Abstract
Reconstructing the 'Tree of Life' is complicated by extensive horizontal gene transfer between diverse groups of organisms. While numerous conceptual and technical obstacles remain, a report in this issue of Journal of Biology from Koonin and colleagues on the largest-scale prokaryotic genomic reconstruction yet attempted shows that such a tree is discernible, although its branches cannot be traced.
The Tree of Life (ToL) is a widely used metaphor to describe the history of life on Earth. While Darwin argued that the 'Coral of Life' may be a more apt description (since only the surface remains alive, supported by the dead generations beneath it), relationships between organisms based on shared characters are best organized using the schematic representation of a tree. Use of molecular markers, in particular small-subunit ribosomal RNA, have allowed this metaphor to be extended to microorganisms; however, this has also presented unique challenges for notions of phylogeny and evolution. One of the most significant challenges is the impact of horizontal gene transfer, which causes genes that coexist in a genome to have different molecular phylogenies [1]. Despite these challenges, the increasing ease with which genomes can be sequenced has reinvigorated attempts to use genomic information to reconstruct the ToL.
Combining datasets: supertree and supermatrix methods
All microbial individuals arise as the result of a fission of a parent individual. Therefore, a vertical line of descent exists, and could theoretically be reconstructed as a purely bifurcating tree (that is, an organismal or cytoplasmic tree). However, while evolution presupposes and requires descent via reproduction, the two are not analogous. Evolution is, by definition, the change in the genetic material within a population of organisms across generations; therefore, any process by which genetic material within a population changes that is unrelated to the reproduction of individuals will show a history that is unrelated to the organismal vertical line of descent. This includes horizontal gene transfer. In many cases, the sum effect of these other genetic processes may completely obfuscate vertical descent, leaving only some measure of 'relatedness' based on overall genetic similarity.
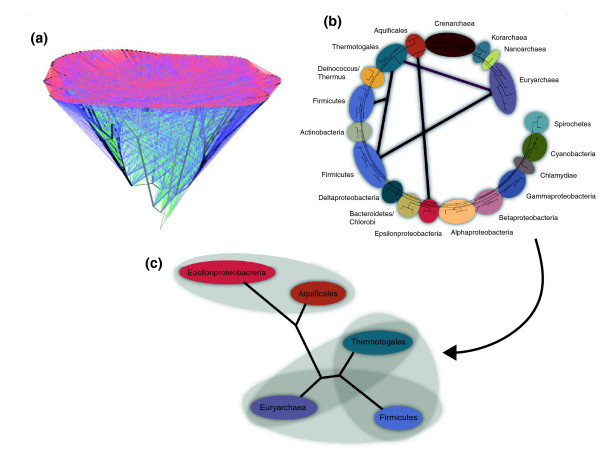
Two common approaches in constructing a genome-based ToL are supermatrix analyses, in which sequence alignments for individual gene families are concatenated into a single dataset that is then used to construct a tree [2], and supertree analyses, in which a consensus phylogeny is constructed from multiple gene trees [3]. In some cases, datasets are generated by finding orthologous genes in all organisms and removing all genes whose conflicting phylogenetic topologies seem to indicate horizontal gene transfer, and then using the remaining genes to reconstruct the presumed vertical lines of descent of the genomes (see, for example, [4-6]). This approach has an obvious shortcoming in that gene transfer and the resulting phylogenetic conflicts can only be inferred if each individual gene has retained sufficient phylogenetic information to enable its origin to be correctly assigned. Furthermore, the absence of evidence for gene transfer does not constitute evidence for the absence of gene transfer. Thus, combining genes with different histories into a single data set will almost certainly result in a phylogeny that represents neither the history of any individual gene, nor the history of the organism as a whole. Another problem with supermatrix and supertree analyses is that they often give equal weight to genes that have different histories of horizontal gene transfer. This results in an average or median phylogeny that may not represent organismal history; if there are 'highways' of gene sharing – that is, large numbers of genes have, for some reason, been shared between specific groups of otherwise phylogenetically distinct organisms – this can easily be mistaken for a consistent signal supporting an organismal tree. For example, because of such highways of gene sharing these types of analyses group members of the order Thermotogales with the Firmicutes, and the members of the Aquificales with the ε-Proteobacteria. In contrast, 16S rRNA gene phylogenies and concatenated ribosomal protein phylogenies strongly support these two orders as deeply branching bacterial lineages [7,8] (Figure 1).
Figure 1.
The Tree of Life as impacted by horizontal gene transfer. (a) Extensive horizontal gene transfer at all phylogenetic levels combine to produce a 'Web of Life' that often obscures the lines of descent between groups (modified from [10]). Copyright (2008) National Academy of Sciences, USA. (b) Major microbial groups as defined by 16S ribosomal RNA phylogeny. Bands represent some avenues of extensive gene sharing involving Thermotogales, Aquificales, and Firmicutes. (c) Impact on relationships between Thermotogales and Aquificales of genome content changes due to extensive horizontal gene transfer. Grey clouds represent groups of shared genes between clades that are non-monophyletic in the 16S tree. The phylogeny based on these 'gene content' clouds is quite distinct from that of 16S or other ribosome-based trees.
Ribosomal trees and the 'genome core'
If stringent criteria are applied to remove or down-weigh transferred genes from supertree or supermatrix analyses, the resulting trees at best represent the history of only a minor fraction of the genome, largely consisting of ribosomal proteins, effectively a 'tree of one percent' [9]. Even if this remaining 'genome core' retains a strong signal of vertical descent, this does not capture the true evolutionary history of genomes; that is, a web where different strands depict the history of different genes. A ribosomal tree of life has other shortcomings, in that within taxonomic orders many recombination and lineage sorting events may occur, and ribosomal genes are so highly conserved that such events at the tips of the tree may not be detectable. However, it can still provide a useful backbone for a reticulated genomic or organismal phylogeny [10,11], especially with respect to sets of genes that clearly have undergone horizontal transfer between more distantly related groups. While ribosomal protein and RNA encoding genes have been transferred in the past (see discussion in [12]), these genes are resistant to transfer [13], with most transfers occurring between close relatives. These properties make a phylogenetic reconstruction using ribosomal RNA and proteins an ideal scaffold upon which to map horizontal gene transfers, clearly depicting their distinct contribution to genomic (and organismal) evolution. Several attempts have been made to capture this web-like genome history (see, for example, [10,11] using ribosomal rRNA as a backbone (Figure 1). Conceptually, this method is distinct from any 'tree of one percent' [9] or genome averaging approach in that rather than being discarded, genes undergoing horizontal transfer are included in the final reconstruction without obscuring the vertical signal, even if that vertical signal is preserved only in a minority of genes.
The Forest of Life
In this issue, Puigbo, Wolf and Koonin [14] present an approach for salvaging the ToL that is a variant on other supertree methods, in which nearly 7,000 phylogenetic trees of prokaryotic genes (a 'Forest of Life') are compared in order to determine a central tendency in their topologies. The trees are built from clusters of orthologous groups of proteins (COGs), and the central tendency is deduced from a set of nearly universal trees (NUTs), defined by Puigbo et al. as those trees generated from a set of COGs that are represented in >90% of the analyzed prokaryote taxa. What distinguishes their approach from earlier supertree analyses – apart from the very large number of genes included in the comparison – is that it does not depend on a concatenation of highly conserved proteins or rRNAs, or on a supertree generated by 'pruning' down to those genes giving a consistent topology, to determine a central tendency. Instead, Puigbo et al. calculate an 'inconsistency score' that is a measure of how representative a particular topology of each tree is to the rest of the trees in the Forest of Life.
In reconstructing the central tendency in such a broad distribution of gene phylogenies, the work by Puigbo et al. also shows the difficulty in resolving deep branches, which often simply collapse into radiations without any topological structure. In confronting this problem, they show that the relationship between phylogenetic depth and resolution supports a tree-like structure for these deep branches. This result is significant in that it suggests that there is no need to postulate exotic 'big bang' radiations early in evolution; rather, deep phylogenies can still be represented as bifurcating evolutionary events, albeit with extremely short branches that can prove difficult (or sometimes impossible) to resolve.
Integrating the vertical descent of organisms and their genomes with the myriad phylogenetic patterns produced by horizontal gene transfer is essential for a truly comprehensive understanding of evolution. A new method that acknowledges and promotes this integration, even if falling short of fully encompassing the intricate details of a complex genome-based biological reality, represents progress towards this goal, and it now appears that a vertical signal can be discerned, if not clearly resolved.
Acknowledgments
Acknowledgements
Work in the authors' lab is supported through the NSF Assembling the Tree of Life (DEB 0830024) and NASA exobiology (NAG5-12367 and NNX07AK15G) programs.
References
- Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR. The evolution of supertrees. Trends Ecol Evol. 2004;19:315–322. doi: 10.1016/j.tree.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Galtier N, Daubin V. Dealing with incongruence in phylogenomic analyses. Philos Trans R Soc Lond B Biol Sci. 2008;363:4023–4029. doi: 10.1098/rstb.2008.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussau B, Gueguen L, Gouy M. Accounting for horizontal gene transfers explains conflicting hypotheses regarding the position of aquificales in the phylogeny of Bacteria. BMC Evol Biol. 2008;8:272. doi: 10.1186/1471-2148-8-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O, Swithers KS, Lapierre P, Fournier GP, Bickhart DM, DeBoy RT, Nelson KE, Nesbø CL, Doolittle WF, Gogarten JP, Noll KM. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc Natl Acad Sci USA. 2009;106:5865–5870. doi: 10.1073/pnas.0901260106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci USA. 2008;105:10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP. The early evolution of cellular life. Trends Ecol Evol. 1995;10:147–151. doi: 10.1016/S0169-5347(00)89024-2. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- Puigbo P, Wolf YI, Koonin EV. Search for a 'Tree of Life' in the thicket of the phylogenetic forest. J Biol. 2009;8:59. doi: 10.1186/jbiol159. [DOI] [PMC free article] [PubMed] [Google Scholar]