Abstract
Using single molecule fluorescence resonance energy transfer, we investigated the interaction between a quadruplex-binding ligand and the human telomeric G-quadruplex. The binding of quinolinecarboxamide macrocycle to telomeric DNA was essentially irreversible and selectively induced and favored one quadruplex conformation. The ligand−quadruplex complex displayed intramolecular dynamics including quadruplex folding and unfolding in the absence of ligand association and dissociation. We report that the G-quadruplex can be stabilized without preventing the intrinsic intramolecular dynamics of telomeric DNA.
G-Quadruplexes are noncanonical structures formed by certain guanine-rich sequences of DNA.(1) The G-quadruplex formed by the human telomeric DNA sequence has been of particular interest, owing to the importance of telomere maintenance for cellular proliferation.(2) Small molecules that bind and stabilize the G-quadruplex can inhibit cell growth via mechanisms that may involve disruption of the telomere and/or the prevention of telomere extension.(3) Consequently, the G-quadruplex formed by telomeric DNA is under investigation as a potential molecular target for anticancer drugs.(4)
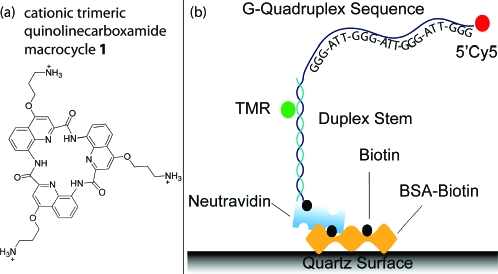
Human telomeric DNA is intrinsically dynamic,(5) and the effect of quadruplex-binding ligands on these dynamics is not known. We studied the effect of quinolinecarboxamide macrocycle 1 (Scheme 1a),(6) a potent quadruplex stabilizing ligand with potential anticancer properties, on the structural dynamics of human telomeric DNA using single molecule fluorescence resonance energy transfer (FRET) spectroscopy.
Scheme 1.
(a) Macrocycle 1. (b) The (GGGTTA)3-GGG oligonucleotide is hybridized to the complementary stem strand and immobilized via a biotin−Neutravidin interaction to a bovine serum albumin-biotin coated quartz surface. TMR (green) and Cy5 (red) are the donor and acceptor fluorophores, respectively.
Single molecule FRET can report on the conformations and dynamics of DNA.(7) A donor (tetramethylrhodamine) and acceptor (Cy5) labeled human telomeric DNA sequence (GGGTTA)3-GGG, h-telo, was tethered to a quartz surface via a biotin−Neutravidin interaction and illuminated using total internal reflection excitation (Scheme 1b).(8) FRET, which is defined as the ratio of acceptor emission to the combined donor and acceptor emission, was calculated for each single molecule. This experimental configuration allowed resolution of the intrinsic structural dynamics between the various conformations adopted by h-telo.5,9
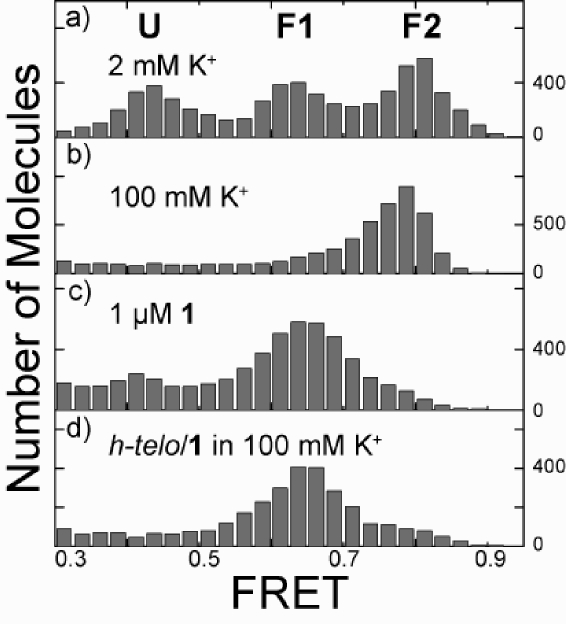
An unfolded h-telo molecule results in the greatest distance between the fluorophores and should display a low FRET value while the decreased distance between the fluorophores in a folded G-quadruplex should result in higher FRET. Monovalent cations, especially potassium, are efficient at inducing and stabilizing folded G-quadruplex structures,(10) and consistent with our previous single molecule FRET studies,(5) unfolded h-telo (U) can be folded into two resolvable conformations, F1 and F2 by 2 mM K+ (FRET efficiency E = 0.43, 0.63, and 0.80 respectively, Figure 1a). At 100 mM K+ concentration, only F2 is observed (Figure 1b). In contrast, at the saturating concentration of 1 (1 μM) and without K+, h-telo is found primarily in F1 (Figure 1c). Therefore, 1 drives the human telomeric DNA to a conformation that is not well populated under physiological K+ concentrations.
Figure 1.
(a) FRET histogram of h-telo with 2 mM K+ showing an unfolded (U) and two distinct intramolecular folded states (F1 and F2). (b) h-telo with 100 mM K+, folded into F2. (c) h-telo/1 (1 μM 1) complex, primarily in F1. (d) h-telo/1 complex in 100 mM K+, 4 h after removal of free 1 (1 μM) (all measurements at 25 °C).
The K+ stabilized h-telo can be completely unfolded in less than 1 min by flushing the sample chamber with a buffer without K+ (data in Supporting Information, SI). In contrast, binding of 1 μM 1 to h-telo was essentially irreversible. After incubating h-telo with 1 μM 1 and obtaining the FRET histogram (Figure 1c), we removed free 1 by repeated flushing of the sample chamber. We imaged h-telo molecules, 4 h after removal of free 1, in a buffer with no 1 or K+ and observed no change in the FRET histogram. We then flowed in 100 mM K+ in an attempt to facilitate the release of 1 and change the h-telo conformation to F2 but did not detect any change (Figure 1d). Though to a lesser extent, the 1 bound to h-telo at lower concentrations (10 and 100 nM 1) was also resistant to immediate removal via buffer exchange (SI).
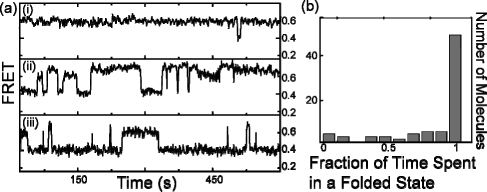
Given the tight binding of 1 to h-telo, we expected each h-telo/1 complex to display a constant FRET value, representing a fixed folded conformation. Instead, we found the h-telo/1 complex (formed by incubating h-telo with 1 μM 1) in a solution with no free 1 to be dynamic (Figure 2a). A histogram of the fraction of time spent folded (Figure 2b, obtained from 10 min time traces of 110 h-telo/1 (1 μM 1) complexes) showed that over 30% of the complexes spent at least 10% of the time in the unfolded conformation. We observed similar dynamics with 100 and 10 nM 1 as well (SI).
Figure 2.
(a) FRET time traces of a h-telo/1 (1 μM 1) complex (with no free 1 in solution) (i) primarily in F1; (ii) transitioning between U, F1, and F2; and (iii) primarily unfolded with transitions to both F1 and F2. (b) A histogram representing the fraction of time spent folded by h-telo/1 complexes.
As the concentration of immobilized h-telo and bound 1 is in the picomolar range, potential rebinding of dissociated 1 in the absence of free 1 in solution is extremely unlikely, and hence, 1 must remain associated with h-telo during the folding and unfolding transitions. That is, h-telo can undergo conformational changes as dramatic as unfolding and refolding without complete dissociation of 1. Although our previous energetics analysis indirectly indicated that a different ligand (hemicyanine-peptide) may allow a partial and transient unraveling of h-telo,(11) our result here demonstrates that h-telo can be completely unfolded for an extended period of time while still being associated with the ligand.
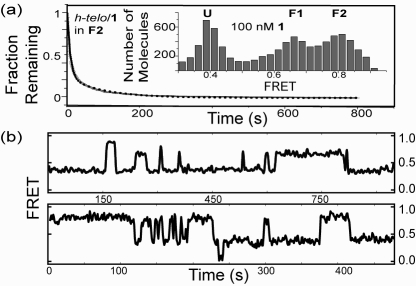
We previously observed six h-telo conformations in the presence of 2 mM K+: long-lived and short-lived species of U, F1, and F2 that differed in their lifetimes by an order of magnitude.(5) To test if the h-telo/1 complex also possessed conformational subspecies, we incubated h-telo with 100 nM 1 (h-telo/1 at this concentration of 1 shows both folded states, Figure 3a inset) and washed away the free ligand. The FRET time traces showed that h-telo/1 complexes can access the same molecular conformations as K+ stabilized h-telo — long- and short-lived U, F1, and F2 (Figure 3b).
Figure 3.
(a) Fraction of h-telo/1 complexes in F2 remaining in F2 after time t. (Inset) h-telo stabilized with 100 nM 1 showing U, F1, and F2. (b) h-telo/1 complexes showing both short (<100 s) and long (>100 s) U, F1, and F2 states in the absence of free 1.
We quantified this by measuring the dwell times of F2 (from 110 individual complexes over 758 transitions) to obtain the survival probability of a complex in F2 remaining in F2 after time t (Figure 3a). Fitting the curve with a double exponential decay function gave decay times of 99.5(±1.3) and 8.5(±0.11) s. These decay times differ by a factor of 12 and show that F2 of the h-telo/1 complex is composed of at least two species with significantly different stabilities. The decay times for F2 stabilized with 2 mM K+ differed by a factor of 9 (188(±2) s and 20(±0.1) s),(5) and the comparable ratio of the F2 decay times for both 1 and K+ stabilized h-telo suggests that K+ and 1 induce h-telo conformational subspecies that are similar in their relative stabilities.
In conclusion, we provide fundamental insights into the dynamic properties of a ligand-stabilized human telomeric DNA G-quadruplex. 1 binds extremely tightly to an unfolded telomeric strand and selectively stabilizes the conformation that is not normally favored under physiological conditions. Without involving full dissociation of the ligand, the ligand-quadruplex complex displays conformational diversity and structural dynamics similar to the G-quadruplex stabilized by potassium. Our observation that 1 promotes G-quadruplex folding without preventing the intrinsic intramolecular dynamics of the telomeric DNA may have implications for this synthetic ligand’s potential to interfere with telomere function in vivo.
Acknowledgments
This work was supported in part by National Institutes of Health Grant GM065367 (to T.H.), Biotechnology and Biological Sciences Research Council (to S.B.), and l’Association de la Recherche contre le Cancer (to K.L.-R.).
Supporting Information Available
Details of the experimental scheme, FRET data analysis method, 1 and K+ removal studies, control experiments with a mutant sequence and time traces of h-telo/1 without free 1 in solution. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Guschlbauer W.; Chantot J. F.; Thiele D. J. Biomol. Struct. Dyn. 1990, 8, 491–511. [DOI] [PubMed] [Google Scholar]
- Blasco M. A. Nat. Rev. Genet. 2005, 6, 611–622. [DOI] [PubMed] [Google Scholar]
- a Gomez D.; Paterski R.; Lemarteleur T.; Shin-ya K.; Mergny J. L.; Riou J. F. J. Biol. Chem. 2004, 279, 41487–41494. [DOI] [PubMed] [Google Scholar]; b Cerone M. A.; Londõo-Vallejo J. A.; Autexier C. Oncogene 2006, 25, 7411–7420. [DOI] [PubMed] [Google Scholar]; c Tera M.; Sohtome Y.; Ishizuka H.; Doi T.; Takagi M.; Shin-ya K.; Nagasawa K. Heterocycles 2006, 69, 505–514. [Google Scholar]; d Dixon I. M.; Lopez F.; Estève J. P.; Tejera A. M.; Blasco M. A.; Pratviel G.; Meunier B. ChemBioChem 2005, 6, 123–132. [DOI] [PubMed] [Google Scholar]; e Minhas G. S.; Pilch D. S.; Kerrigan J. E.; LaVoie E. J.; Rice J. E. Bioorg. Med. Chem. Lett. 2006, 16, 3891–3895. [DOI] [PubMed] [Google Scholar]
- Patel D. J.; Phan A. T.; Kuryavyi V. Nucleic Acids Res. 2007, 35, 7429–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y.; Okumus B.; Kim D. S.; Ha T. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 18938–18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirude P. S.; Gillies E. R.; Ladame S.; Godde F.; Shin-ya K.; Huc I.; Balasubramanian S. J. Am. Chem. Soc. 2007, 129, 11890–11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T. Biochemistry 2004, 43, 4055–4063. [DOI] [PubMed] [Google Scholar]
- Roy R.; Hohng S.; Ha T. Nat. Methods 2008, 5, 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Ja Y. L.; Yoon J.; Hyun W. K.; Kim D. S. Biochemistry 2008, 47, 3389–3396. [DOI] [PubMed] [Google Scholar]; b Ying L.; Green J. J.; Li H.; Klenerman D.; Balasubramanian S. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 14629–14634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A. T.; Patel D. J. J. Am. Chem. Soc. 2003, 125, 15021–15027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. J.; Ladame S.; Ying L.; Klenerman D.; Balasubramanian S. J. Am. Chem. Soc. 2006, 128, 9809–9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.