Abstract
Adult stable flies are blood feeders, a nuisance, and mechanical vectors of veterinary diseases. To enable efficient feeding, blood sucking insects have evolved a sophisticated array of salivary compounds to disarm their host's hemostasis and inflammatory reaction. While the sialomes of several blood sucking Nematocera flies have been described, no thorough description has been made so far of any Brachycera, except for a detailed proteome analysis of a tabanid (Xu et al., 2008). In this work we provide an insight into the sialome of the muscid Stomoxys calcitrans, revealing a complex mixture of serine proteases, endonucleases, Kazal-containing peptides, anti-thrombins, antigen-5 related proteins, antimicrobial peptides, and the usual finding of mysterious secreted peptides that have no known partners, and may reflect the very fast evolution of salivary proteins due to the vertebrate host immune pressure. Supplemental tables S1 and S2 can be downloaded from http://exon.niaid.nih.gov/transcriptome/S_calcitrans/T1/Sc-tb1-web.xls and http://exon.niaid.nih.gov/transcriptome/S_calcitrans/T2/Sc-tb2-web.xls.
Keywords: Salivary glands, stable fly, hematophagy, sialome, cDNA library, proteome
1. Introduction
The habit of blood feeding evolved independently several times in Diptera (Grimaldi and Engel, 2005), including the Nematocera and Brachycera. Evolution to blood feeding is associated with the expression of specialized saliva that is pharmacologically active against vertebrate hemostasis and inflammation. Because inflammation and hemostasis are complex and redundant phenomena, the salivary potion of blood feeders is also complex, containing vasodilators, anti-clotting substances and enzymes that destroy vertebrate agonists and matrix components (Ribeiro, 1995). Sialotranscriptome analysis (from the Greek, Sialo=Saliva) of mosquitoes, sand flies, biting midges and black flies are revealing a vast repertoire of polypeptides recruited to serve this function (Andersen et al., 2009; Arca et al., 2007; Arca et al., 2005; Calvo et al., 2004; Campbell et al., 2005; Ribeiro et al., 2004; Valenzuela et al., 2003).
It has been proposed that all blood sucking Nematocera, except for sand flies, have a common blood feeding ancestor (Grimaldi and Engel, 2005). While the blood feeding Nematocera salivary proteins have some common protein families, such as the very divergent D7 protein family also found in sand flies (Valenzuela et al., 2002a), most other protein families are family or genus specific. For example, the salivary anticlotting protein of Aedes aegypti is a member of the serpin family (Stark and James, 1998), while a novel peptide named anophelin is an anti-thrombin in Anopheles albimanus (Valenzuela et al., 1999). As another example, while salivary apyrase activity, responsible for ADP hydrolysis, is found in both sand flies and mosquitoes, the gene families recruited for these functions are completely different in these two organisms (Champagne et al., 1995; Valenzuela et al., 2001). While the Brachycera contain diverse families of blood sucking arthropods of medical and veterinary importance including the tsetse, tabanids, keds and stable flies, no detailed transcriptome analysis has been done so far from any of these flies, although a detailed sialoproteome analysis has been made from the tabanid Tabanus yao (Xu et al., 2008).
The stable fly, Stomoxys calcitrans is an important pest of cattle, their larvae growing in decaying faeces and straw, the adults biting and blood feeding on mammals (Campbell et al., 2001; Taylor and Berkebile, 2006). They can also vector pathogens by mechanical transmission. It is the aim of this study to provide a preliminary description of the sialome of S. calcitrans.
2. Materials and methods
2.1. Insects
Stable flies were reared from a colony collected in Manhattan, KS in 1990. Eggs were collected with a wet black cloth wick. The larvae were reared in a fermented mixture of wheat bran (Farmers' Cooperative Association, Frederick, MD), vermiculite (Hummert International, St. Louis, MO), and Calf Manna (Manna PRO Corporation, Chesterfield, MO). Adult flies were fed with 1 M sucrose or Gatorade. For reproduction, the flies were fed daily with citrated bovine blood supplied in saturated cotton pads. The flies were kept at 26 ± 1°C, a photoperiod of 16:8 h (L:D), and at 65 ± 5% relative humidity. Flies from this colony maintain the ability to feed well on cattle (A. Broce, unpublished observations).
2.2. Salivary gland preparation
The salivary glands were dissected from male and female adults (not blood-fed) between 1 and 7 days after eclosion. The glands were immediately transferred to either TRI Reagent (Sigma) on ice for mRNA purification or to 10 mM Hepes, pH 7.0, 150 mM NaCl (HBS) on ice for extraction of proteins. Usually, groups of 20 salivary glands were placed in 20 μl of HBS in a 0.5 ml centrifuge tube. The salivary glands in TRI Reagent or HBS were stored at -80°C.
2.3. cDNA library construction and sequencing
Total mRNA was purified using TRI Reagent (Sigma) according to the manufacturer's protocol. A long distance polymerase chain reaction (PCR)-based cDNA library was constructed in a λ Triple Ex2 vector following the procedures from the SMART™ cDNA Library Construction Kit (Clontech, USA). Briefly, 1.1 μg of total RNA purified from salivary glands of male and female adult stable flies was used to prepare first–strand cDNA in a 10 μl reaction volume at 42°C for 1 hour with PowerScript™ Reverse Transcriptase and CDS III primer (Clontech, USA). Half of the above first strand cDNA reaction volume (5 μl) was used as template for double stranded cDNA amplification in a reaction volume of 100 μl by long distance PCR using SMART IV oligonucleotide as forward primer and CDS III primer as reverse primer. The two primers incorporated Sfi IA and Sfi IB sites at the two ends of cDNAs suitable for digestion with Sfi I. PCR was conducted on a Perkin Elmer 9600 thermal cycler (Perkin Elmer, USA) using a Clontech Advantage 2 PCR kit (Clontech, USA) with denaturation at 95°C for 20 s, followed by 20 cycles of 95°C for 5 s and 68°C for 6 min.
The double stranded cDNA was treated with proteinase K at 45°C for 20 min and then extracted with phenol:chloroform:isoamyl alcohol. The cDNA was digested with Sfi I, size fractionated with a CHROMA SPIN-400 column (Clontech), and ligated into the λ TripleEx2 vector (Clontech) containing digested asymmetrical Sfi I sites. A packaging reaction was performed with Gigapack III Gold Packaging Extract (Stratagene) according to manufacturer's instructions. Titer of the unamplified cDNA library was determined, and the library was then amplified by plating 4 × 106 plaques and eluting resulting phage with 0.01% gelatin, 100 mM NaCl, 10 mM MgSO4, 35 mM Tris· Cl, pH 7.5.
In an initial test, 106 plaques were randomly selected for sequencing. The λ TripleEx2 phage was converted to pTripleEx2 plasmid by incubating 150 μl of the eluted phage with 200 μl overnight culture of E.coli BM25.8 (OD600nm at 1.1) at 31°C. Each culture was spread onto an LB/Ampicillin plate and grown at 31°C overnight. One colony from each plate was grown in 3 ml LB/ampicillin medium. Plasmids were purified from the cultures using QIAprep Spin Miniprep Kit (Qiagen) according to manufacturer's protocol. These DNA samples were then analyzed by digestion with BstXI and SphI or EcoRI and XhoI, followed by electrophoresis on a 1.1% agarose gel to assess insert size. Plasmid DNA samples were sequenced at the DNA Sequencing Facility in the Department of Plant Pathology, Kansas State University, using the following primers from the SMART™ cDNA Library Construction Kit (Clontech, USA): 5′-seqencing primer TCCGAGATCTGGACGAGC and 3′-sequencing primer TAATACGACTCACTATAGGG. Larger scale sequencing of the cDNA library was performed as described elsewhere (Francischetti et al., 2002; Valenzuela et al., 2002b).
2.4. Bioinformatic tools used
ESTs were trimmed of primer and vector sequences, clusterized, and compared to other databases as described before (Valenzuela et al., 2003). The blast tool (Altschul et al., 1997) and CAP3 assembler (Huang, 1992) were used for the EST clusterization, by using a decreasing word size inclusion strategy. First, all ESTs were blasted against all ESTs with a word size of 100. All matches were fed as input to the CAP3 assembler, and a fasta of the assembled data was obtained, including quality files. This assembly done with word size of 100 was in turn used for the next assembly round, but now using a word size of 80, then 60 then 40 to produce the final assembly shown in supplemental table S1. This assembly strategy is easy to parallelize, allowing for very large data sets to be clusterized. The final output was piped into a tab delimited file that was imported into an Excel spreadsheet. These operations were automated by a program written in Visual Basic named Megacluster. We submitted all translated sequences (starting with a Met) to the Signal P (Nielsen et al., 1997) server to help identify putative secreted peptides. Deduced coding sequences (CDS) were also sent to the above prediction server, as well as to the SecretomeP (Bendtsen et al., 2004) to identify non-classical secreted proteins, to the TMHMM server (Sonnhammer et al., 1998) to detect membrane helices, the NETOGLYC server to detect possible mucin-type galactosylations (Hansen et al., 1998) and to the ProP server (Duckert et al., 2004) to identify putative furin processed protein cleavage sites. BLAST searches were done locally from executables obtained at the NCBI FTP site (ftp://ftp.ncbi.nih.gov/blast/executables/) against the non redundant protein database of the NCBI, the gene ontology fasta subset (Lewis et al., 2000), the Conserved Domains Database of NCBI (Marchler-Bauer et al., 2002) containing the KOG (Tatusov et al., 2003), Pfam (Bateman et al., 2000) and Smart (Schultz et al., 2000) motifs, and to custom downloaded databases containing mitochondrial and rRNA nucleotide sequences available at the NCBI.
2.5. Analysis of proteins in salivary gland extracts
To obtain salivary gland extracts (SGE), 20 salivary glands in 20 μl HBS were quickly frozen in liquid nitrogen and thawed in a 37°C water bath, and the procedure was repeated for another 3 cycles. After centrifuging (13, 000 × g, 2 min), the supernatant was analyzed by electrophoresis. The SGE sample was treated with NuPAGE 4 × LDS sample buffer (Invitrogen) without adding reducing agent and heated at 80°C for 10 min. The sample containing approximately 20 μg protein was applied to a NuPAGE® Novex 12% Bis-Tris Gel 1.0 mm, 10 well (Invitrogen). SeeBlue™ molecular weight standards (Invitrogen) were applied to an adjacent well. Electrophoresis was carried out in MOPS buffer (Invitrogen) at 200 V for 30 min. The proteins were transferred to a polyvinylidene difluoride (PVDF) membrane using 10 mM CAPS buffer, pH 11.0, 10% methanol as transfer buffer on a mini trans-blot electrophoretic transfer cell (Bio-Rad). Immediately before the transfer, the gel was pre-equilibrated with the transfer buffer for 1 min, and the membrane was first washed with 100% methanol for 30 s and then washed with water for 1 min. The transfer was conducted with constant current at 250 mA for 2 h. After transfer, the membrane was washed three times with deionized water, 5 min per wash. The membrane was stained for 5 min with 0.025% Coomassie Blue in 40% methanol. It was destained for 10 min in 50% methanol and then washed with water several times and air-dried. Stained protein bands were excised and analyzed by Edman degradation sequencing using a Procise sequencer (Perkin Elmer).
Amino-terminal de-blocking with pyroglutamate aminopeptidase was carried out similarly to a published protocol (Coligan et al., 1995) with minor modification. Briefly, the PVDF membrane was incubated in 200 μl 0.5% polyvinylpyrrolidone (PVP-40, Sigma) in 0.1 M acetic acid for 30 min at 37°C and then rinsed with water. Twenty microliters (2 mU) pyroglutamate aminopeptidase (PGAP, TaKaRa) and 80 μl freshly prepared 1 × PGAP buffer consisting of 50 mM sodium phosphate, 10 mM DTT, 1 mM EDTA, pH 7.0 was added to the tube, which was flushed with nitrogen and incubated at 50°C for 5 h. The membrane was rinsed three times with water and then air dried. Protein on the membrane was sequenced by Edman degradation using an Applied Biosystem Procise 492 Protein Sequencer at the Kansas State University Biotechnology Core Facility.
To identify cDNA sequences that matched the amino-terminal sequence of protein bands from the salivary gland extracts, a search program written by Dr. Ribeiro (Francischetti et al., 2002; Valenzuela et al., 2002b) was used to check the N-terminal sequencing information against three possible protein translations of each cDNA from cDNA library sequencing. The program was also used to resolve complex amino-terminal sequence information from a protein band that contained a mixture of proteins.
3. Results and Discussion
3.1 Overview of the assembled salivary EST set
A total of 820 cDNA clones were used to assemble a database (Supplemental Table S1) that yielded 240 clusters of related sequences, 187 of which contained only one EST. The 240 clusters were compared, using the programs blastx, blastn, or RPS-BLAST (Altschul et al., 1997), to the nonredundant (NR) protein database of the National Center of Biological Information (NCBI), National Library of Medicine, NIH, to a gene ontology database (Ashburner et al., 2000), to the conserved domains database of the NCBI (Marchler-Bauer et al., 2002), and to a custom prepared subset of the NCBI nucleotide database containing either mitochondrial or rRNA sequences.
Manual annotation of the contigs resulted in four broad categories of expressed genes (Fig. 1). The putatively secreted (S) category contained 61% of the sequences, the housekeeping (H) category had 29%, 0.73 % derived from transposable elements and 9% of the ESTs could not be classified and belong to the unknown (U) class. The transcripts of the U class could represent novel proteins or derive from the less conserved 3/ or 5/ untranslated regions of genes, as was indicated for the sialotranscriptome of An. gambiae (Arca et al., 2005). The fourth class of transcripts were associated with transposable elements (TE). They may represent either the presence of active transposition in the fly, or more likely, the expression of sequences suppressing transposition. Low level expression of TE sequences have been a relatively common finding in previous sialotranscriptomes. It is possible that amplification of the library led to decreased complexity or altered representation of sequences in the original mRNA. However, sequences most highly represented in the cDNA library were also the most abundant proteins detected in salivary protein extracts (described below), suggesting that amplification did not account for over-representation of these sequences.
Figure 1.
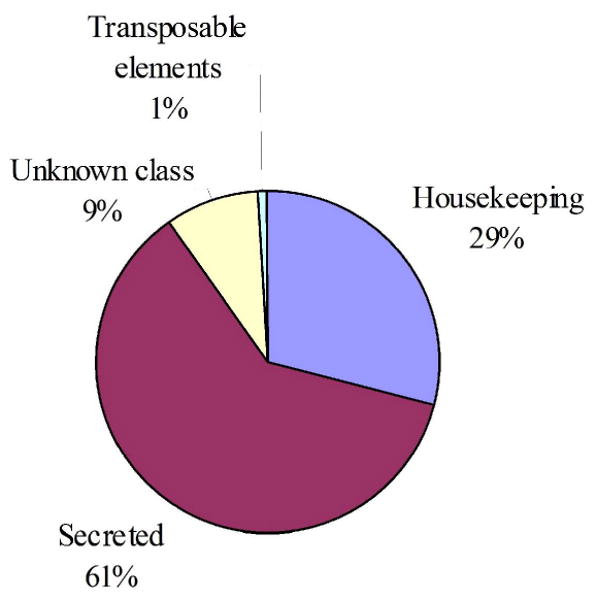
Distribution of the transcripts from the salivary gland cDNA library of S. calcitrans according to functional class.
3.1.1 Housekeeping (H) genes
The 240 ESTs attributed to H genes expressed in the salivary glands (SGs) of S. calcitrans were further characterized into 16 subgroups according to function (Table 1 and Supplemental Table S1). Transcripts associated with the protein synthesis machinery represented 45% of all transcripts associated with a housekeeping function, an expected result for the secretory nature of the organ. Energy metabolism accounted for 16.7% of the transcripts. Twenty percent of the transcripts were classified as either ‘Hypothetical conserved’ or ‘Conserved secreted’ proteins. These represent highly conserved proteins of unknown function, presumably associated with cellular function but still uncharacterized.
Table 1.
Classification of transcripts that are associated with housekeeping function
| Class | Number of transcripts | Percent of housekeeping group |
|---|---|---|
| Protein synthesis machinery | 108 | 45.0 |
| Energy metabolism | 40 | 16.7 |
| Transcription machinery | 18 | 7.5 |
| Hypothetical conserved | 16 | 6.7 |
| Protein modification machinery | 13 | 5.4 |
| Protein export machinery | 11 | 4.6 |
| Cytoskeletal | 8 | 3.3 |
| Signal transduction | 7 | 2.9 |
| Transcription factors | 5 | 2.1 |
| Lipid metabolism | 4 | 1.7 |
| Nuclear regulation | 2 | 0.8 |
| Carbohydrate metabolism | 2 | 0.8 |
| Oxidant metabolism | 2 | 0.8 |
| Transporters/storage | 2 | 0.8 |
| Nucleotide metabolism | 1 | 0.4 |
| Detoxication metabolism | 1 | 0.4 |
| Total | 240 | |
3.1.2 Possibly secreted (S) class of expressed genes
A total of 498 ESTs represent putative S. calcitrans salivary components (Table 2 and Supplemental Table S1). These include previously known gene families, such as enzymes, antigen 5 proteins and antimicrobial peptides. However, several putative proteins, including some that appear to be multigenic, have no similarities to known proteins, as will be described further below.
Table 2.
Classification of transcripts that are associated with secreted products
| Class | Number of transcripts | Percent of secreted group |
|---|---|---|
| Putative secreted peptides of unknown function | 200 | 40.2 |
| Antigen 5 | 133 | 26.7 |
| Protease inhibitors | 142 | 28.6 |
| Serine proteases | 13 | 2.6 |
| Nucleases | 4 | 0.8 |
| Antimicrobial peptides | 6 | 1.2 |
| Total | 498 | |
3.2 Characterization of the stable fly salivary gland proteome
In addition to obtaining transcriptome information, we tried to identify the major proteins expressed in the salivary gland. Proteins extracted from 20 salivary glands were analyzed by SDS-PAGE. The proteins were transferred to PVDF membrane, and Coomassie blue stained bands were excised for Edman degradation sequencing. The gel showed about 20 clearly visible protein bands, with a predominant doublet at ∼26-27 kDa. Amino-terminal sequencing information was successfully obtained for nine proteins bands. Sequences for the predominant double bands at 26-27 kDa were obtained only after deblocking with pyroglutamine aminopeptidase. The nine sequences obtained by Edman degradation matched predicted amino acid sequences from clusters of the cDNA database in the S category (Fig 2). Other bands did not yield useful information either because of their low signal, or because of mixed sequencing information that could not be assigned to any of the deduced amino acid sequence from the clusters of the cDNA database. The polypeptide sequences associated with amino-terminal sequences found by Edman degradation had EST representation between three to one hundred and thirty three sequences, with an average of 52.2 EST's sequences per polypeptide (Supplemental table S2), while the remaining proteins have an average of 2.98 sequences per polypeptide. Accordingly, the predominant proteins in the salivary gland correlated with the mRNA abundance in the transcriptome. This was also observed in other proteome analysis of salivary glands in Anopheles stephensi (Valenzuela et al., 2003).
Figure 2.
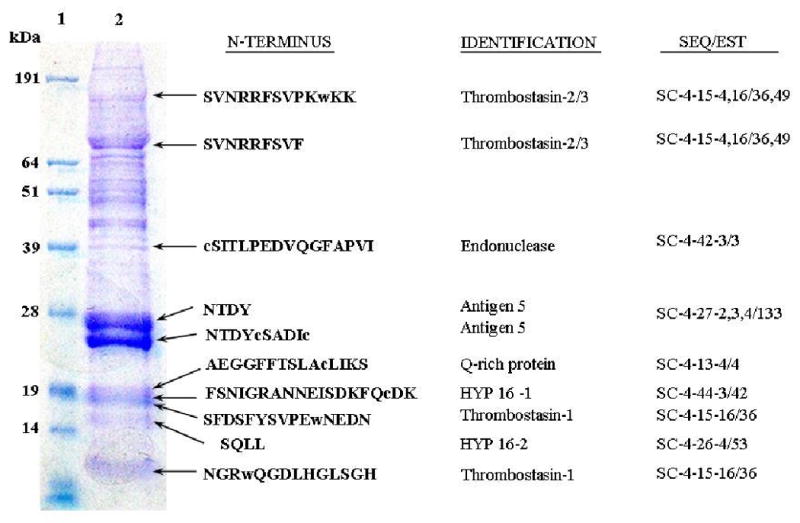
SDS-PAGE analysis of stable fly salivary gland extract. Lane 1, molecular weight standards; Lane 2, salivary gland extract. Ten putative secreted proteins were identified by N-terminal sequencing after separating proteins extracted from 20 salivary glands by SDS-PAGE. Molecular weight standards and SGE protein bands are shown on the left side. Next are the amino terminal sequences of bands determined by Edman degradation sequencing, and best matches to known proteins. The lower case amino acids represent missing Edman products cysteine and tryptophan. On the right side are the corresponding coding sequence names matching the N-terminal sequence of the protein bands and the number of the EST sequences associated with them. The two bands labeled NTDY… were obtained after removal of pyroglutamate.
Of the nine amino terminal sequences, four peptides related to thrombostasin were found; their position in the gel indicates that these proteins may be processed to smaller peptides, as described further in section 3.2.1.3. The predominant double bands at 26-27 kDa were related to the antigen 5 (Ag5) family of proteins. A 39 kDa protein related to an endonuclease of Culex pipiens quinquefasciatus was found. Two proteins at about 19 kDa were a glutamine-rich protein and member of the Hyp-16 family. Their function in blood feeding is unknown.
3.2 The salivary secretome of S. calcitrans
From the sequenced cDNAs, a total of 74 protein sequences was derived, 27 of which encode putative secreted products (Table 2, Table 3, and Supplemental Table S2). Table 3 presents a summary of the secreted subset, with links to GenBank. The following presentation is a guide for browsing Supplemental Table S2.
Table 3. Coding sequences associated with putative secreted salivary polypeptides.
| Sequence name | NCBI number | Description |
|---|---|---|
| Putative secreted proteins | ||
| Enzymes | ||
| Serine proteases | ||
| SC-4-11-3 | gi|32395297 | chymotrypsin |
| SC-4-17-3 | gi|15554312 | serine protease Ssp3 |
| SC-4-4-4 | gi|68500400 | serine protease Ssp3-2 precursor |
| SC-4-9-2 | gi|32395299 | Trypsin |
| SC-4-5-4 | gi|224924442 | Salivary trypsin - truncated at 3 prime |
| SC-4-19-4 | gi|224924362 | Trypsin - truncated at 5 prime |
| Endonuclease | ||
| SC-4-42-3 | gi|224924418 | Salivary endonuclease - truncated at 3 prime |
| Thrombostasin | ||
| SC-4-14-2-SEQ | gi|37778968 | Thrombostasin |
| SC-4-15-16 | gi|224924356 | Thrombostasin-2 |
| SC-4-15-4 | gi|224924448 | Thrombostasin-3 |
| Kazal family | ||
| SC-4-17-4 | gi|68500439 | Kazal protease inhibitor |
| SC-4-21-3 | gi|63148854 | putative 6.3 kDa secreted salivary gland protein |
| Antigen 5 family | ||
| SC-4-27-2 | gi|32395295 | antigen 5 precursor |
| SC-4-27-3 | gi|32395295 | antigen 5 precursor II |
| SC-4-27-4 | gi|224924378 | antigen 5 precursor III |
| Antimicrobial peptides | ||
| SC-4-22-3 | gi|224924368 | hypothetical secreted peptide wity GGY motifs |
| SC-4-43-6 | gi|68500429 | stomoxyn 2 |
| SC-4-11-1 | gi|68500452 | diptericin A |
| Hyp 16 family of mosquito proteins | ||
| SC-4-26-4 | gi|63148846 | putative 13.7 kDa secreted salivary gland protein |
| SC-4-44-3 | gi|63148848 | putative 15.6 kDa secreted salivary gland protein |
| Secreted proteins of unknown classes | ||
| 11.4 kDa conserved family | ||
| SC-4-11-7 | gi|63148852 | putative 11.4 kDa secreted salivary gland protein |
| SC-4-9-5 | gi|224924446 | hypothetical conserved secreted protein |
| Q-rich secreted protein | ||
| SC-4-13-4 | gi|68500357 | putative 13.0 kDa secreted salivary gland protein |
| SC-4-47-2 | gi|68500386 | putative 7.4 kDa secreted salivary gland protein 2 |
| Orphan secreted peptides | ||
| SC-4-1-3 | gi|224924350 | hypothetical conserved Diptera protein |
| SC-4-22-4 | gi|63148856 | putative 10.9 kDa secreted salivary gland protein |
| SC-4-39-1 | gi|63148850 | putative 11.5 kDa secreted salivary gland protein |
3.2.1 Proteins with presumed or experimentally validated function
3.2.1.1 Serine proteases
Six different serine proteases are presented in Table 3, 4 of which are full length and 2 are fragments. When compared to the NR protein database, they provide best matches to Drosophila proteins. The proteins named SC-4-4-4 and SC-4-17-3 are most closely related, approaching 60 % similarity. These enzymes may be associated with fibrinogenolytic activity, as demonstrated for Tabanus flies (Xu et al., 2008), but may also be related to digestion of host extracellular matrix components.
3.2.1.2 Nucleases
A secreted endonuclease similar to a salivary homologue from tsetse is described. A previously described salivary endonuclease of Culex quinquefasciatus was suggested to function by decreasing the viscosity of the feeding matrix and producing pharmacologically active DNA fragments that might assist blood feeding (Calvo and Ribeiro, 2006).
3.2.1.3 Proteins containing serine protease inhibitor domains
Thrombostasin (Zhang et al., 2002) is a protein with anti-thrombin activity obtained from the salivary gland of horn flies. The S. calcitrans thrombostasin homologue has only 41% sequence similarity to the horn fly protein, and presents only low complexity matches to other unrelated proteins due to the repeating charged amino acids that it contains. The sequences of two additional Stomoxys thrombostasin family members are also presented, thrombostasin-2 having only 37 % identity to its paralogue, and no predicted similarity to the Haematobia protein. The full length sequence of thrombostasin-3 is also presented in supplemental table S2. It has numerous QHDGESNEESDE repeats, which gives an acidic pI of 3.9 for the predicted protein. Such acidic residues may interact with the anion binding exosites of thrombin and Factor Xa. Edman degradation products for the 3 proteins were found in the PAGE experiment (Fig 2). Similarly to the horn fly thrombostasin, the Stomoxys homologue is also processed from a preproprotein. Indeed the ProP server predicts several furin putative cleavage sites, including at the regions found by the Edman degradation products (see column AI on supplemental file S2). The Stomoxys thrombostasins are abundantly expressed, being represented by 98, 36 and 49 EST's for thrombostasins-1-3, respectively.
The Kazal domain is ubiquitously found in plants and animals and is normally associated with protease inhibitors (Kanost, 1999; Schlott et al., 2002). Exceptionally, the vasodilator of tabanids was identified as belonging to this protein family (Takac et al., 2006; Xu et al., 2008). Table 3 presents the sequences of two polypeptides of this family, both producing highest similarities to Drosophila peptides. They have relatively small EST representation, with 1 and 5 ESTs. The function of these proteins as vasodilators remains to be confirmed.
3.2.1.4 Antigen 5 protein family
The antigen 5 family, also known as CAP or CRISP families, was originally described in the venom of wasps and belong to a ubiquitous family of proteins found in plants and animals (Schreiber et al., 1997). Members of these families function in diverse ways, being toxins and ion channel blockers in lizard and snake venom (Nobile et al., 1996; Yamazaki et al., 2003; Yamazaki and Morita, 2004), or proteases in Conus snails (Milne et al., 2003). Antigen-5 related proteins have been described from virtually all sialotranscriptomes from hematophagous arthropods done to date. Three very closely related proteins of this family are described in Table 3, which may be products of the same gene as alleles or splice variants. One variant possibly lacking an exon produces a product 3 kDa smaller that could explain the two bands with same Edman product found in the PAGE experiment (Fig 2). The mature protein starts with a glutamine, and treatment with pyroglutamase was necessary to produce the Edman products. Overall these proteins have a total of 133 matching ESTs and represent the most abundantly expressed gene in the salivary glands of S. calcitrans. Recently a recombinant member of this family was shown to bind the Fc portion of immunoglobulins, and may function as a complement inhibitor (Ameri et al., 2008).
3.2.1.5 Antimicrobial peptides
Three unrelated peptides that might serve an antimicrobial function are reported in their full length form (Table 3). SC-4-22-3 is 53% similar to a Drosophila protein containing GGY repeats, which was shown in C. elegans to have antimicrobial function (Couillault et al., 2004). Stomoxin is a member of the cecropin family previously reported from the gut of the stable fly (Boulanger et al., 2002; Landon et al., 2006), and is here reported to be found in the insect salivary glands. The third AMP has the PFAM Attacin C-terminal region, being a member of the Diptericin family. Truncated fragments of a homolog of sarcotoxin were also found (Supplemental table S1).
3.2.2. Secreted proteins of unknown class
Hyp 16 family found in mosquito sialotranscriptomes: Two distantly related salivary peptides from S. calcitrans, producing mature peptides of 13.7 and 15.6 kDa are also distantly related to mosquito proteins. Alignment of these two peptides with the mosquito proteins indicates a conserved amino acid backbone G-x(2)-N-x(2)-I-x(6)-C-D-x(3)-C-P-x(5)-C-x(3)-K-x(15)-C-x(4)-G-x(4)-K-x(8,9)-P-x(8)-E (Fig 3). Interestingly, two of the mosquito proteins, deriving from An. stephensi and from Ae. albopictus, were found in previously described sialotranscriptomes (Arca et al., 2007; Valenzuela et al., 2003). The molecular model database (MMDB) of NCBI (Wang et al., 2007) has structural matches for both proteins indicating these to be related to the kissing bug salivary protein named nitrophorin I, which is a lipocalin carrier of heme, NO, and binds histamine (Andersen et al., 2005), suggesting this family derives from a lipocalin structure and may function by sequestering agonists of inflammation or hemostasis, as do many tick and kissing bug lipocalins, as well as the D7 family of mosquitoes (Calvo et al., 2006). The Stomoxys proteins are represented by 53 and 42 EST's each, and their predicted amino terminal sequences were found in the PAGE experiment (Fig 2). Their abundant expression is compatible with an agonist sequestration function.
Figure 3.
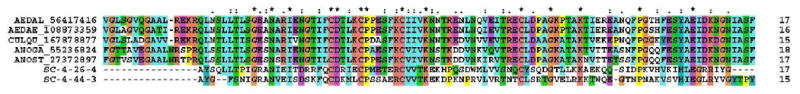
Alignment of the Hyp 16 family of Stomoxys calcitrans and other related mosquito salivary proteins. The mosquito species are indicated by 5 letters, the first three being from the genus and the last two from the species names, thus ANOGA for An. gambiae. The number following the letters refers to their GenBank accession number.
11.4 kDa conserved family: Two related proteins similar to other secreted insect proteins of unknown function were deduced from the S. calcitrans sialotranscriptome. The closest D. melanogaster relative, gi:24653255 is annotated in Flybase as an unknown protein that is highly expressed in larval and adult midguts, as indicated in flyatlas. They may function in immunity or in some conserved function associated with the endoplasmic reticulum or Golgi.
Q-rich secreted polypeptides: Two proteins containing repeats involving the amino acid motif QDNGA are described. These may derive from alternate splicing of the same gene. Their repeat nature suggests interaction with matrix proteins, possibly collagen. Sequences with large contents of glutamine were also found abundantly expressed in black fly sialomes (Andersen et al., 2009). A total of 7 transcripts are associated with the 2 Q-rich polypeptides. Evidence for secretion of SC-4-13-4 was found in the PAGE experiment (Fig 2).
Orphan secreted proteins: Table 3 additionally depicts the full length sequence of 3 putative secreted peptides. SC-4-1-3 is 98% identical to the tsetse Fb12 protein, but has very weak similarity to Drosophila proteins of the same size, both of unknown function. The remaining 2 proteins do not match any other protein of the NR database in a significant way, and include the relatively well expressed protein encoded by SC-4-39-1, which was assembled from 16 ESTs. Unique proteins such a these are commonly found in sialotranscriptomes of blood sucking arthropods and possibly reflect the very fast pace of evolution observed for these proteins possibly due to host immune pressure.
3.3 Housekeeping proteins and transposable element
The EST set acquired in this study allowed for the description of 46 coding sequences, mostly full length, associated with housekeeping functions, including a set of conserved hypothetical proteins that might be related with protein synthesis or protein modification. A CDS fragment coding for a protein similar to insect proteins annotated as transposable elements is also presented.
Conclusions
From the analysis of previous sialotranscriptomes, it is becoming clear that the “generic” salivary potion of any blood feeding arthropod, even those not descending from a common blood sucker ancestor, consists of the somewhat unrelated classes of enzymes, protease inhibitors, vasodilator agonists, serotonin- and histamine-binding proteins, antigen 5 proteins, antimicrobial peptides and a large group of mysterious, unknown secreted proteins. This disjointed list is found in triatomines, mosquitoes, sand flies, biting midges and ticks.
Among the enzymes, ATP-diphosphohydrolases are commonly found in the saliva of hematophagous arthropods feeding on mammals, but not on those feeding on birds or reptiles (Ribeiro, 2000; Ribeiro et al., 1989; Ribeiro et al., 1985). They degrade ATP and ADP to AMP thus eliminating important agonists of neutrophil and platelet aggregation. Different protein families can perform this function. Mosquito and triatomine bugs of the genus Triatoma have recruited members of the 5′-nucleotidase family (Champagne et al., 1995; Faudry et al., 2004; Sun et al., 2006), while the bed bug Cimex lectularius and sand flies have recruited a unique enzyme, the Cimex type of apyrase (Valenzuela et al., 2001; Valenzuela et al., 1998), now found to be ubiquitous in animals. In these insects, transcripts for these enzymes are found abundantly, but in this Stomoxys sialotranscriptome we found no evidence for the expression of any protein of these families. Previous work with salivary gland homogenates of Haematobia irritans, also a blood feeding Muscidae, did not identify salivary apyrase activity or anti-platelet activity (Cupp et al., 1998; Kerlin and Hughes, 1992), perhaps explaining the absence of transcripts coding for known ATPdiphosphohydrolases in Stomoxys. This negative finding was interpreted as resulting from bovine platelets being exceptional in their response to ADP. Bovine platelets respond with a monophasic, not diphasic wave of platelet aggregation by ADP, and require higher agonist concentrations, when compared to human platelets, for example (Bondy and Gentry, 1989; Dodds, 1977).
Serine proteases are also commonly found in the saliva of blood feeding arthropods, but only in Tabanus were they characterized to have a fibrinogenolytic, and thus anti-clotting, action (Xu et al., 2008). The S. calcitrans sialotranscriptome revealed transcripts coding for an endonuclease, which could help to decrease the skin viscosity, as does hyaluronidase. Hyaluronidase activity, however, was measured, and not detected, in stable fly salivary gland homogenates (Volfova et al., 2008).
The S. calcitrans sialotranscriptome reveals only one family of putative proteins containing domains associated with protease inhibitors, namely for the Kazal domain. However, Kazal domain containing peptides were verified to be vasodilatory peptides in horse flies (Takac et al., 2006; Xu et al., 2008), suggesting the S. calcitrans peptides could be performing a similar function. On the other hand, the S. calcitrans sialotranscriptome abounds with transcripts coding for homologues of the horn fly thrombin inhibitor named thrombostasin (Zhang et al., 2002). Without the previous discovery of the Haematobia thrombin inhibitor we would have no clue for the function of this protein in Stomoxys. We also call the attention for the bioinformatic tool ProP (Duckert et al., 2004) that could identify correctly the furin cleavage and processing of the S. calcitrans thrombostasin peptides as detected by Edman degradation (Fig 2) (Duckert et al., 2004). Future sialotranscriptome analysis may benefit from this tool, mainly when attempting to predict the function of relatively large proteins containing repetitive domains.
Antigen-5 related proteins are ubiquitously found in sialotrancriptomes, but little is known about their function in the saliva of blood-feeding arthropods. However, the S. calcitrans protein here described has immunoglobulin binding capacity (Ameri et al., 2008) and may inhibit the classical pathway of complement activation. In Tabanus yao members of this protein family acquired an RGD motif and inhibit platelet aggregation by interfering with the platelet receptor for fibrinogen, which causes platelet cross-linking (Xu et al., 2008).
Three antimicrobial peptides were discovered in the S. calcitrans sialotranscriptome, one of which, named stomoxyn, was previously discovered in the fly's gut (Boulanger et al., 2002; Landon et al., 2006). Salivary antimicrobial peptides, when ingested with the blood meal may prevent microorganism growth in the meal, and when left in the host skin may prevent wound infection, a good husbandry practice on the part of the stable fly.
Regarding biogenic amine binders, the MMDB indicates lipocalin matches for the Hyp16 members, which are highly expressed. The 11.4 kDa conserved family member SC-4-47-2 is also highly expressed. These proteins are strong candidates as biogenic amine binders, based on their high expression levels and the observation that other salivary proteins in this family are always associated with this function (Calvo et al., 2006).
We additionally obtained the CDS for glutamine-rich proteins possibly associated with matrix (collagen?) binding and three additional proteins for which we have no clue for their function, characterizing the mysterious group always associated with sialotranscriptomes.
Finally, when comparing the complexity of both the sialotranscriptome and the SDS gel of S. calcitrans with that of mosquitoes, or triatomine bugs, the lower complexity of Stomoxys is apparent. While mosquitoes have been blood feeders for over 150 MY, when Aedes and Anopheles shared a common blood feeding ancestor (Grimaldi and Engel, 2005; Krzywinski et al., 2006), it is likely that stable flies are relative newcomers to the art, and still have some evolutionary time ahead to perfect their “magic potion” as well as their feeding apparatus, as evidenced by their cruder mouthparts that inflict a quite painful bite. Indeed horn and stable flies appear to be opportunistically migrating into this feeding mode after emergence of large mammals that are relatively helpless in defending themselves. This contrasts with the more sophisticated potion of Tabanids, elegantly demonstrated by Xu et. al (Xu et al., 2008). Tabanids are vectors of apicomplexan parasites to turtles (DeGiusti et al., 1973) indicative of a long association with feeding on blood of vertebrates, possibly starting before the radiation of mammals, ∼60 MYA. Similarly, tsetse, also a Muscidae, appears to have an ancient origin as evidenced by the preference for some species to feed on reptiles, by their efficient transmission of Trypanosoma protozoa and their fine feeding apparatus (Gordon and Crewe, 1948; Krafsur, 2009; Okiwelu and Maiga, 1981).
Supplementary Material
Additional file 1 Supplemental Table S1: Hyperlinked Excel spreadsheet and associated files with EST assembly results. NOTE: This is a compressed ZIP file that should be expanded to a new directory. After this is done, start Excel and then open the file ending in .xls so the hyperlinks will work.
Additional file 2 Supplemental Table S2: Hyperlinked Excel spreadsheet with deducted protein sequences. See note above.
Acknowledgments
This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, NIH Grant GM41247, and Kansas State University Agricultural Experimental Station Grant, 418343. We thank Kent Hampton for insect rearing.
Because JMCR is a government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
Abbreviations
- aa
amino acid
- AMP
antimicrobial peptide
- AG5
antigen 5 family
- EST
expressed sequence tag
- H class
housekeeping
- NR
nonredundant
- MMDB
Molecular Model database
- OBP
odorant-binding protein
- S class
secreted
- SG
salivary gland
- SMART
switching mechanism at 5/ end of RNA transcript
- U class
unknown function
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameri M, Wang X, Wilkerson MJ, Kanost MR, Broce AB. An immunoglobulin binding protein (antigen 5) of the stable fly (Diptera: Muscidae) salivary gland stimulates bovine immune responses. J Med Entomol. 2008;45:94–101. doi: 10.1603/0022-2585(2008)45[94:aibpao]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JF, Gudderra NP, Francischetti IM, Ribeiro JM. The role of salivary lipocalins in blood feeding by Rhodnius prolixus. Arch Insect Biochem Physiol. 2005;58:97–105. doi: 10.1002/arch.20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JF, Pham VM, Meng Z, Champagne DE, Ribeiro JM. Insight into the Sialome of the Black Fly, Simulium vittatum. J Proteome Res. 2009 doi: 10.1021/pr8008429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Francischetti IM, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JM. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem Mol Biol. 2007;37:107–27. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, Ribeiro JM. An updated catalogue of salivary gland transcripts in the adult female mosquito. Anopheles gambiae J Exp Biol. 2005;208:3971–86. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–6. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17:349–56. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- Bondy GS, Gentry PA. Characterization of the normal bovine platelet aggregation response. Comp Biochem Physiol C. 1989;92:67–72. doi: 10.1016/0742-8413(89)90204-1. [DOI] [PubMed] [Google Scholar]
- Boulanger N, Munks RJ, Hamilton JV, Vovelle F, Brun R, Lehane MJ, Bulet P. Epithelial innate immunity. A novel antimicrobial peptide with antiparasitic activity in the blood-sucking insect Stomoxys calcitrans. J Biol Chem. 2002;277:49921–6. doi: 10.1074/jbc.M206296200. [DOI] [PubMed] [Google Scholar]
- Calvo E, Andersen J, Francischetti IM, de LCM, deBianchi AG, James AA, Ribeiro JM, Marinotti O. The transcriptome of adult female Anopheles darlingi salivary glands. Insect Mol Biol. 2004;13:73–88. doi: 10.1111/j.1365-2583.2004.00463.x. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Andersen JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J Biol Chem. 2006;281:1935–42. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Calvo E, Ribeiro JM. A novel secreted endonuclease from Culex quinquefasciatus salivary glands. J Exp Biol. 2006;209:2651–9. doi: 10.1242/jeb.02267. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Vandyke KA, Letchworth GJ, Drolet BS, Hanekamp T, Wilson WC. Midgut and salivary gland transcriptomes of the arbovirus vector Culicoides sonorensis (Diptera: Ceratopogonidae) Insect Mol Biol. 2005;14:121–36. doi: 10.1111/j.1365-2583.2004.00537.x. [DOI] [PubMed] [Google Scholar]
- Campbell JB, Skoda SR, Berkebile DR, Boxler DJ, Thomas GD, Adams DC, Davis R. Effects of stable flies (Diptera: Muscidae) on weight gains of grazing yearling cattle. J Econ Entomol. 2001;94:780–3. doi: 10.1603/0022-0493-94.3.780. [DOI] [PubMed] [Google Scholar]
- Champagne DE, Smartt CT, Ribeiro JM, James AA. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5′-nucleotidase family. Proc Natl Acad Sci U S A. 1995;92:694–8. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coligan JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT. Current Protocols in Protein Science. Vol. 1. John Wiley & Sons, Inc.; 1995. pp. 11.7.3–11.7.4. [Google Scholar]
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–94. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Cupp EW, Cupp MS, Ribeiro JM, Kunz SE. Blood-feeding strategy of Haematobia irritans (Diptera: Muscidae) J Med Entomol. 1998;35:591–5. doi: 10.1093/jmedent/35.4.591. [DOI] [PubMed] [Google Scholar]
- DeGiusti DL, Sterling CR, Dobrzechowski D. Transmission of the chelonian haemoproteid Haemoproteus metchnikovi by a Tabanid fly Chrysops callidus. Nature. 1973;242:50–1. doi: 10.1038/242050a0. [DOI] [PubMed] [Google Scholar]
- Dodds WJ. Platelet function in animals: species specificities. In: D G, Garattini S, editors. Platelets: a multidisciplinary approach. New York: Raven; 1977. pp. 45–59. [Google Scholar]
- Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17:107–12. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- Faudry E, Lozzi SP, Santana JM, D'Souza-Ault M, Kieffer S, Felix CR, Ricart CA, Sousa MV, Vernet T, Teixeira AR. Triatoma infestans apyrases belong to the 5′-nucleotidase family. J Biol Chem. 2004;279:19607–13. doi: 10.1074/jbc.M401681200. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector. Anopheles gambiae J Exp Biol. 2002;205:2429–51. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- Gordon RM, Crewe W. The mechanism by which mosquitoes and tsetse flies obtain their blood meal, the histology of the lesions produced, and the subsequent reactions of the mammalian host, together with some observations on the feeding of Chrisops and Cimex. Ann trop Med Parasitol. 1948;42:334–356. doi: 10.1080/00034983.1948.11685382. [DOI] [PubMed] [Google Scholar]
- Grimaldi D, Engel M. Evolution of the insects. New York: Cambridge University Press; 2005. [Google Scholar]
- Hansen JE, Lund O, Tolstrup N, Gooley AA, Williams KL, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj J. 1998;15:115–30. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- Huang X. A contig assembly program based on sensitive detection of fragment overlaps. Genomics. 1992;14:18–25. doi: 10.1016/s0888-7543(05)80277-0. [DOI] [PubMed] [Google Scholar]
- Kanost MR. Serine proteinase inhibitors in arthropod immunity. Dev Comp Immunol. 1999;23:291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- Kerlin RL, Hughes S. Enzymes in saliva from four parasitic arthropods. Med Vet Entomol. 1992;6:121–126. doi: 10.1111/j.1365-2915.1992.tb00587.x. [DOI] [PubMed] [Google Scholar]
- Krafsur ES. Tsetse flies: genetics, evolution, and role as vectors. Infect Genet Evol. 2009;9:124–41. doi: 10.1016/j.meegid.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski J, Grushko OG, Besansky NJ. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol Phylogenet Evol. 2006;39:417–23. doi: 10.1016/j.ympev.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Landon C, Meudal H, Boulanger N, Bulet P, Vovelle F. Solution structures of stomoxyn and spinigerin, two insect antimicrobial peptides with an alpha-helical conformation. Biopolymers. 2006;81:92–103. doi: 10.1002/bip.20370. [DOI] [PubMed] [Google Scholar]
- Lewis S, Ashburner M, Reese MG. Annotating eukaryote genomes. Curr Opin Struct Biol. 2000;10:349–54. doi: 10.1016/s0959-440x(00)00095-6. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–3. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TJ, Abbenante G, Tyndall JD, Halliday J, Lewis RJ. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J Biol Chem. 2003;278:31105–10. doi: 10.1074/jbc.M304843200. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nobile M, Noceti F, Prestipino G, Possani LD. Helothermine, a lizard venom toxin, inhibits calcium current in cerebellar granules. Exp Brain Res. 1996;110:15–20. doi: 10.1007/BF00241369. [DOI] [PubMed] [Google Scholar]
- Okiwelu N, Maiga S. Natural hosts of Glossina morsitans submorsitans News tead and Glossina pallpalis gambiensis Vanderplank in the Republic of Mali. Cah ORSTOM ser Ent med et Parasitol. 1981;19:179–186. [Google Scholar]
- Ribeiro JM. Blood-feeding in mosquitoes: probing time and salivary gland anti- haemostatic activities in representatives of three genera (Aedes, Anopheles, Culex) Med Vet Entomol. 2000;14:142–8. doi: 10.1046/j.1365-2915.2000.00227.x. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem Mol Biol. 2004;34:543–63. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC. Blood-feeding arthropods: Live syringes or invertebrate pharmacologists. Infect Agents Dis. 1995;4:143–152. [PubMed] [Google Scholar]
- Ribeiro JMC, Modi GB, Resh RB. Salivary apyrase activity of some old world phlebotomine sand flies. Insect Biochem. 1989;19:409–412. [Google Scholar]
- Ribeiro JMC, Rossignol PA, Spielman A. Salivary gland apyrase determines probing time in anopheline mosquitoes. J Insect Physiol. 1985;9:551–560. [Google Scholar]
- Schlott B, Wohnert J, Icke C, Hartmann M, Ramachandran R, Guhrs KH, Glusa E, Flemming J, Gorlach M, Grosse F, et al. Interaction of Kazal-type inhibitor domains with serine proteinases: biochemical and structural studies. J Mol Biol. 2002;318:533–46. doi: 10.1016/S0022-2836(02)00014-1. [DOI] [PubMed] [Google Scholar]
- Schreiber MC, Karlo JC, Kovalick GE. A novel cDNA from Drosophila encoding a protein with similarity to mammalian cysteine-rich secretory proteins, wasp venom antigen 5, and plant group 1 pathogenesis-related proteins. Gene. 1997;191:135–41. doi: 10.1016/s0378-1119(97)00010-3. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–4. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- Stark KR, James AA. Isolation and characterization of the gene encoding a novel factor Xa-directed anticoagulant from the yellow fever mosquito Aedes aegypti. J Biol Chem. 1998;273:20802–9. doi: 10.1074/jbc.273.33.20802. [DOI] [PubMed] [Google Scholar]
- Sun D, McNicol A, James AA, Peng Z. Expression of functional recombinant mosquito salivary apyrase: A potential therapeutic platelet aggregation inhibitor. Platelets. 2006;17:178–84. doi: 10.1080/09537100500460234. [DOI] [PubMed] [Google Scholar]
- Takac P, Nunn MA, Meszaros J, Pechanova O, Vrbjar N, Vlasakova P, Kozanek M, Kazimirova M, Hart G, Nuttall PA, et al. Vasotab, a vasoactive peptide from horse fly Hybomitra bimaculata (Diptera, Tabanidae) salivary glands. J Exp Biol. 2006;209:343–52. doi: 10.1242/jeb.02003. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DB, Berkebile D. Comparative efficiency of six stable fly (Diptera: Muscidae) traps. J Econ Entomol. 2006;99:1415–9. doi: 10.1603/0022-0493-99.4.1415. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Belkaid Y, Rowton E, Ribeiro JM. The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. J Exp Biol. 2001;204:229–37. doi: 10.1242/jeb.204.2.229. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Charlab R, Galperin MY, Ribeiro JM. Purification, cloning, and expression of an apyrase from the bed bug Cimex lectularius. A new type of nucleotide-binding enzyme. J Biol Chem. 1998;273:30583–90. doi: 10.1074/jbc.273.46.30583. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Charlab R, Gonzalez EC, Miranda-Santos IKF, Marinotti O, Francischetti IM, Ribeiro JMC. The D7 family of salivary proteins in blood sucking Diptera. Insect Mol Biol. 2002a;11:149–55. doi: 10.1046/j.1365-2583.2002.00319.x. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Pham VM, Garfield MK, Francischetti IM, Ribeiro JMC. Toward a description of the sialome of the adult female mosquito Aedes aegypti. Insect Biochem Mol Biol. 2002b;32:1101–1122. doi: 10.1016/s0965-1748(02)00047-4. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003;33:717–32. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Ribeiro JM. Purification, cloning, and synthesis of a novel salivary anti-thrombin from the mosquito Anopheles albimanus. Biochemistry. 1999;38:11209–11215. doi: 10.1021/bi990761i. [DOI] [PubMed] [Google Scholar]
- Volfova V, Hostomska J, Cerny M, Votypka J, Volf P. Hyaluronidase of bloodsucking insects and its enhancing effect on leishmania infection in mice. PLoS Negl Trop Dis. 2008;2:e294. doi: 10.1371/journal.pntd.0000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Addess KJ, Chen J, Geer LY, He J, He S, Lu S, Madej T, Marchler-Bauer A, Thiessen PA, et al. MMDB: annotating protein sequences with Entrez's 3D-structure database. Nucleic Acids Res. 2007;35:D298–300. doi: 10.1093/nar/gkl952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yang H, Ma D, Wu J, Wang Y, Song Y, Wang X, Lu Y, Yang J, Lai R. Toward an understanding of the molecular mechanism for successful blood feeding by coupling proteomics analysis with pharmacological testing of horsefly salivary glands. Mol Cell Proteomics. 2008;7:582–90. doi: 10.1074/mcp.M700497-MCP200. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Hyodo F, Morita T. Wide distribution of cysteine-rich secretory proteins in snake venoms: isolation and cloning of novel snake venom cysteine-rich secretory proteins. Arch Biochem Biophys. 2003;412:133–41. doi: 10.1016/s0003-9861(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Morita T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon. 2004;44:227–31. doi: 10.1016/j.toxicon.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Zhang D, Cupp MS, Cupp EW. Thrombostasin: purification, molecular cloning and expression of a novel anti-thrombin protein from horn fly saliva. Insect Biochem Mol Biol. 2002;32:321–30. doi: 10.1016/s0965-1748(01)00093-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Supplemental Table S1: Hyperlinked Excel spreadsheet and associated files with EST assembly results. NOTE: This is a compressed ZIP file that should be expanded to a new directory. After this is done, start Excel and then open the file ending in .xls so the hyperlinks will work.
Additional file 2 Supplemental Table S2: Hyperlinked Excel spreadsheet with deducted protein sequences. See note above.


