Abstract
Purpose
Over the past few years, the alkylating agent temozolomide (TMZ) has become the standard-of-care therapy for patients with glioblastoma, the most common brain tumor. Recently, large-scale cancer genome sequencing efforts have identified a hypermutation phenotype and inactivating MSH6 mismatch repair gene mutations in recurrent, post-TMZ glioblastomas, particularly those growing more rapidly during TMZ treatment. This study aimed to clarify the timing and role of MSH6 mutations in mediating glioblastoma TMZ resistance.
Experimental Design
MSH6 sequence and microsatellite instability (MSI) status were determined in matched pre- and post-chemotherapy glioblastomas identified by The Cancer Genome Atlas (TCGA) as having post-treatment MSH6 mutations. TMZ-resistant lines were derived in vitro via selective growth under TMZ and the MSH6 gene was sequenced in resistant clones. The role of MSH6 inactivation in mediating resistance was explored using lentiviral shRNA knockdown and MSH6 reconstitution.
Results
MSH6 mutations were confirmed in post-treatment TCGA glioblastomas but absent in matched pre-treatment tumors. The post-treatment hypermutation phenotype displayed a signature bias toward CpC transitions and was not associated with MSI. In vitro modeling via exposure of an MSH6-wildtype glioblastoma line to TMZ resulted in resistant clones; one clone showed an MSH6 mutation, Thr1219Ile, that had been independently noted in two treated TCGA glioblastomas. Knockdown of MSH6 in the glioblastoma line U251 increased resistance to TMZ cytotoxicity and reconstitution restored cytotoxicity in MSH6-null glioma cells.
Conclusions
MSH6 mutations are selected for in glioblastomas during TMZ therapy both in vitro and in vivo, and are causally associated with TMZ resistance.
Keywords: glioblastoma, MSH6, mismatch repair, resistance, temozolomide
Introduction
The standard of care for newly diagnosed glioblastomas involves surgery and external beam radiation therapy (XRT) in conjunction with the alkylating agent temozolomide (TMZ) (1). Benefit from TMZ is most noted in patients whose tumors have transcriptional silencing of the O6-methylguanine methyltransferase (MGMT) gene mediated by promoter methylation (2), which occurs in approximately half of tumors (3). Nonetheless, the prognosis for patients with glioblastoma remains bleak. Virtually all patients recur after initial therapy and average survival remains around twelve months (4).
Our knowledge of the genetic changes underlying glioblastoma, although considerable, is largely confined to pre-treatment cases (5). Given that all glioblastomas recur and that the recurrent lesions invariably lead to patient death, there is a pressing need to understand the molecular changes that occur during treatment and that characterize the therapeutically resistant recurrences (6). In this regard, we first identified inactivating somatic mutations in the mismatch repair gene MSH6 in two recurrent glioblastomas treated with TMZ (7). Survey of the genome in these two tumors revealed large numbers of somatic mutations with mutational signatures consistent with those resulting from defects in DNA mismatch repair. Of note, studies in normal and neoplastic cells had shown inactivation of MSH6 results in resistance to cytotoxicity mediated by alkylating agents (8–12) and we therefore proposed that MSH6 inactivation may be one mechanism underlying TMZ resistance in glioblastomas. In a follow-up study, we examined a larger series of pre- and post-treatment glioblastomas for MSH6 mutations and MSH6 expression (13). MSH6 alterations (mutations and/or absent expression) were not found in any pre-treatment glioblastomas and not in any post-treatment glioblastomas given only XRT, but were detected in approximately half of the recurrent glioblastomas treated with TMZ and XRT. Furthermore, temporal measurements of three-dimensional reconstructed magnetic resonance images showed that MSH6-negative glioblastomas demonstrated more rapid radiological progression while under TMZ treatment compared to MSH6-positive tumors. These data supported a role for MSH6 inactivation in the emergence of TMZ resistance in glioblastoma patients.
Two other studies have now reported MSH6 mutation in glioblastomas following alkylating agent chemotherapy. The Cancer Genome Atlas (TCGA) reported an analysis of 91 glioblastomas with matched peripheral blood, of which 19 cases were recurrent glioblastomas that had received alkylating agent chemotherapy (14). In keeping with our findings, the TCGA reported non-synonymous MSH6 mutations in five of the recurrent glioblastomas (26% of recurrent tumors), all with a hypermutation phenotype consistent with mismatch repair defects. Two other tumors had a hypermutation phenotype, one with a mutation in another mismatch repair gene. In addition, Maxwell et al. demonstrated seven non-synonymous and therefore putative MSH6 mutations (two of them truncating) out of 27 post-TMZ samples (26%); of note, this series included some malignant gliomas other than glioblastoma, such as oligodendroglial tumors that in our experience to date do not undergo frequent MSH6 alterations (15). Unfortunately, only 2 cases with sequence variations had available matched pre-treatment samples, and these both had only a common MSH6 polymorphism rather than non-synonymous mutations. Thus, while the TCGA and Maxwell et al. reports confirm that MSH6 alterations are common in those recurrent glioblastomas exposed to alkylating agents, neither had access to matched pre-treatment samples to assess the timing of these mutations.
To pursue this hypothesis further, we undertook additional studies to clarify the timing of MSH6 inactivation in the TCGA clinical cases, as well as to model such inactivation in vitro and to evaluate directly the role of MSH6 in TMZ resistance in vitro. The results of these studies support the hypothesis that MSH6 inactivation occurs during alkylating agent chemotherapy and that, at least in the common setting of MGMT inactivation, MSH6 inactivation is directly related to therapeutic resistance.
Materials and Methods
TCGA Tissue samples and DNA stocks
Unstained slides from formalin-fixed paraffin-embedded (FFPE), anonymous, matched pre- and post-chemotherapy glioblastomas were obtained from M.D. Anderson Cancer Center, Houston, TX. The post-treatment samples were the same ones used by TCGA for genomic and epigenomic profiling and were identified as displaying the hypermutation phenotype and somatic MSH6 mutations (14). Tumor tissue from unstained FFPE glass slides was deparaffinized in xylene followed by immersion in graded alcohols until rehydration and genomic DNA was extracted using the Gentra PureGene Kit (Qiagen, Hilden, Germany). DNA quantitation was performed using a Nanodrop ND-1000 UV-Vis spectrophotometer (Nanodrop Technologies, Wilmington, DE).
Mutation targeted PCR and sequencing
Targeted polymerase chain reaction or PCR primers were designed to amplify MSH6 mutations initially identified by the TCGA consortium and posted in the publicly accessible database1 (14). PCR was performed as described previously (13). Targeted sequencing was performed using the standard Sanger method in both forward and reverse directions. Each individual sequencing reaction was repeated for confirmation. Analysis of DNA tracings was performed using Mutation Surveyor version 3.2 (Softgenetics, State College, PA).
Microsatellite instability testing
PCR was conducted in 20 μl volumes using 1X Platinum Taq PCR buffer, 200 mM dNTPs, 2.0 mM MgCl2, 0.4 mM primers, 1.0 U of Platinum Taq polymerase (all from Invitrogen (Carlsbad, CA)) with 40 ng of tumor DNA as template. Primer sets comprised the 5 reference panel markers recommended by the National Cancer Institute, with 5’ phosphoramidite fluorescent labeling of forward primers as follows: BAT-25 (NED), BAT-26 (6-FAM), D5S346 (VIC), D17S250 (6-FAM), and D2S123 (VIC). The primer sequences for D2S123 were 5’-AACATTGCTGGAAGTTCTGG-3’ (forward) and 5’-GTGTCTTGACTTTCCACCTATGGGACTG-3’ (reverse). Primer sequences for the remaining loci were identical to those previously described (16) except that a 5’ GTGTCTT sequence was added to each reverse primer to facilitate non-template adenylation of the 3’ end of the forward strand.
PCR was performed in a Mastercycler PCR machine (Eppendorf, Hamburg, Germany) with an initial denaturing step at 94°C for 5 min; followed by 38 cycles of denaturing at 94°C for 30 s, annealing at either 50°C or 55°C for 30 s, and extension at 72°C for 30 s; and a final elongation step at 72°C for 10 min. PCR products were pooled and fractionated by size using an Applied Biosystems (Foster City, CA) 3130 DNA Analyzer with GeneMapper software. Microsatellite loci at which tumor DNA showed a novel allele profile not present in the corresponding normal DNA were classified as MSI.
Tissue culture
The human glioblastoma cell lines A172 and U251 were originally obtained from the American Tissue Culture Collection (Manassas, VA). The primary human glioblastoma cell culture Gli60 was established from recurrent tumor xT3162 post-TMZ and radiotherapy; the tumor has the somatic mutation p.Val809X in MSH6, which results in premature termination, and has lost the remaining copy of chromosome 2, leading to null expression of the protein (7, 13). TMZ was purchased from Sequoia Research Products Limited (Pangbourne, United Kingdom), reconstituted to a stock concentration of 100 mmol/L and stored at − 80°C. O6-benzylguanine (O6-BG) was obtained from Sigma-Aldrich (St Louis, MO), reconstituted to stock concentration of 320 mmol/L with DMSO and stored at − 80°C. A172, U251, and Gli60 cells were maintained in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal calf serum, 1% L-glutamine (Invitrogen) and grown at 37°C humidified atmosphere containing 5% CO2. Cells were confirmed to be free of mycoplasma using the Lonza MycoAlert detection kit (Hopkinton, MA).
Generation of TMZ-resistant glioblastoma subclones
The glioblastoma cell line A172 was treated with TMZ at 100 μmol/L or DMSO solvent control at a final concentraion of 0.1% for three weeks. To generate TMZ-resistant glioma sublines, A172 cells were cultured in 6-well plates and allowed to adhere overnight at 37°C incubator. Control groups were treated with 0.1% DMSO alone. Cell treatment was repeated every 24 h for five consecutive days and then exposure to the fresh TMZ every three days to a total of three weeks. Each single clone was grown up to derive stable resistant cell lines for subsequent study. MSH6 and MGMT expression was measured by western blot using the cell lysates treated with TMZ.
Reconstitution of MSH6 expression in Gli60 using lentiviral approach
The vector backbone for lentiviral reconstitution of Gli60, as well as the viral packaging procedure, have been described previously (17, 18). Briefly, cDNA for MSH6 was cloned into the construct under the control of the cytomegalovirus (CMV) promoter and cDNA for the fluorescence marker GFP was under the control of internal ribosomal entry sequence (IRES). We used empty constructs expressing only GFP as an infection control. Gli60 cells were infected with lentiviral particles in the presence of protamine sulfate (Sigma) at a multiplicity of infection of 10:1. Successful infection was confirmed by appearance of green fluorescing cells and western blot evaluation for MSH6 protein.
Generation of lentiviral constructs for shRNA- mediated MSH6 knockdown
Bacterial plasmid constructs containing shRNA sequence candidates against MSH6 mRNA were obtained from the MGH shRNA core (Dr Toshi Shioda, MGH). Subsequent steps in the production of lentiviral particles and infection of glioblastoma cells were performed according to protocols listed in the TRC/Broad website2. Lentiviral supernatants were removed from U251 cells after overnight incubation and the cells were allowed to grow in DMEM with 10% FCS for 48 hours prior to selection with increasing doses of puromycin from 0.5 to 2.0 μg/ml (Sigma-Aldrich). MSH6 and MGMT protein levels in U251 cells stably infected with the five candidate shRNA constructs were assessed by western blot analysis.
Cell viability assay
To compare cell viability between parental glioblastoma cell lines and TMZ-treated, resistant or MSH6 knockdown cell lines, we used the MTS cytotoxicity assay measuring absorbance at 490 nm (Promega, Madison, WI). Briefly, 1000 cells were seeded per well in 96-well tissue culture plates before the drug treatment. Cells were treated daily with fresh TMZ (100 μmol/L) for 7 days. U251 and U251 Sh1 cells were additionally treated with TMZ in the presence of O6-benzylguanine (40 μmol/L). Gli60 cells reconstituted with wildtype MSH6 and with empty control vector were incubated with various concentrations of TMZ for five days and cell growth was evaluated by MTS assay.
Western blot analysis
Antibodies were obtained from the following sources: MSH6 monoclonal antibody (BD Biosciences, San Diego, CA); MGMT (Lab Vision Co., Fremont, CA); β-Actin monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Cells were lysed in RIPA protein extraction buffer (Sigma-Aldrich, St Louis, MO) together with protease and phosphatase inhibitors at 1:100 dilutions (Sigma-Aldrich) and then centrifuged 10 min at 4°C to harvest the supernatant. The concentration of the extracted protein was determined using the Bio-Rad protein assay kit (Bio-Rad Laboratories, New York, NY). Twenty micrograms per lane of the extracted protein were loaded onto 4–12% Tris-Glycine gels (Invitrogen) for electrophoresis, and electro-transferred to Pure Nitrocellulose Membrane (Bio-Rad Laboratories, Hercules, CA). The membrane was blocked with 5% nonfat dry milk in TBS-T buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween-20) for 1 h at room temperature, followed by incubation with different antibodies in the blocking buffer over-night at 4°C. After washing with TBS-T buffer, the membrane was incubated with horseradish peroxidase-conjugated anti-mouse IgG antibody (Promega Co., Madison, WI) at a dilution of 1:2000 in blocking buffer for 2 h at room temperature. The membrane was developed using the enhanced chemiluminescence system (Perkin Elmer Inc., Boston, MA) and exposed to Biomax XAR film (Kodak, Rochester, NY).
Analysis Methods
GraphPad Prism 3.0 (La Jolla, CA) was used to determine statistically significant differences between cytotoxicity curves. Best fit curves for growth of A172/A172TR3 lines, U251 line and U251-Sh1 were calculated using the program-derived “Exponential growth” nonlinear regression equation with the starting points of all curves held constant at the average OD value for all Day 1 points. Second order polynomial nonlinear regression was used for the analysis of survival fractions at the end of five- day incubation with TMZ in Gli60 cells reconstituted with MSH6 and control vector. To determine significance, only the mean Y value was considered for each replicate point and rate constants of the calculated best fit curves were compared using the t-test. Due to multiple testing within each cytotoxicity experiment, statistical significance was defined as p<0.01.
Results & Discussion
Genetic analysis of matched TCGA glioblastoma samples: MSH6 mutations, hypermutation phenotype and MSI status
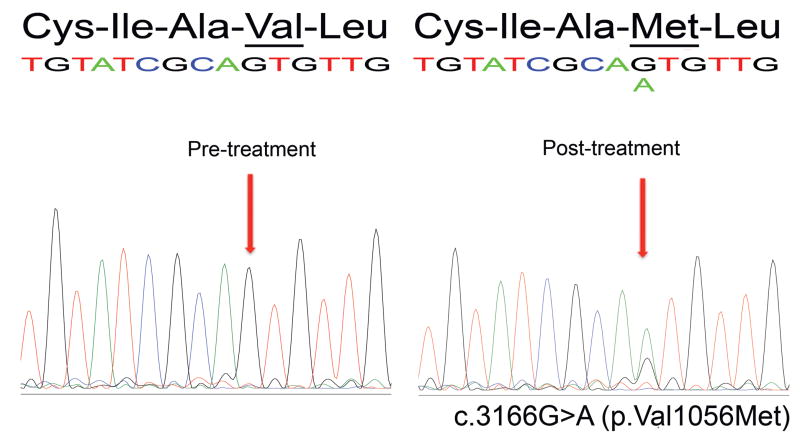
We have previously reported somatic MSH6 mutations in three of 11 recurrent glioblastomas treated with alkylating agents and XRT, but in no pre-treatment tumors or in post-treatment tumors treated with XRT only, suggesting that MSH6 mutations arise specifically after alkylating agent chemotherapy (13). Given that the TCGA has now found somatic MSH6 mutations in five of 19 recurrent glioblastomas, we sought to determine the timing of these genetic changes. We obtained matched pre- and post-treatment unstained FFPE-tumor tissue sections from four of the five cases identified by TCGA as having somatic MSH6 mutations (TCGA-02-0043, -02-0083, -02-0099, -02-0114). To demonstrate that sequencing from FFPE tissues was of adequate sensitivity and specificity to detect mutations, we sequenced the post-treatment samples and confirmed all five MSH6 mutations reported by TCGA in the four cases (the TCGA-02-0114 post-treatment specimen had two distinct MSH6 somatic mutations). Notably, however, sequencing of the matched pre-treatment glioblastomas (including two pre-chemotherapy specimens from TCGA-02-0114) showed no mutations (Table 1 and Figure 1). The absence of germline and of somatic pre-treatment MSH6 mutations strongly suggests that MSH6 mutations do not contribute to the development of glioblastoma. Rather, the presence of MSH6 mutations in the post-treatment samples is consistent with de novo alterations in the tumor cell genome in association with treatment.
Table 1.
MSH6 mutations in pre- and post-treatment TCGA glioblastoma cases.
| Case ID | Treatment status | Chemotx | MSH6 somatic mutations | amino acid change |
|---|---|---|---|---|
| TCGA-02-0043 | Pre-treatment | - | none | none |
| Post-treatment | CCNU1 | c.3656C>T | p.Thr1219Ile | |
| TCGA-02-0083 | Pre-treatment | - | none | none |
| Post-treatment | Temozolomide | c.3166G>A | p.Val1056Met | |
| TCGA-02-0099 | Pre-treatment | - | none | none |
| Post-treatment | PCV2 | c.3656C>T | p.Thr1219Ile | |
| TCGA-02-0114 | Pre-treatment | - | none | none |
| Post-treatment | PCV | c.1450G>A, c.2294G>A | p.Glu484Lys, p.Cys765Tyr | |
CCNU= Lomustine
PCV= procarbazine, CCNU, vincristine.
Figure 1.
MSH6 mutation c.3166G>A (p.Val1056Met) is present in the post-treatment TCGA-02-0083 glioblastoma (right) but not in the matched pre-treatment sample (left).
The TCGA report noted a hypermutation phenotype in all four of these MSH6-mutant glioblastomas (14). However, the published data did not clarify the sequence context of the mutations beyond classifying them as CpG or non-CpG. We therefore undertook a detailed analysis of the somatic mutation data of these four cases, and confirmed our prior findings of a hypermutation phenotype with a preponderance of C:G>T:A transitions at CpC dinucleotides that is striking when compared to recurrent, post-TMZ glioblastomas without MSH6 mutations (Table 2) (7). For example, TCGA-02-0083 contains 94 C>T somatic mutations, of which 60 are within the context of CpC dinucleotides. Interestingly this recurrent tumor also harbors somatic mutations in two other mismatch repair genes, MSH2 and MLH1, which could account for the larger number of such mutations compared to the other hypermutant cases. The other three post-treatment TCGA cases in this series (TCGA-02-0043, -0099, -0114) also contain markedly higher numbers of somatic mutations and a preponderance of mutations in the context of CpC dinucleotides. On the other hand, these cases did not show high microsatellite instability (MSI); we did not detect MSI-high (≥ 3/5 unstable loci) in any cases and detected only minor shifts in <2 loci in two cases (TCGA-02-0043 and -02-0083, data not shown). These findings are in keeping with a recent evaluation of the role of MSI as a surrogate marker for MSH6 inactivation in recurrent malignant gliomas; Maxwell et al. showed no correlation between MSH6 mutations and MSI, as assessed by a panel of five mononucleotide loci (15). That study, however, did not determine whether the recurrent tumors with MSH6 mutations had a hypermutation phenotype. In our study, the overwhelming number of somatic mutations in these tumors, in conjunction with MSH6 mutations, are wholly consistent with MSH6 inactivation causing the hypermutation phenotype, and the absence of MSI is consistent with known MSH6 function (7, 13, 19).
Table 2.
Mutational burden and sequence context of C:G>T:A mutations in the four post-treatment TCGA glioblastomas with hypermutation phenotype (top), compared to four non-hypermutation post-treatment glioblastomas (bottom).
| CpA | CpC | CpG | CpT | TOTAL C:G>T:A mutations |
Mutations in CpC context |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case ID | ACA | CCA | GCA | TCA | ACC | CCC | GCC | TCC | ACG | CCG | GCG | TCG | ACT | CCT | GCT | TCT | |||
| MSH6 mutated post-treatment cases with hypermutation | TCGA-02-0043 | 0 | 0 | 0 | 0 | 8 | 9 | 6 | 4 | 0 | 0 | 3 | 2 | 2 | 2 | 2 | 0 | 38 | 27 |
| TCGA-02-0083 | 1 | 2 | 3 | 0 | 10 | 19 | 15 | 16 | 2 | 3 | 2 | 1 | 4 | 9 | 4 | 3 | 94 | 60 | |
| TCGA-02-0099 | 0 | 1 | 0 | 0 | 2 | 3 | 0 | 7 | 1 | 2 | 1 | 1 | 3 | 2 | 3 | 3 | 29 | 12 | |
| TCGA-02-0114 | 1 | 2 | 5 | 0 | 10 | 11 | 12 | 15 | 0 | 2 | 3 | 1 | 6 | 3 | 8 | 6 | 85 | 48 | |
| MSH6 wildtype post-treatment cases without hypermutation | TCGA-02-0021 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 6 | 1 |
| TCGA-02-0024 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 5 | 1 | |
| TCGA-02-0058 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 5 | 2 | |
| TCGA-02-0116 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 1 | 0 | 2 | 9 | 1 | |
Molecular characterization of in vitro derived TMZ resistant glioblastoma cell line
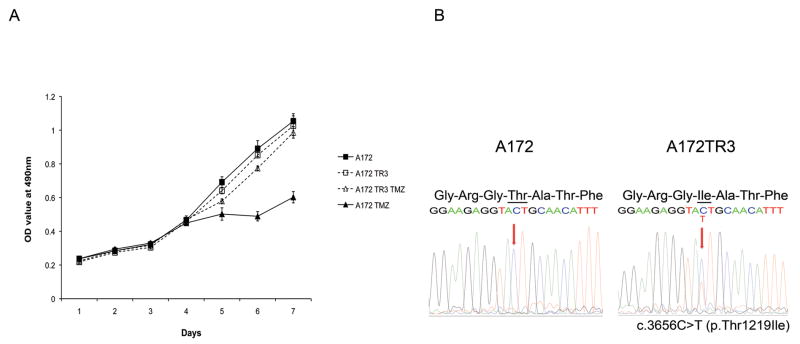
We next sought to model the phenomenon of MSH6 inactivation in vitro. To do so, we exposed the human glioblastoma cell line A172 to TMZ to generate drug resistant clones and found reduced MSH6 protein level in a highly resistant line, A172TR3. There was no difference in growth rate between parental A172 and A172TR3; however, the resistant clone demonstrated significantly enhanced survival in the presence of 100μM TMZ compared to the parental A172 (p<0.01) (Figure 2a). We further reasoned that, similar to the situation in human glioblastomas, the development of a TMZ-resistant clone with reduced MSH6 protein could result from in vitro somatic MSH6 mutation. In fact, sequencing of MSH6 in the resistant line identified a novel MSH6 alteration that was not present in the parental A172 cells. This clone, A172TR3, had a c.3656C>T MSH6 somatic mutation, altering threonine to isoleucine at amino acid position 1219 (Figure 2b). This same mutation has been identified in two TCGA recurrent glioblastomas with the hypermutation phenotype (TCGA-02-0043 and -02-0099), in the malignant melanoma cell line MZ7-mel derived from a post-chemotherapy splenic metastasis (Welcome Trust Sanger Institute, COSMIC database, The Cancer Genome Project3), and as a germline mutation in a colorectal cancer and a case of complex non-atypical endometrial hyperplasia (20). Notably, the MZ7-mel cell line also contains a significant number of somatic mutations consistent with that of the hypermutation phenotype. The Thr1219Ile mutation is therefore likely important to MSH6 function, and suggests that the in vitro induced mutation of Thr1219Ile in the TMZ-resistant A172TR3 cells is biologically and functionally significant.
Figure 2.
a: The parental A172 and the TMZ-resistant subclone A172TR3 exhibit similar growth in the absence of TMZ (solid and dashed lines, square markers). Parental A172 (solid line, triangle marker) is sensitive to 100μM TMZ for seven days, whereas of A172TR3 growth (dashed line, triangle marker) is minimally affected at 100μM TMZ (p<0.01). b: The TMZ-resistant subclone TR3 (left) has a c.3656C>T MSH6 mutation, resulting in a Thr to Ile amino acid change at codon 1219, in comparison to the TMZ-sensitive, parental glioblastoma A172 line (right).
It is also noteworthy that we were able to derive the MSH6 Thr1219Ile mutation after TMZ exposure since the same mutation has been reported independently in glioblastomas that failed therapy with other alkylating agents. TCGA-02-0043 was treated with the alkylating agent CCNU (lomustine) and TCGA-02-0099 was treated with the “PCV” combination therapy (procarbazine, lomustine, and vincristine). In this regard, it is interesting to note that TMZ is a SN1-type methylating agent that mediates guanine modification resulting in base pair mismatch during replication, whereas CCNU causes interstrand crosslinking (12, 21, 22). Moreover, whereas the role of the mismatch repair pathway in facilitating TMZ-mediated cell death is well understood (12), the role of mismatch repair proteins in mediating CCNU cytotoxicity is less clear. These results may suggest a separable convergence in downstream signaling function from DNA damage recognition, as has been hypothesized (19, 23).
MSH6 inactivation may be heterozygous and is expectedly not associated with high MSI
Some of the recurrent TCGA glioblastomas with a hypermutation phenotype contain heterozygous somatic MSH6 mutations, rather than biallelic inactivation. In addition, A172TR3 demonstrated a heterozygous Thr1219Ile MSH6 mutation that was clearly associated with TMZ resistance in vitro. In addition, patients with colorectal cancers and MSH6 missense mutations often have preserved MSH6 immunoreactivity; in fact, colorectal cancer cells and cells from endometrial complex non-atypical hyperplasia with the germline Thr1219Ile missense mutation have been reported to have preserved MSH6 immunoreactivity (20). It would therefore appear that partial MSH6 inactivations, both in vivo and in vitro, may be associated with functional effects.
MMR proteins function in multimeric complexes; MSH6 dimerizes with MSH2 to form the eukaryotic equivalent of the bacterial mutSα complex (19). The MSH6:MSH2 dimer functions to detect single nucleotide mismatches and subsequent corrective actions are undertaken by complexes of other MMR family members. In this regard, it has been reported that compromise of MMR function can occur following mutation of one allele. This may occur through a dominant-negative affect via “soaking-up” of the normal binding partner – MSH2 in the case of MSH6 (24). Alternatively, inactivation of one copy of MSH6, as seen in the TCGA recurrent tumors and by us, could contribute to compromised MMR function via a “gene-dosage” effect, which could be exaggerated in an environment with strong selective pressures, as in the tumor microenvironment in the presence of TMZ. Notably, analyses of changes in MSH6 protein tertiary structure secondary to the missense mutations in the 4 recurrent TCGA glioblastoma demonstrate significant alterations of protein folding in functional domains – in the case of Thr1219Ile, a putative protein-protein interaction domain.4 These data suggest that the elucidation of the differential effects of homozygous versus heterozygous MSH6 inactivation in glioblastoma therapeutic resistance will be an important subject for further inquiry.
As noted, inactivation of MSH6 is typically associated with the MSI-L phenotype, not with high MSI (25, 26). Moreover, for MSH6, there exists functional dichotomy between mismatch repair (or surveillance) function and apoptotic signaling secondary to cytotoxic agents (24). This might further explain the lack of correlation between missense MSH6 mutations and level of MSI in recurrent glioblastomas. There also exists a substantial body of literature on the role of MSH6 in mediating somatic hypermutation in the generation of antibody gene diversity (27, 28). This is not surprising since MSH6 functions solely in the survey of the genome for single base-pair mismatches and does not participate in their subsequent repair (29). These observations are in keeping with our findings and argue that the role of MSH6 in TMZ response is not dependent on MSI.
Genetic knockdown of MSH6 in U251 glioblastoma cell line and correlation with TMZ resistance
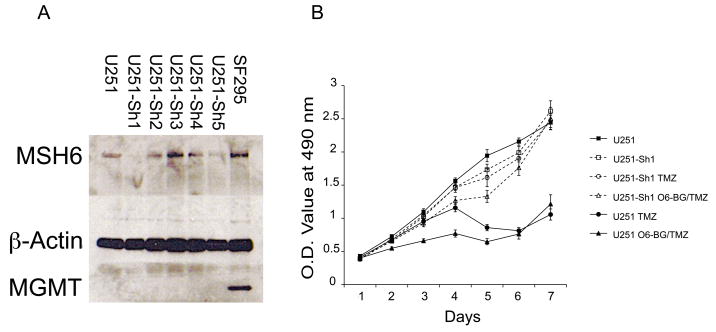
To investigate the specific functional role of MSH6 in glioblastomas, particularly in response to TMZ treatment, we knocked down expression of wildtype MSH6 protein in the human glioblastoma cell line U251 via a lentiviral-mediated shRNA approach. We obtained clones of shRNA against the human MSH6 gene from the TRC/BROAD consortium and generated lentiviral constructs of the five candidate clones (Table 2, Supplemental data). Infection of U251 cells by lentiviral constructs expressing the five MSH6 shRNA candidates were followed by puromycin selection of successfully infected cells. We then performed western blot analyses of the MSH6 protein to identify the shRNA candidates with most efficiently downregulated MSH6 protein (Figure 3a). The shRNA construct#1 (U251-Sh1) demonstrated 90% knockdown of MSH6 compared to 50% in construct#5 (U251-Sh5). We proceeded to examine the response of these cells to TMZ in in vitro cytotoxicity assays. Both U251-Sh1 and U251-Sh5 are significantly more resistant to TMZ compared to parental controls when exposed to 100μM TMZ for seven days (Figure 3a, data for U251-Sh5 not shown) (p<0.01). Nonetheless, U251-Sh1 and U251-Sh5 exhibited similar proliferation indices to parental U251 cells, demonstrating that enhanced TMZ resistance in the MSH6-knockdown cells is not due to alteration in cell growth kinetics. These results confirm a role for MSH6 in mediating TMZ cytotoxicity.
Figure 3.
a: Western blot for MSH6 protein in U251 cells stably infected with five candidate shRNA lentiviral constructs show variable reduction of MSH6 expression, with Sh1 demonstrating maximal inhibition. MGMT is not expressed by any of the knockdown clones or the parental U251 cells (positive glioblastoma control SF295). b: Parental U251 and MSH6-knockdown U251-Sh1 cells exhibit similar growth in the absence of TMZ (solid and dashed lines, square markers). Whereas growth of parental U251 is significantly reduced in the presence of 100μM TMZ for seven days (solid line, closed circle), U251-Sh1 appear resistant to 100μM TMZ (dashed line, open circle) (p<0.01). Addition of 40μmol/l of O6-BG had no effect on TMZ cytotoxicity in both parental U251 and MSH6-knockdown U251-Sh1 cells (solid and dashed lines, triangle markers).
Restoration of TMZ cytotoxicity in Gli60 with reconstitition of MSH6 expression
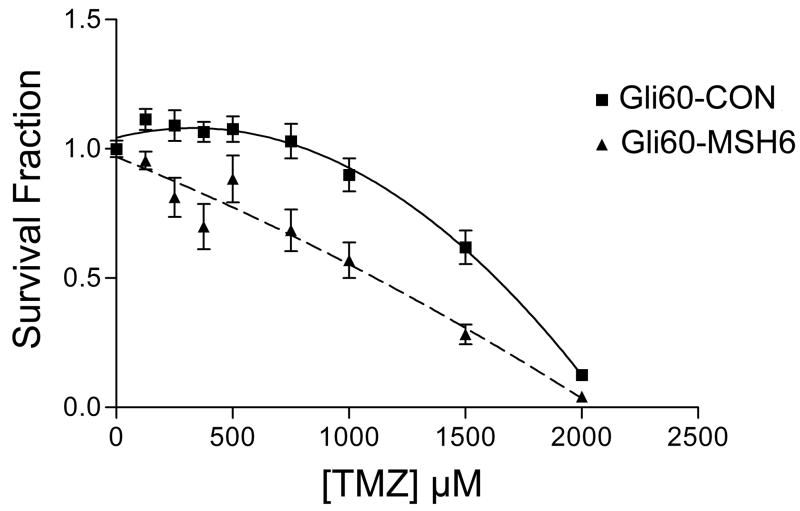
We had previously derived and characterized the primary glioblastoma cell culture Gli60 from a recurrent glioblastoma in a patient who had recurred quickly during treatment with TMZ and radiotherapy. Gli60 has the somatic MSH6 mutation delG2425, which results in premature protein termination, and loss of the remaining wildtype MSH6 gene, with resulting null expression of MSH6 on western blotting; Gli60 also exhibits the characteristic hypermutation phenotype of C→T transitions preferentially at CpC dinucleotides (7, 13). Gli60 thus represents a unique resource for the study of post-TMZ recurrent glioblastoma. To support our hypothesis that somatic inactivating MSH6 mutations result in a survival advantage to these cells, we restored MSH6 expression in Gli60 and examined the subsequent response to TMZ. Gli60 cells were infected with a lentiviral construct expressing MSH6 under the control of the CMV promoter (Gli60-MSH6) or with an empty control vector (Gli60-Con vector). We then incubated the cells with different concentrations of TMZ and examined the cytotoxicity using the MTS assay. Gli60-MSH6 demonstrated restored TMZ sensitivity compared to Gli60-CON (p<0.01) (Figure 4). This confirms that restoration of MSH6 expression in glioma cells from a patient who failed TMZ treatment conferred TMZ sensitivity in vitro, and functionally links MSH6 function to TMZ sensitivity in a glioblastoma that harbors the characteristic hypermutation phenotype.
Figure 4.
The Gli60 primary glioblastoma, which is MSH6-null secondary to nonsense mutation and loss of the remaining chromosome 2, infected with control empty lentiviral vector (Gli60-Con vector) and with MSH6-vector (Gli60-MSH6) showed differential sensitivity to TMZ after five days of drug exposure. Gli60 cells with restored MSH6 expression are significantly more sensitive to TMZ than cells infected with control vectors (p<0.01).
Relationship of MSH6 alterations to MGMT status
TMZ acts by adding methyl groups to the O6 position of guanine nucleotides. The first line of response to repair this chemotherapeutic event is mediated by MGMT, which removes these methyl groups (30). However, a substantial proportion of glioblastomas have transcriptional silencing of the MGMT gene via promoter hypermethylation and cases with intact MGMT expression can have MGMT downregulated by the MGMT inhibitor O6-benylguanine (O6-BG) (31, 32). In such cases, the mismatch repair pathway becomes a primary mediator of O6-methylguanine cytotoxicity, and defects in the mismatch repair pathway can therefore serve as an alternate resistance mechanism for cancer cells (12, 33). As a result, to develop resistance after TMZ exposure, cancer cells that cannot upregulate MGMT expression would be expected to inactivate this mismatch repair pathway.
In the present experiments, in keeping with what has been found in primary human glioblastomas, both the TCGA cases and the studied cell lines had inactivation of MGMT. All four of the TCGA cases had methylation of the MGMT promoter (14) which has been correlated with decreased expression of MGMT and improved response to TMZ in initially treated primary tumors (2, 34). The A172 cell line does not express MGMT, nor does the TMZ-treated resistant clone A172TR3 (data not shown). The U251 cell line does not express MGMT, and MGMT expression is not induced in the knockdown cells (Figure 3a). Nonetheless, to confirm that MGMT did not play a role in the U251 knockdown experiments, we also added the irreversible MGMT inhibitor O6-BG to both parental U251 and the MSH6-knockdown U251-Sh1 and U251–Sh5 clones, and this did not alter drug sensitivity in both the parental and the two MSH6-knockdown clones (p<0.01) (Figure 3b). Thus, the difference in sensitivity to TMZ between the parental and the genetically-modified clones was due principally to MSH6. At the same time, O6-BG is being considered as an adjuvant in TMZ therapy due to its ability to deplete cellular MGMT (31, 35). In this regard, pharmacological inhibition of MGMT by O6-BG in vivo or escalation of TMZ dosing via “dose dense” treatment scheduling could result in accelerated selection pressure to develop alternate escape mechanisms to TMZ-mediated cytotoxicity (33) one of which would be somatic mutations of a mismatch repair gene such as MSH6. Future adjuvant therapies aiming at overcoming MGMT activity could therefore potentially increase the frequency we observe alternate escape pathways such as mismatch repair inactivation (36).
In summary, MSH6 mutations are frequent in recurrent glioblastomas that have been treated with alkylating agents, but have not been found in any pre-chemotherapy glioblastomas. Combining our data (3/11, 27%) with those of the TCGA (5/19, 26%) and Maxwell et al. (7/27, 26%), MSH6 mutations have now been found in 15 of 57 (26%) glioblastomas post-alkylating agent chemotherapy, with similar incidences found in the three series (13–15). We have also demonstrated that in vitro derivation of TMZ-resistant cells can be associated with MSH6 inactivation and mutation. Moreover, in vitro inactivation of wildtype MSH6 protein in glioblastoma cells can result in increased TMZ resistance, and in vitro reconstitution of MSH6 expression can restore TMZ sensitivity in glioblastomas lacking MSH6. These multiple approaches support an integral role for MSH6 inactivation in mediating TMZ resistance in glioblastomas. It is also likely that defects in other mismatch repair proteins could play similar roles: as mentioned above, one case from the TCGA data (TCGA-02-0083) had somatic mutations in two other mismatch repair genes, MSH2 and MLH1; and, in another large-scale genome-wide study of glioblastomas, one recurrent tumor post-TMZ/XRT had the hypermutation phenotype, but MSH6 mutations were not found in this tumor (37). Thus, mismatch repair defects may be a common resistance pathway for treated glioblastomas that have already inactivated MGMT.
Supplementary Material
Acknowledgments
Supported by NIH CA57683 (CLN, DNL) and Clinician Investigator Fellowship from The Royal College of Physicians and Surgeons of Canada (SY), and a Burroughs-Wellcome Career Award in the Medical Sciences (DPC).
Footnotes
Translational relevance
Glioblastomas are highly malignant brain tumors. Current standard therapy utilizes temozolomide (TMZ) and radiation. Previously, we showed that the mismatch repair gene MSH6 is mutated in some recurrent, post-TMZ glioblastomas, and recent data from The Cancer Genome Atlas (TCGA) project has confirmed this observation. We further demonstrate mutations of MSH6 in the post- and not in the pre-treatment TCGA tumors, and have modeled this situation in vitro. Chronic exposure of a glioblastoma line to TMZ generated multiple resistant clones, with one clone harboring an MSH6 mutation. Knockdown of MSH6 expression enhanced survival with cytotoxic doses of TMZ, and MSH6 reconstitution restored TMZ sensitivity in MSH6-null glioblastoma cells. These results indicate that MSH6 is an important mediator of TMZ cytotoxicity and its inactivation is associated with treatment failure in glioblastomas.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 3.Hau P, Stupp R, Hegi ME. MGMT methylation status: the advent of stratified therapy in glioblastoma? Dis Markers. 2007;23:97–104. doi: 10.1155/2007/159242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bondy ML, Scheurer ME, Malmer B, et al. Brain tumor epidemiology: Consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008;113:1953–68. doi: 10.1002/cncr.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 6.Sarkaria JN, Kitange GJ, James CD, et al. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14:2900–8. doi: 10.1158/1078-0432.CCR-07-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter C, Smith R, Cahill DP, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–91. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kat A, Thilly WG, Fang WH, Longley MJ, Li GM, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc Natl Acad Sci U S A. 1993;90:6424–8. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Umar A, Koi M, Risinger JI, et al. Correction of hypermutability, N-methyl-N'-nitro-N-nitrosoguanidine resistance, and defective DNA mismatch repair by introducing chromosome 2 into human tumor cells with mutations in MSH2 and MSH6. Cancer Res. 1997;57:3949–55. [PubMed] [Google Scholar]
- 10.Levati L, Marra G, Lettieri T, et al. Mutation of the mismatch repair gene hMSH2 and hMSH6 in a human T-cell leukemia line tolerant to methylating agents. Genes Chromosomes Cancer. 1998;23:159–66. [PubMed] [Google Scholar]
- 11.Hickman MJ, Samson LD. Apoptotic signaling in response to a single type of DNA lesion, O(6)-methylguanine. Mol Cell. 2004;14:105–16. doi: 10.1016/s1097-2765(04)00162-5. [DOI] [PubMed] [Google Scholar]
- 12.Allan JM, Travis LB. Mechanisms of therapy-related carcinogenesis. Nat Rev Cancer. 2005;5:943–55. doi: 10.1038/nrc1749. [DOI] [PubMed] [Google Scholar]
- 13.Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–45. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Network TCGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 doi: 10.1038/nature11903. [DOI] [PubMed] [Google Scholar]
- 15.Maxwell JA, Johnson SP, McLendon RE, et al. Mismatch repair deficiency does not mediate clinical resistance to temozolomide in malignant glioma. Clin Cancer Res. 2008;14:4859–68. doi: 10.1158/1078-0432.CCR-07-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loukola A, Eklin K, Laiho P, et al. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC) Cancer Res. 2001;61:4545–9. [PubMed] [Google Scholar]
- 17.Sena-Esteves M, Tebbets JC, Steffens S, Crombleholme T, Flake AW. Optimized large-scale production of high titer lentivirus vector pseudotypes. J Virol Methods. 2004;122:131–9. doi: 10.1016/j.jviromet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Kock N, Kasmieh R, Weissleder R, Shah K. Tumor therapy mediated by lentiviral expression of shBcl-2 and S-TRAIL. Neoplasia. 2007;9:435–42. doi: 10.1593/neo.07223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–46. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 20.Berends MJ, Wu Y, Sijmons RH, et al. Molecular and clinical characteristics of MSH6 variants: an analysis of 25 index carriers of a germline variant. Am J Hum Genet. 2002;70:26–37. doi: 10.1086/337944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gombar CT, Tong WP, Ludlum DB. Mechanism of action of the nitrosoureas--IV. Reactions of bis-chloroethyl nitrosourea and chloroethyl cyclohexyl nitrosourea with deoxyribonucleic acid. Biochem Pharmacol. 1980;29:2639–43. doi: 10.1016/0006-2952(80)90079-9. [DOI] [PubMed] [Google Scholar]
- 22.Goerne R, Bogdahn U, Hau P. Procarbazine--a traditional drug in the treatment of malignant gliomas. Curr Med Chem. 2008;15:1376–87. doi: 10.2174/092986708784567707. [DOI] [PubMed] [Google Scholar]
- 23.Clark AB, Deterding L, Tomer KB, Kunkel TA. Multiple functions for the N-terminal region of Msh6. Nucleic Acids Res. 2007;35:4114–23. doi: 10.1093/nar/gkm409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang G, Scherer SJ, Shell SS, et al. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6:139–50. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y, Berends MJ, Mensink RG, et al. Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am J Hum Genet. 1999;65:1291–8. doi: 10.1086/302612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verma L, Kane MF, Brassett C, et al. Mononucleotide microsatellite instability and germline MSH6 mutation analysis in early onset colorectal cancer. J Med Genet. 1999;36:678–82. [PMC free article] [PubMed] [Google Scholar]
- 27.Martomo SA, Yang WW, Gearhart PJ. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J Exp Med. 2004;200:61–8. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slean MM, Panigrahi GB, Ranum LP, Pearson CE. Mutagenic roles of DNA "repair" proteins in antibody diversity and disease-associated trinucleotide repeat instability. DNA Repair (Amst) 2008;7:1135–54. doi: 10.1016/j.dnarep.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 30.Silber JR, Bobola MS, Ghatan S, Blank A, Kolstoe DD, Berger MS. O6-methylguanine-DNA methyltransferase activity in adult gliomas: relation to patient and tumor characteristics. Cancer Res. 1998;58:1068–73. [PubMed] [Google Scholar]
- 31.Quinn JA, Desjardins A, Weingart J, et al. Phase I trial of temozolomide plus O6-benzylguanine for patients with recurrent or progressive malignant glioma. J Clin Oncol. 2005;23:7178–87. doi: 10.1200/JCO.2005.06.502. [DOI] [PubMed] [Google Scholar]
- 32.Hegi ME, Liu L, Herman JG, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–99. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 33.Liu L, Markowitz S, Gerson SL. Mismatch repair mutations override alkyltransferase in conferring resistance to temozolomide but not to 1,3-bis(2-chloroethyl)nitrosourea. Cancer Res. 1996;56:5375–9. [PubMed] [Google Scholar]
- 34.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 35.Koch D, Hundsberger T, Boor S, Kaina B. Local intracerebral administration of O(6)-benzylguanine combined with systemic chemotherapy with temozolomide of a patient suffering from a recurrent glioblastoma. J Neurooncol. 2006 doi: 10.1007/s11060-006-9244-8. [DOI] [PubMed] [Google Scholar]
- 36.Alvino E, Castiglia D, Caporali S, et al. A single cycle of treatment with temozolomide, alone or combined with O(6)-benzylguanine, induces strong chemoresistance in melanoma cell clones in vitro: role of O(6)-methylguanine-DNA methyltransferase and the mismatch repair system. Int J Oncol. 2006;29:785–97. [PubMed] [Google Scholar]
- 37.Parsons DW, Jones S, Zhang X, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008 doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.