expressed in Pdx1+ cells inhibits pancreatic development.
A, C, E In E17.5 embryos from indicated genotypes, the pancreas (white spot, C, E; arrows) was seen in controls but not in bigenic (A). B, D, F Show the dissected viscera from the A, C, and E embryos containing dorsal pancreas (d), ventral pancreas (v), duodenum (du), spleen (sp) and stomach (s). Bigenic mice had a significantly reduced size of pancreas. G-N The expression of transgene from the initiation of Pdx1 expression until E17.5 inhibited differentiation of endocrine cells. Bigenic mice had significant reduction in hormone-expressing cells as seen with synaptophysin (green, GH), glucagon (green, IJ), somatostatin (green, KL), ghrelin (green, MN) and insulin (red, I-N), while organization of amylase-expressing (red, GH) cells appeared similar in both bigenic and control pancreas. Bar: 20 μm. O-T Reduced expression of key transcription factors accompanies impaired endocrine differentiation. In bigenic pancreas only a few insulin+ (red) cells and Pdx1high (green) cells (O) were seen, whereas both Pdx1high (insulin+) and Pdx1low (insulin-) cells were seen in control pancreas (P). Similarly, the number of cells expressing other transcription factors, Nkx2.2 (green, QR), Nkx6.1 (green, ST), Ngn3 (green, UV) was reduced in Pdx1tTA/+;tetOMafA pancreas compared to controls. A few transcription factor+ cells present in the bigenic pancreas were observed within (arrows) or near (arrowheads) the branching epithelium. Bar: 50 μm.
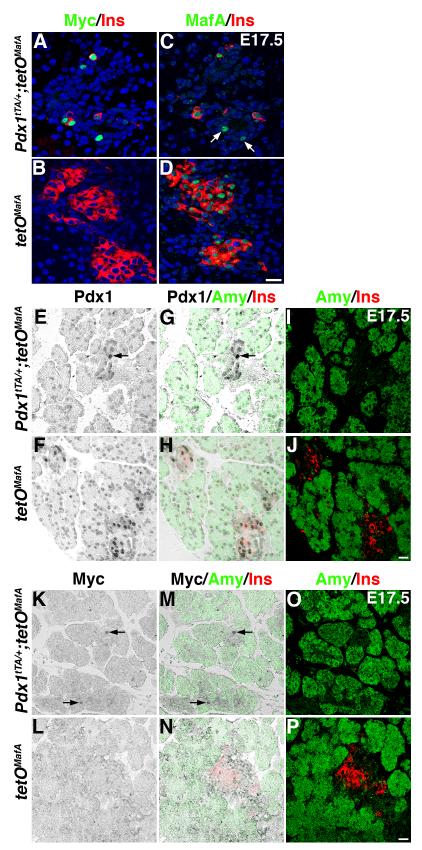
At E17.5, bigenic mice had reduced numbers of insulin or Pdx1 expressing cells (Fig. 3), hence we examined whether at this stage transgene expression was maintained in the bigenic pancreas. In bigenics, only a few insulin or Myc expressing cells were seen, and the majority of Myc+ cells (∼75%) were insulin- (Fig. 4A, 2E), which is consistent with a dramatic decrease in Pdx1+insulin+ cells in bigenics (∼45% of total Pdx1+ cells, Fig. 2F, 3O) compared to controls (∼85% of total Pdx1+ cells, Fig. 3P). No Myc expression was detected in insulin+ cells of control littermates (Fig. 4B). There were a few MafA+insulin- cells in Pdx1tTA/+;tetOMafA pancreas (Fig. 4C) but MafA+insulin- cells were never observed in control littermates (Fig. 4D). To determine the cell-type expressing transgene at E17.5 in bigenic pancreas, pancreatic sections were stained to detect the expression of Pdx1, amylase and insulin (Fig. 4E-J) or Myc, amylase and insulin (Fig. 4K-P). As expected, insulin+ cells and occasional cells in the ducts in controls were Pdx1high, while acinar cells and the majority of ductal cells were Pdx1low. In bigenics, only a few insulin+ cells were seen and some of those were Pdx1high (Fig. 2F), but the majority of insulin-Pdx1high cells seen in the bigenics were ductal cells (Fig 4E, G, I). Consistent with this expression pattern of Pdx1high in E17.5 bigenic pancreas, transgene (Myc+) expression was also seen in occasional ductal cells (Fig. 4 K, M, O). These observations demonstrate that at E17.5 in bigenic pancreas transgene was not expressed in Pdx1low ductal and acinar cells but was restricted to a few insulin- Pdx1high ductal cells and insulin+ Pdx1high cells.
Figure 4. Transgene+ insulin- cells in E17.5 bigenic pancreas are mostly “ductal” cells.
A, B Some Myc+ (green) cells co-expressed insulin (red) while others did not in pancreas of Pdx1tTA/+;tetOMafA, but no Myc expression was seen in control pancreas (tetOMafA). C, D Some MafA+ (green) cells, like Myc+ cells in A, did not express insulin (red) in Pdx1tTA/+;tetOMafA pancreas (arrows) whereas all MafA+ cells were insulin+ in control tetOMafA. Peroxidase staining was performed to clearly show the location of cells in the pancreas (EFKL) that express Pdx1high or Myc, and sections were then stained to detect insulin+ or amylase- cells (IJOP). Merged images (GHMN) show that in the bigenic pancreas Pdx1high insulin- or amylase- cells and Myc+ insulin- or amylase- cells were primarily present in the embryonic ducts, while in the control pancreas there were many Pdx1high+ cells expressing insulin (HJ) but no Myc+ cells (NP) were observed. Bars: 20 μm.
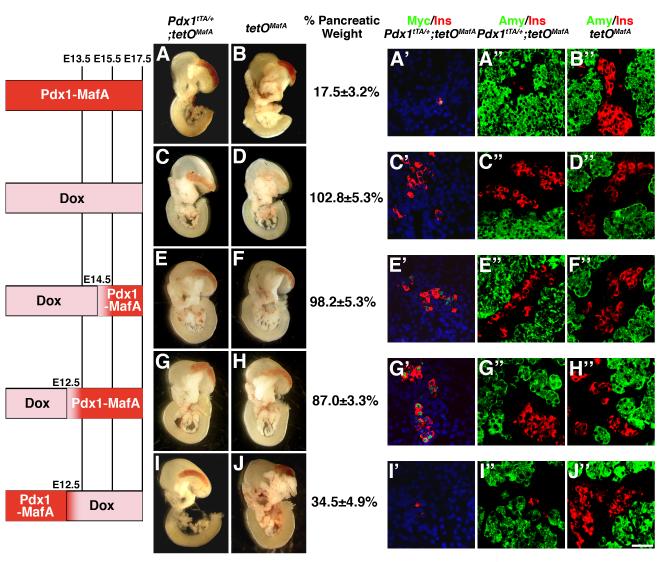
To determine the mechanism responsible for the reduction in pancreatic mass and endocrine cells in bigenic mice, expression of tetOMafA was regulated for different durations by doxycycline (Dox) administration. In the absence of Dox, continuous expression of MafA in Pdx1+ cells until E17.5 inhibited pancreatic mass by nearly 4 to 5-fold (17.5%±3.2% of control, p=0.002) (Fig. 5A,B) and preferentially reduced insulin-expressing cells in the remaining pancreas (Fig. 5A”,B”). As a control, administration of Dox from conception until E17.5 resulted in pancreas of normal appearance and weight (Fig. 5C,D). Similarly, administration of Dox until E14.5 or E12.5 prevented the reduction of pancreatic mass (Fig. 5E-H), while Dox treatment from E12.5 to E17.5, permitting misexpression of MafA in the Pdx1+ pancreatic progenitors until ∼E12.5, was sufficient to reduce the pancreatic mass (p=0.009 for comparison of pancreatic weights of bigenic Fig. 5I and control Fig. 5J littermates). This reduction was not different from that observed in the bigenics that did not receive any Dox (p=0.21 for comparison of pancreatic weights of bigenic Fig. 5I and bigenic Fig. 5A embryos). Thus, misexpression of MafA transgene in Pdx1+ cells until E12.5 was necessary and sufficient to reduce pancreatic weight.
Figure 5. Expression of transgene until E12.5 is sufficient to inhibit pancreatic mass and endocrine differentiation in Pdx1tTA/+;tetOMafA mice.
Viscera from E17.5 embryos without treatment (A, B) or after doxycycline administration (C-J) for the indicated durations (CD from E0.5 to E17.5, EF from E0.5 to E14.5, GH E0.5 to E12.5, IJ from E12.5 to E17.5). Pancreatic weights are presented as % pancreatic mass ± s.e.m. of the bigenic relative to that of control littermates (100%), from at least 5 embryos from each group. A representative image of the pancreatic sections from each time point stained for Myc (green) and insulin (red) (A’C’E’G’I’) or amylase (green) and insulin (red) (A”-J”) is shown. Bar: 50 μm.
Pancreases from mice receiving different Dox treatments were stained to detect the expression of transgene (Myc) and insulin in bigenics, and insulin and amylase in both bigenics and controls (Fig. 5A’, A”, B” to I’, I”, J”). With Dox treatment until E17.5, E14.5 or E12.5, pancreases from bigenics had comparable volumes of insulin+, amylase+ and glucagon+ cells as in the controls (Table 1, data not shown); however, only those receiving Dox until E14.5 or E12.5 had Myc+ cells, most of which co-expressed insulin. In pancreases from mice not given Dox or given Dox only after E12.5, relative volume of insulin+, glucagon + (data not shown) and Myc+ cells were dramatically reduced; there was some increase in the relative volume of amylase+ cells but it was not significantly different than that in controls (Table 1). Thus, these results demonstrate that the Pdx1-promoter dependent expression of transgene in the bigenic embryos until E12.5 is sufficient to reduce both pancreatic weight and endocrine cells, while the expression of transgene after E12.5 did not affect the pancreatic mass and the formation of endocrine cells.
Table I.
Relative Volume of Insulin+, Glucagon+ and Amylase+ Cells at E17.5
| Control | No Dox | Dox until E17.5 |
Dox until E14.5 |
Dox until E12.5 |
Dox from E12.5 |
|
|---|---|---|---|---|---|---|
| Insulin+ | 3.8±0.5% | 0.1±0.0%* | 3.8±0.3% | 3.6±0.1% | 3.4±0.2% | 0.2±0.0%* |
| Glucagon+ | 4.5±0.7% | 0.2±0.0%* | 4.2±0.1% | 4.4±0.1% | 4.3±0.1% | 0.3±0.1%* |
| Amylase+ | 82.3±1.2% | 93.5±4.9% | 85.6±3.6% | 85.2±3.3% | 88.7±4.1% | 91.7±6.7% |
% Area relative to pancreatic epithelial area. Average±s.e.m. from three embryos each.
p<0.05.
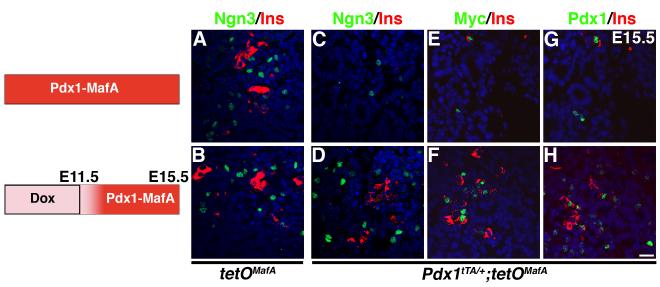
The finding that expression of transgene until E12.5 was sufficient to significantly reduce the endocrine cells (Fig. 5) suggested that the Pdx1+ progenitors present in the E12.5 epithelium gave rise to the majority of endocrine cells seen at E17.5. Interestingly, at E12.5 the number of Ngn3+ cells present in the normal pancreatic epithelium is low, reaching a peak by E14.5/E15.5 (White et al. 2008). Hence, one would expect that a significant proportion of Ngn3+ endocrine progenitors seen at E15.5 were derived from cells committed to this fate before E12.5 and that transgene expression after E12.5 should not affect the number of these progenitors. To test this hypothesis, we examined whether transgene expression in Pdx1+ cells after E12.5 affected Ngn3 expression in bigenic embryos at E15.5. Pregnant females were treated with or without Dox until E11.5, and at E15.5 pancreases were analyzed for Ngn3, Myc, Pdx1 and insulin. Dox did not affect the numbers of Ngn3 or insulin expressing cells in pancreases from tetOMafA controls (Fig. 6A,B). Similarly, in bigenics, expression of the transgene from ∼E12 to E15.5 did not affect the numbers of Ngn3+ and insulin+ cells (Fig. 6D, Table 2). Since at this stage not all insulin+ cells are Pdx1high, and not all Pdx1high cells are insulin+ (Nishimura et. al., 2006), we observed in bigenics expressing transgene from ∼E12 to E15.5 only some insulin+ cells expressing both the transgene and Pdx1high while some Myc+ and Pdx1high cells did not express insulin (Fig. 6F,H). However, expression of MafA transgene from conception until E15.5 significantly reduced Ngn3+ and insulin+ cells (Fig. 6C,E,G, Table 2), demonstrating that a significant proportion of Ngn3+ cells seen at E15.5 are derived from the Pdx1+ progenitor cells committed to endocrine fate before E12.5. These results further support our conclusion that the expression of MafA transgene in Pdx1+ cells only until E12.5 is detrimental to both pancreatic development and endocrine differentiation while its expression after E12.5 is not.
Figure 6. Expression of transgene before E12.5 reduces numbers of endocrine progenitors.
The schematic shows the duration of inhibition of transgene expression by Dox in bigenic mice expressing transgene from conception (ACEG), or in mice receiving doxycycline (to inhibit transgene expression) until E11.5 (BDFH). Ngn3 expression was observed in control tetOMafA pancreas at E15.5 (AB). In Pdx1tTA/+;tetOMafA embryos, expression of Ngn3 was inhibited (C). However, no reduction in Ngn3+ (D), insulin+ (DFH), cells was seen in pancreas expressing transgene from ∼E12 to E15.5. Pancreases from embryos with transgene expression from ∼E12 to E15.5 also showed insulin+ and insulin- cells expressing Myc and Pdx1 (FH), Ngn3 (green ABCD), Myc (green EF), Pdx1 (green GH) and insulin (red). Bars: 20 μm.
Table II.
Proportion of Ngn3+ and Insulin+ Cells Compared to tetOMafA Control at E15.5
| No Dox | Dox from E11.5 to E15.5 | |||
|---|---|---|---|---|
| tetOMafA | Pdx1tTA/+;tetOMafA | tetOMafA | Pdx1tTA/+;tetOMafA | |
| Ngn3+ Cells | 100.0±13.0% | 17.7±3.2%* | 100.8±5.8% | 107.3±15.3% |
| Insulin+ Cells | 100.0±2.2% | 7.7±2.2%* | 112.8±9.2% | 114.1±6.4% |
Ngn3+ cells counted from three embryos each: n=248 (tetOMafA w/o Dox), 44 (Pdx1tTA/+;tetOMafA w/o Dox), 250 (tetOMafA w/Dox), 266 (Pdx1tTA/+;tetOMafA w/Dox).
p<0.05.
Expression of MafA in Pdx1+ pancreatic progenitors inhibits their proliferation and differentiation
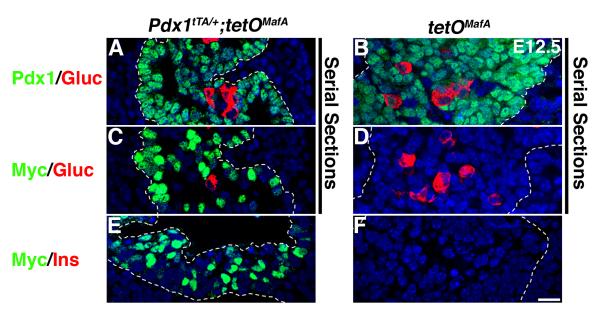
The reduced pancreatic mass and number of endocrine cells in response to transgene expression until E12.5 could result from precocious differentiation or from impaired proliferation and differentiation of pancreatic progenitors. Cells expressing insulin and MafA are rarely detected at E11.5-E12.5 in wild type pancreas (Artner et al., 2006; Nishimura et al., 2006); similarly we did not observe MafA+ cells or MafA mRNA in E12.5 pancreases from control tetOMafA embryos (Fig. 1D, data not shown). However, in serial sections from E12.5 pancreases, the majority of cells in both bigenic and control embryos expressed Pdx1 and 82.3±6.7% of Pdx1+ cells in Pdx1tTA/+;tetOMafA embryos also expressed Myc (Figs. 2, 7ACE, S2). It is important to note that the transgene expression in Pdx1+ progenitor cells at E12.5 was not sufficient to induce the expression of insulin or glucagon (Fig. 7C,E). Similarly, misexpression of tetOMafA in pancreatic progenitors did not trigger precocious differentiation into the other pancreatic cell types such as amylase+ acinar (Fig. S3) or DBA+ ductal cells (data not shown).
Figure 7. TetOMafA expression in E12.5 Pdx1+ cells is not sufficient to induce expression of hormones.
At E12.5 in bigenic mice a significant proportion of Pdx1+ cells (A) expressed transgene (Myc+) (C) but this transgene expression was not sufficient to promote expression of glucagon (C) or insulin (E). In controls (B, D, F) no transgene expression was seen. Pdx1 (green AB), Myc (green CDEF), glucagon (red A-D) and insulin (red EF). Dashed lines demarcate pancreatic tissue. Bar: 20 μm.
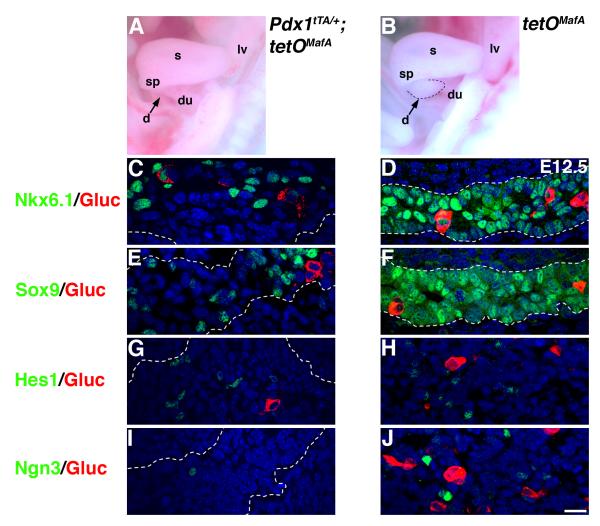
Since reduced pancreatic mass in bigenic embryos could not be accounted by premature differentiation of progenitors, we examined whether expression of tetOMafA affected transcription factors (Sox9, Hes1, Nkx6.1 and Ngn3) required for proliferation of progenitors and their endocrine differentiation (Gradwohl et al., 2000; Jensen et al., 2000; Sander et al., 2000; Seymour et al., 2007). At E12.5, most cells in the control littermates expressed Nkx6.1 and Sox9, and a smaller sub-population of cells expressed Hes1 or Ngn3 (Fig. 8D, F, H, J). In contrast, in Pdx1tTA/+;tetOMafA embryos, pancreatic buds were smaller (Fig. 8A) and the proportion of Pdx1+ cells expressing Sox9 was significantly reduced (21.8±2.6% of control), but that expressing Hes1 remained unchanged (96.5±15.5% of control) (Fig. 8E,G). Significant reductions in cells expressing Nkx6.1 or Ngn3 were also observed in bigenic embryos (Fig. 8C,I). These results suggest that early expression of MafA inhibited genes regulating proliferation and differentiation of pancreatic progenitors.
Figure 8. Early expression of MafA in Pdx1+ cells inhibits expression of Nkx6.1, Sox9 and Ngn3 but not Hes1.
A, B The viscera from E12.5 embryos of each genotype contained dorsal pancreas (d), duodenum (du), spleen (sp), stomach (s) and liver (lv). Dashed line demarcates dorsal pancreatic bud (arrow) in control and the corresponding region in the viscera of bigenic mice had smaller and not clearly defined pancreatic bud (arrow). C-J At E12.5 in bigenic pancreas the number of cells expressing Nkx6.1, Sox9 or Ngn3 was reduced, but the number of Hes1+ cells was comparable to that in controls. Nkx6.1 (green CD), Sox9 (green EF), Hes1 (green GH), Ngn3 (green IJ) and glucagon (red). Dashed lines demarcate pancreatic tissue. Bar: 20 μm.
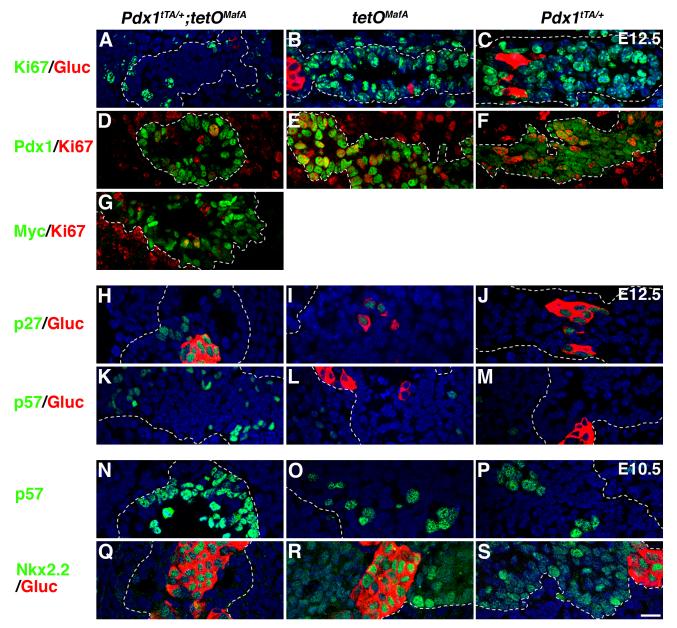
The impaired pancreatic growth by misexpressed tetOMafA could result from enhanced apoptosis and/or inhibition of proliferation of pancreatic progenitors. TUNEL staining was performed on E12.5 pancreas to examine if transgene expression induced cell death. Very few apoptotic cells were seen in either bigenic or control pancreas (Fig. S4), minimizing the role of increased apoptosis in the reduction of pancreatic mass. We next investigated whether transgene expression inhibited proliferation of pancreatic progenitors. At E12.5 (Fig. 9A-F) a dramatically reduced number of Pdx1+ cells co-expressed Ki67 (14.0±0.3%) in Pdx1tTA/+;tetOMafA pancreases compared with controls Pdx1tTA/+ (86.1±4.0%) and tetOMafA (83.6±1.0%). Consistent with these results, very few Myc+ cells in bigenic pancreas expressed Ki67 (Fig. 9G). We examined the expression of cyclin kinase inhibitors (CKIs) (p21, p27, and p57) that have been implicated in regulating pancreatic growth. In E12.5 bigenic pancreases the number of cells expressing p27 was increased compared to controls (Fig. 9H-J). Only glucagon-expressing cells in control littermates expressed p27, while in bigenic embryos both glucagon+ and glucagon- cells expressed p27. Expression of p21 was very low in both control and Pdx1tTA/+;tetOMafA pancreas (data not shown). However, a key effect of misexpression of MafA in pancreatic progenitors was an increase in the number of cells expressing p57 in bigenic mice (Fig. 9K-M). In wild type embryos p57 is expressed in some pancreatic progenitors and inhibits their cell cycle progression (Georgia et al., 2006), so an increase of p57 expression in bigenic progenitors should significantly inhibit their proliferation. To examine whether expression of transgene did initiate changes to cell-cycle regulation and differentiation of pancreatic epithelium before E12.5, we examined E10.5 pancreatic sections from bigenics and controls (Fig. 9N-S). At E10.5, p57 expression can be seen in some pancreatic progenitors in controls, but a dramatically higher proportion of bigenic progenitors cells expressed p57. Interestingly, Nkx2.2 that is broadly expressed in the control pancreatic epithelium at E10.5 was restricted only to glucagon+ cells. Our results show that misexpression of transgene in early progenitors did not trigger apoptosis nor induced premature differentiation into endocrine, acinar or ductal cells. Rather, it enhanced the number of progenitor cells expressing p27 and p57 and reduced the proportion of cells expressing transcription factors associated with endocrine differentiation. We suggest that such changes should impede proliferation and differentiation of progenitors thereby reducing pancreatic mass and proportion of endocrine cells (Fig. 3-5).
Figure 9. Expression of MafA in early pancreatic epithelium inhibits proliferation and induces expression of cyclin kinase inhibitors (CKIs).
At E12.5 a significant reduction in pancreatic cells expressing Ki67 (green, ABC; red, DEFG) was seen in bigenic but not in controls (tetOMafA or Pdx1tTA/+), resulting in a reduced proportion of Pdx1+Ki67+ (D) and Myc+Ki67+ (G) cells in bigenic pancreas. In bigenic pancreas an increased proportion of cells expressed p27 (H) and p57 (K) than in the controls (IJLM). Quantification of Pdx1+ cells co-expressing Ki67 in Pdx1tTA/+;tetOMafA (14.0±0.3%), tetOMafA (83.6±1.0%) and Pdx1tTA/+ (86.1±4.0%) demonstrated a significant reduction in number of proliferating cells of bigenic embryos at E12.5. Total number of Pdx1+ cells counted from 3 embryos: 347 (Pdx1tTA/+;tetOMafA), 360 (tetOMafA), 250 (Pdx1tTA/+). Number of cells expressing p57 (N) was already increased by E10.5, while the number of cells expressing Nkx2.2 (Q) was reduced compared to control (OPRS). Pdx1 (green, DEF), p27 (green, HIJ), p57 (green, K-P), Myc (green, M), Nkx2.2 (green, Q-S) and glucagon (red, A-C, H-M, Q-S). Dashed lines demarcate pancreatic tissue. Bar: 20 μm.
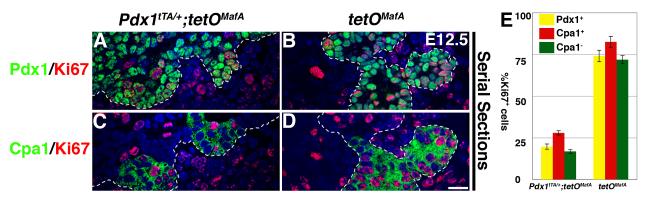
Of interest, transgene-dependent inhibition of pancreatic mass did not equally inhibit the proportions of endocrine and acinar cells (Figs. 3-5). Recent results suggest that the pancreatic mass is determined by the size of initial pancreatic progenitor pool (Stanger et al., 2007) and that the endocrine, acinar and ductal cells are derived from multipotent progenitors marked by the expression of Pdx1+Ptf1a+Myc+Cpa1+, present during branching morphogenesis (Zhou et al., 2007). Cpa1+ cells give rise to Cpa1- trunk cells that form endocrine and ductal cells, and by E14.5, Cpa1+ cells undergo a developmental switch limiting their fate to only acinar cells. So, we examined whether misexpression of MafA transgene differentially regulated proliferation of Pdx1+, Cpa1+ and Cpa1- cells at E12.5. Bigenic pancreas had a significant (p=0.004) reduction of cells co-expressing Pdx1 and proliferation marker Ki67 (Fig. 10A,B). Similarly, the number of cells expressing Cpa1+Ki67+ or Cpa1-Ki67+ was significantly lower (p=0.002 and 0.003) in bigenic than in control pancreases (Fig. 10C,D). Bigenic pancreases showed 66.1±2.8% reduction in Cpa1+Ki67+ cells compared to controls, which was significantly less than the reduction in Cpa1-Ki67+ cells (76.7±1.2%, p=0.02). Moreover, we observed that in Pdx1+ pancreatic epithelium, Cpa1+ cells were present at a higher proportion in bigenic (30.8±4.8%) than in control (19.9±2.8%) mice. These observations suggest that misexpression of MafA in early pancreatic epithelium causes a greater inhibition in proliferation of endocrine and duct progenitors (Cpa1-) than acinar progenitors (Cpa1+), which in turn may cause a disproportionate reduction of endocrine cells observed in Pdx1tTA/+;tetOMafA embryonic pancreas.
Figure 10. Expression of tetOMafA in Pdx1+ cells until E12.5 results in a greater reduction in the proliferation of Cpa1- than of Cpa1+ progenitors.
At E12.5 there was a clear reduction in the proliferating (Ki67+, red) Pdx1+ (green, AB) and Cpa1+ (green, CD) cells in bigenic pancreas. E. Quantification determined the % of Pdx1+, Cpa1+ and Cpa1- cells in the Pdx1+ areas (demarcated by dashed line) that co-expressed Ki67. Total number of Pdx1+ cells counted from 3 embryos each from bigenic: 841, control: 1892. Bar: 20 μm.
DISCUSSION
In this study, we investigated the consequences of MafA misexpression in the early pancreatic epithelium in order to examine whether MafA can induce specification of β-cell during pancreatic development. We showed that expression of MafA in Pdx1+ cells was not sufficient to specify insulin+ cells but did result in significantly reduced pancreatic mass with a preferential reduction in endocrine over acinar cells in the remaining pancreas. In E12.5 pancreatic epithelium a majority of cells expressed Pdx1 and the transgene, but this co-expression did not result in increased proportion of endocrine cells. The transgene expression increased the number of cells expressing cyclin kinase inhibitors p27 and p57, without changing the number of apoptotic cells. Importantly, transgene expression in early pancreatic epithelium also reduced the number of cells expressing Sox9, Nkx6.1, Nkx2.2 and Ngn3, factors that are implicated in growth and differentiation of pancreatic progenitors. Furthermore, unlike at E12.5, when the majority of cells in the pancreatic epithelium expressed both Pdx1high and the transgene, at E15.5 and E17.5 the number of cells expressing these genes was reduced in the bigenic pancreas. We suggest that this reduction in the number of cells expressing Pdx1high and the transgene is due to the inhibition of endocrine differentiation by the early expression of transgene; this inhibition limits the induction of Pdx1high expression during secondary transition (Fig. S5). Thus, our results demonstrate that the expression of MafA transgene in the pancreatic progenitors inhibits their proliferation without enhancing their precocious differentiation into pancreatic cell types.
In chicken and quail, MafA/L-Maf is essential for lens development and triggers the lens differentiation program (Ogino and Yasuda, 1998). Additionally previous data suggested that MafA can also initiate specification of insulin+ cells. Stable expression of MafA in pancreatic α-cell lines induced expression of endogenous insulin gene (Matsuoka et al., 2004). Electroporation of MafA, but not MafB, in chicken gut epithelium in ovo induced insulin-expressing cells (Artner et al., 2008). MafA, in conjunction with Pdx1 and NeuroD1, induced expression of endogenous insulin gene in liver, with MafA being essential for the induction of mouse Insulin1 expression (Kaneto et al., 2005). Similarly, MafA expression, together with Pdx1 and Ngn3, could reprogram pancreatic acinar cells into insulin+ cells (Zhou et al., 2008). These results suggested that MafA has functions in addition to its role as a β-cell maturation factor (Nishimura et al., 2006; Wang et al., 2007) and may regulate specification of insulin+ cells. Although, our results do not rule out the possibility that in vivo co-expression of Ngn3, Pdx1 and MafA can specify insulin+ cells, our finding clearly demonstrates that early expression of MafA in the pancreatic epithelium does not result in precocious differentiation of progenitors into endocrine/β-cells. Furthermore, our observation that not all cells expressing Pdx1 and MafA/Myc at different stages during pancreatic development (E12.5, E15.5 and E17.5) expressed insulin suggests that the expression of MafA and Pdx1 in pancreatic cells is not sufficient to trigger insulin expression. Consistent with our observation, adenoviral-mediated expression of Pdx1 and MafA (or combination of any two factors of Ngn3, Pdx1 and MafA) in adult acinar cells was not sufficient to convert them into insulin+ cells (Zhou et al., 2008). Yet the requirement of MafA in reprogramming acinar cells into insulin+ cells contrasts with our conclusions that MafA is not required for the specification of insulin+ cells. MafB, a downstream target of Ngn3 (Juhl et al., 2008; White et al., 2008) that can regulate specification of insulin+ cells (Artner et al., 2007; Nishimura et al., 2008), is likely to have been induced by Ngn3 in the acinar cells. It is likely that Ngn3 can trigger MafB-dependent expression of insulin in acinar cells and expression of Pdx1 and MafA are required for survival/proliferation and maturation of β-cells. The ability of MafA, but not MafB, to specify insulin+ cells in chicken gut epithelium (Artner et al., 2008), could be due to differences in embryonic pancreas and gut, and/or due to some species-specific differences.
Glucagon+ cells do not express Pdx1, so at present we cannot rule out the role of MafA in specifying insulin expression in glucagon+ and other endocrine cells. Future analysis of mice expressing MafA in different endocrine cell types will address this issue. Our results are consistent with other studies in that the formation of insulin+ cells requires regulated expression of several transcription factors (Oliver-Krasinski and Stoffers, 2008) and that MafA is primarily involved in regulating the maturation of insulin+ cells (Nishimura et al., 2006). Such role of MafA is distinct from its role in the lens development where MafA (L-Maf) is expressed earlier than cMaf, followed by the expression of MafB (Reza et al., 2007b). Electroporation of MafA in chick embryos induced expression of both c-Maf and MafB, while electroporated c-Maf induced expression of MafB but not MafA. Furthermore, electroporation of MafB resulted in down regulation of both MafA and c-Maf expression. These results suggest that different Maf factors may have different roles at different stages of development, and their order of expression may determine their role in differentiation of the cell type. Hence, we suggest that the different order of expression of Maf factors [MafA before MafB in chicken lens (Reza et al., 2007b) but MafB before MafA in pancreatic endocrine cells (Artner et al., 2006; Nishimura et al., 2006)], may explain the different function of MafA during lens and pancreatic development.
The reduced pancreatic mass in bigenic mice resulting from reduced proliferation of pancreatic progenitors is consistent with the recent finding that pancreatic size is determined by the size of embryonic pancreatic progenitor pool (Stanger et al., 2007). The reduced pancreatic mass in both studies was achieved using the same Pdx1tTA/+ mice by inducing tetO-mediated expression of a transgene, diphtheria toxin (DTA) (Stanger et al., 2007) or MafA (Fig. 3, 5). However, the relative inhibition of pancreatic weight by these two effectors was different: expression of DTA resulted in almost complete agenesis of pancreas while MafA misexpression led to only a ∼4-5 fold reduction in pancreatic mass. This difference may result from the ability of DTA to directly inhibit protein synthesis leading to cell death while MafA may inhibit growth by, either directly or indirectly, inducing genes such as CKIs p57 and p27. The almost complete loss of pancreas in DTA transgenic mice suggests that Pdx1 promoter drives tTA-mediated expression of the tetO-regulated transgene in most, if not all, cells of the pancreatic epithelium. Interestingly, the first visible effect of DTA expression was seen only at E10.5, suggesting that ∼24 h are needed to see the induction of DTA and its action (Stanger et al., 2007). At E10.5 and E12.5 we observed MafA transgene expression in 91.4±2.5% and 82.3±6.7% of Pdx1 expressing cells, respectively. We suggest that the lack of transgene expression in all Pdx1+ cells is most likely due to the delay in the expression of transgene in newly proliferated Pdx1+ cells and/or the difference in detection sensitivity of anti-Pdx1 and anti-Myc antibodies.
Additionally, our studies reveal that the majority of endocrine cells seen at E17.5 are derived from endocrine progenitors that are committed before E12.5. MafA expression in Pdx1+ cells until E12.5 almost completely inhibited formation of endocrine cells even though E17.5 pancreas of bigenic mice had a normal proportion of acinar cells. A recent study suggested that before E14 Cpa1+ cells were the primary, if not the only, multipotent progenitors and that after E14, these cells differentiated into acinar cells only (Zhou et al., 2007). However, the conversion of Cpa1+ cells into acinar cells is not synchronous as one can detect acinar cells before E14.5. At E12.5, a higher proportion of Cpa1+ (tip) cells is proliferating than of the Cpa1- (trunk) cells (Fig. 10), so this differential proliferation would increase the proportion of acinar cells after E14. Consistent with the results from Zhou et. al. 2007, we observed that at E12.5 control pancreas had more replicating Cpa1+ cells than Cpa1- cells (82.6±3.2% Cpa1+ vs. 71.9±2.4%, Cpa1-). In bigenic mice, misexpression of MafA significantly inhibited the proliferation of both tip (Cpa1+ p=0.002) and trunk (Cpa1- p=0.003) cells, yet the proportion of Cpa1+ cells in cell cycle (28.0±1.3% Cpa1+Ki67+) was 166% of the Cpa1- cells (16.8±1.1% Cpa1-Ki67+). A consequence of this differential proliferation of Cpa1+ and Cpa1- cells in control and bigenic mice was that in the bigenic pancreas at E12.5, there were a higher proportion of tip cells (Cpa1+ cells, 30.8±4.8% in bigenic vs. 19.9±2.8% in control). Thus, it is likely that inhibiting proliferation of early pancreatic progenitor will more greatly reduce the proliferation of cells that will give rise to endocrine cells (Cpa1-) than to acinar cells (Cpa1+). Although we did not observe any amylase+ cells in bigenic pancreas at E12.5 (Fig. S3), we cannot rule out the possibility that the increase in Cpa1+ cells in bigenic pancreas could result from enhanced differentiation of progenitors towards acinar pathway.
Transcription factors, Sox9, Nkx6.1, Nkx2.2, Ngn3 and Hes1, and cyclin kinase inhibitors p57 and p27 play an important role in regulating proliferation and differentiation of pancreatic progenitors and endocrine cells (Seymour et al., 2007; Jensen et al., 2000; Georgia et al., 2006a; Georgia et al.,, 2006b; Doyle et al., 2007; Gradwohl et al., 2000). It is likely that the reduction in the expression of Sox9, Nkx6.1 and Nkx2.2 may contribute to the reduction in Ngn3+ endocrine progenitors and their subsequent differentiation into endocrine cells. In our study MafA expression in ∼80-90% of early pancreatic progenitors enhanced the proportion of cells expressing p57 and p27, inhibited the number of cells expressing Sox9 but did not alter the number of cells expressing Hes1 (Figs 9J,G, 8C,E). Unlike Sox9 and Hes1 knockout mice (Georgia et al., 2006; Seymour et al., 2007), this expression of MafA did not trigger premature differentiation of precursors into endocrine cells, most likely due to the continued expression of Hes1 in MafA-expressing progenitors. Since we observed an increase in p57+ cells without a significant change in Hes1+ cells, our results also suggest the presence of a Hes1-independent mechanism for regulating p57 expression in pancreatic progenitors. Interestingly, p27 expression, which is normally restricted to endocrine cells (Georgia and Bhushan, 2006), was enhanced in bigenic pancreatic progenitors. This observation suggests that induction of p27 expression in progenitors either prevents initiation and/or completion of the endocrine differentiation. MafA can activate expression from p27 promoter, which is implicated in regulating lens development in chicken (Reza et al., 2007a). We examined 10 Kb of 5′ flanking region of p57 and found 6 putative MAREs with complete conservation of the core sequence and a Transfac matrix match value of >0.94. The sites are located at ∼ 1.0, 2.5, 3.7, 5.5, 7.0, and 8.7 Kb upstream from the transcription start site. Hence, it is possible that in bigenic mice MafA may directly regulate the expression of p27 and p57 in Pdx1+ progenitors. Increased expression of p27 and p57 in bigenic pancreas also raises the possible involvement of MafA in regulating the proliferation of β-cells. However, the loss of MafA function did not increase the proportion of β-cells at birth in MafA knockout mice (Zhang et al., 2005) suggesting that MafA alone is not sufficient to regulate proliferation of β-cells during embryonic period, but it may have some role in regulating proliferation of β-cells after birth. This observation, together with the increased β-cell proliferation in p27 knockout mice (Georgia and Bhushan, 2006), suggests that MafA may not be the only factor regulating expression of p27. Even so, MafA-mediated induction of p27 may delay the reentry of β-cells into cell-cycle, a step possibly needed to regulate genes required for the maturation of β-cells; such a mechanism would be consistent with the proposed role of MafA in maturation rather than in differentiation of β-cells (Kroon et al., 2008; Nishimura et al., 2006; Wang et al., 2007; Zhang et al., 2005).
In summary, misexpression of MafA in pancreatic progenitors prevented their proliferation, leading to a significantly reduced pancreatic mass. Furthermore, such expression of MafA reduced the number of cells expressing Sox9, Nkx6.1, Nkx2.2 and Ngn3 and did not trigger premature induction of insulin+ or even endocrine cells in the pancreatic epithelium. We showed that early pancreatic progenitors disproportionately contributed to the differentiation of endocrine cells. Additionally, we show that MafA can directly or indirectly induce the expression of p27 and p57 in pancreatic progenitors. One of the major outcomes of this study is the unambiguous demonstration of the lack of differentiation potential of MafA in the mammalian pancreatic epithelium. We suggest that approaches relying on the potential of MafA to specify insulin+ cells from mammalian stem/progenitor cells are flawed since MafA expression may inhibit the proliferation of these cells without initiating their differentiation into insulin+ cells. Hence, a better strategy is to use MafA to enhance the maturation of insulin+ cells derived by a coordinated expression of key factors that regulate specification of insulin+ cells during normal development.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. O. Madsen, P. Serup and NIH-funded Beta Cell Biology Consortium for anti-Nkx6.1 and anti-Ngn3 antibodies, Dr. M. Appel for anti-glucagon antibody and Dr. J. Slack for anti-Pdx1 antibody; J. Stockton and JDRF NOD transgenic mice core at Joslin Diabetes Center for generation of the mice; Dr. K. Juhl for critical reading of this manuscript. This study was supported by research grants from NIH (RO1 DK060127), Harvard Stem Cell Institute and Juvenile Diabetes Research Foundation (JDRF) to AS, JDRF postdoctoral fellowship and Mary K. Iacocca Fellowship to WN, and Media and Advanced Microscopy Cores of Joslin Diabetes Endocrinology Research Center (NIH DK-36836).
Footnotes
Effect of early expression of MafA in pancreatic progenitors
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Artner I, Blanchi B, Raum JC, Guo M, Kaneko T, Cordes S, Sieweke M, Stein R. MafB is required for islet beta cell maturation. Proc. Natl. Acad. Sci. USA. 2007;104:3853–3858. doi: 10.1073/pnas.0700013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artner I, Hang Y, Guo M, Gu G, Stein R. MaFA is a dedicated activator of the insulin gene in vivo. J. Endocrinol. 2008;198:271–279. doi: 10.1677/JOE-08-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artner I, Le Lay J, Hang Y, Elghazi L, Schisler JC, Henderson E, Sosa-Pineda B, Stein R. MafB: an activator of the glucagon gene expressed in developing islet alpha- and beta-cells. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- Benkhelifa S, Provot S, Lecoq O, Pouponnot C, Calothy G, Felder-Schmittbuhl MP. mafA, a novel member of the maf proto-oncogene family, displays developmental regulation and mitogenic capacity in avian neuroretina cells. Oncogene. 1998;17:247–254. doi: 10.1038/sj.onc.1201898. [DOI] [PubMed] [Google Scholar]
- Cordes SP, Barsh GS. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994;79:1025–1034. doi: 10.1016/0092-8674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Doyle MJ, Loomis ZL, Sussel L. Nkx2.2-repressor activity is sufficient to specify alpha-cells and a small number of beta-cells in the pancreatic islet. Development. 2007;134:515–523. doi: 10.1242/dev.02763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgia S, Bhushan A. p27 regulates the transition of beta-cells from quiescence to proliferation. Diabetes. 2006;55:2950–2956. doi: 10.2337/db06-0249. [DOI] [PubMed] [Google Scholar]
- Georgia S, Soliz R, Li M, Zhang P, Bhushan A. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Dev. Biol. 2006;298:22–31. doi: 10.1016/j.ydbio.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMuer M, Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci.USA. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- Holland AM, Hale MA, Kagami H, Hammer RE, MacDonald RJ. Experimental control of pancreatic development and maintenance. Proc. Natl. Acad. Sci. USA. 2002;99:12236–12241. doi: 10.1073/pnas.192255099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat. Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Juhl K, Sarkar SA, Wong R, Jensen J, Hutton JC. The mouse pancreatic endocrine cell transcriptome defined in the embryonic Ngn3 null mouse. Diabetes. 2008;57:2755–2761. doi: 10.2337/db07-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajihara M, Kawauchi S, Kobayashi M, Ogino H, Takahashi S, Yasuda K. Isolation, characterization, and expression analysis of zebrafish large Mafs. J. Biochem. (Tokyo) 2001;129:139–146. doi: 10.1093/oxfordjournals.jbchem.a002825. [DOI] [PubMed] [Google Scholar]
- Kaneto H, Matsuoka TA, Nakatani Y, Miyatsuka T, Matsuhisa M, Hori M, Yamasaki Y. A crucial role of MafA as a novel therapeutic target for diabetes. J. Biol. Chem. 2005;280:15047–15052. doi: 10.1074/jbc.M412013200. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Han SI, Shioda S, Hirai M, Nishizawa M, Handa H. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J. Biol. Chem. 2002;277:49903–49910. doi: 10.1074/jbc.M206796200. [DOI] [PubMed] [Google Scholar]
- Kato T, Shimano H, Yamamoto T, Yokoo T, Endo Y, Ishikawa M, Matsuzaka T, Nakagawa Y, Kumadaki S, Yahagi N, Takahashi A, Sone H, Suzuki H, Toyoshima H, Hasty AH, Takahashi S, Gomi H, Izumi T, Yamada N. Granuphilin is activated by SREBP-1c and involved in impaired insulin secretion in diabetic mice. Cell. Metab. 2006;4:143–154. doi: 10.1016/j.cmet.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Kim JI, Li T, Ho IC, Grusby MJ, Glimcher LH. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc. Natl. Acad. Sci. USA. 1999;96:3781–3785. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, Agulnick AD, D’Amour KA, Carpenter MK, Baetge EE. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Matsuoka TA, Artner I, Henderson E, Means A, Sander M, Stein R. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc. Natl. Acad. Sci. USA. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka TA, Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol. Cell. Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura W, Kondo T, Salameh T, El Khattabi I, Dodge R, Bonner-Weir S, Sharma A. A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev. Biol. 2006;293:526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura W, Rowan S, Salameh T, Maas RL, Bonner-Weir S, Sell SM, Sharma A. Preferential reduction of β-cells derived from Pax6-MafB pathway in MafB deficient mice. Dev. Biol. 2008;314:443–456. doi: 10.1016/j.ydbio.2007.12.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H, Yasuda K. Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science. 1998;280:115–118. doi: 10.1126/science.280.5360.115. [DOI] [PubMed] [Google Scholar]
- Olbrot M, Rud J, Moss LG, Sharma A. Identification of beta -cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc. Natl. Acad. Sci. USA. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Krasinski JM, Stoffers DA. On the origin of the β cell. Genes. Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza HM, Nishi H, Kataoka K, Takahashi Y, Yasuda K. L-Maf regulates p27kip1 expression during chick lens fiber differentiation. Differentiation. 2007a;75:737–744. doi: 10.1111/j.1432-0436.2007.00171.x. [DOI] [PubMed] [Google Scholar]
- Reza HM, Urano A, Shimada N, Yasuda K. Sequential and combinatorial roles of maf family genes define proper lens development. Mol. Vis. 2007b;13:18–30. [PMC free article] [PubMed] [Google Scholar]
- Reza HM, Yasuda K. Roles of Maf family proteins in lens development. Dev. Dyn. 2004;229:440–448. doi: 10.1002/dvdy.10467. [DOI] [PubMed] [Google Scholar]
- Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- Sadl V, Jin F, Yu J, Cui S, Holmyard D, Quaggin S, Barsh G, Cordes S. The mouse Kreisler (Krml1/MafB) segmentation gene is required for differentiation of glomerular visceral epithelial cells. Dev. Biol. 2002;249:16–29. doi: 10.1006/dbio.2002.0751. [DOI] [PubMed] [Google Scholar]
- Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, Sander M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc. Natl. Acad. Sci. USA. 2007;104:1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieweke MH, Tekotte H, Frampton J, Graf T. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445:886–91. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- Wang H, Brun T, Kataoka K, Sharma AJ, Wollheim CB. MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia. 2007;50:348–58. doi: 10.1007/s00125-006-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P, May CL, Lamounier RN, Brestelli JE, Kaestner KH. Defining pancreatic endocrine precursors and their descendants. Diabetes. 2008;57:654–668. doi: 10.2337/db07-1362. [DOI] [PubMed] [Google Scholar]
- Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, Kudo T, Engel JD, Yamamoto M, Takahashi S. MafA is a key regulator of glucose-stimulated insulin secretion. Mol. Cell. Biol. 2005;25:4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell. 2007;13:103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.