Abstract
During catalysis, the heme in nitric oxide synthase (NOS) binds NO before releasing it to the environment. Oxidation of the NOS ferrous heme-NO complex by O2 is key for catalytic cycling, but the mechanism is unclear. We utilized stopped-flow methods to study reaction of O2 with ferrous heme-NO complexes of the inducible and neuronal NOS enzymes. We found that the reaction does not involve heme-NO dissociation, but instead proceeds by a rapid, direct reaction of O2 with the ferrous heme-NO complex. This behavior is novel and may distinguish heme-thiolate enzymes like NOS from related heme proteins.
Keywords: Heme protein, heme-thiolate, nitric oxide, redox, enzyme mechanism
Introduction
Nitric oxide (NO) is a signaling and effector molecule in the neural, vascular, and immune systems [1]. Three related NO synthases (NOS, EC 1.14.13.39) generate NO from L-arginine in mammals; inducible NOS (iNOS), endothelial NOS (eNOS), and neuronal NOS (nNOS) [2–5]. NOS-like enzymes also exist in some gram-positive bacteria [6;7]. All NOS are homodimers, and each subunit consists of an N-terminal oxygenase domain that binds iron protoporphyrin IX (heme), 6R-tetrahydrobiopterin (H4B), and L-arginine (L-Arg) and a C-terminal reductase domain that binds FMN, FAD, and NADPH. The two domains are connected to one another by an intervening calmodulin binding sequence [2].
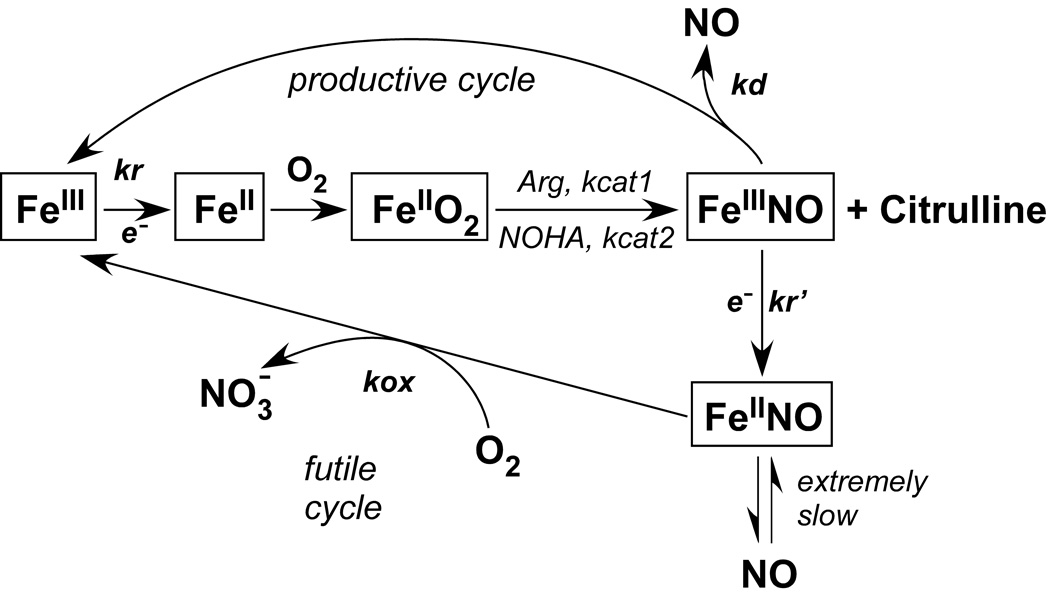
An interesting feature of NOS catalysis is that the newly-synthesized NO binds to the heme prior to release from the enzyme [8;9]. Therefore, the end product of the reaction is not NO but heme FeIII-NO. During catalysis, this product complex can become reduced by the NOS reductase domain to heme FeII-NO at a rate comparable to that of FeIII-NO dissociation. This leads to two possible fates for each new NO molecule: release from the heme FeIII-NO complex into solution (productive cycle) or oxidation through the reaction of the heme FeII-NO complex with O2 (futile cycle). We have proposed a global mechanism for NOS catalysis that takes these facets into account (Fig. 1) [10;11]. Kinetic measurements of the individual steps revealed that the catalytic behavior of any NOS is primarily characterized by the interplay of three kinetic parameters; namely kr (the rate of FeIII (or FeIII-NO)-heme reduction by the reductase domain), kd (the dissociation rate of NO from the heme FeIII-NO complex) and kox (the oxidation rate of the heme FeII-NO complex) [3;10;11] (Fig. 1). Computer simulations of the global mechanism, and characterization of various eNOS and nNOS mutants that have altered kinetic parameters, has shown that the NO synthesis activity and apparent KMO2 of a NOS enzyme can be significantly dependent on the kox rates [12].
Figure 1.
Despite the importance of kox, the reaction of the NOS heme FeII-NO complex with O2 has not been the object of specific studies, and kox rates have only been determined for NOS enzymes at 140 µM oxygen (half air-saturated conditions). Here, we utilized stopped-flow spectroscopy to study the oxidation reaction of the iNOS and nNOS heme FeII-NO complexes at different O2 concentrations. Our results show: (i) NOS heme FeII-NO complexes oxidize at much higher rates than those reported for any other heme protein, (ii) the NOS oxidation mechanism does not involve NO dissociation from the heme, and instead involves a direct reaction of O2 with heme-bound NO. These novel facets of heme-NO reactivity fundamentally distinguish heme-thiolate enzymes like NOS from hemoglobins and related enzymes.
Results
Heme FeII-NO Dissociation rates
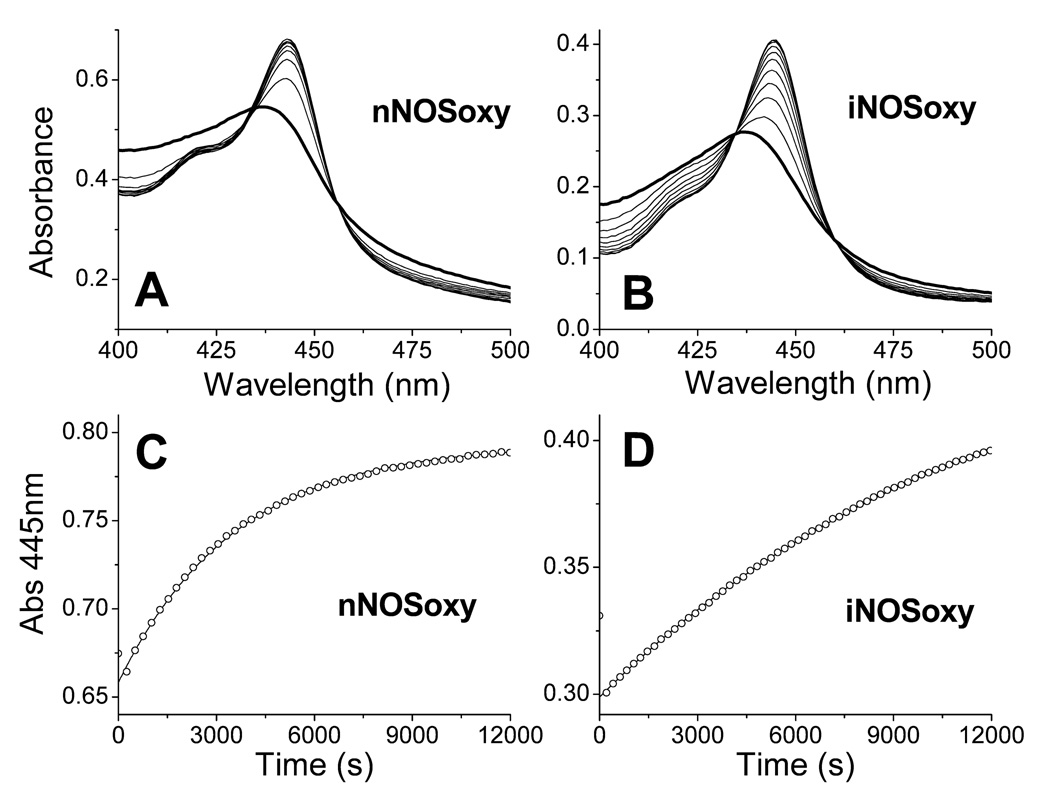
We studied NO dissociation from pre-formed ferrous heme-NO complexes of iNOSoxy and nNOSoxy, using sodium dithionite as an NO scavenger in the presence of CO. When NO dissociates from the NOS heme, CO binds quickly [20–23] to form a heme FeII-CO complex, which can be followed by a shift in the heme Soret absorbance from 436 nm to 445 nm. Spectral analysis of the NO dissociation reactions for nNOSoxy and iNOSoxy are shown in Figure 2. In both cases the absorbance increase at 445 nm (Figures 2C–D) fit well to a single exponential equation and the transitions show isosbestic points consistent with a single step process (Figures 2A–B). The NO dissociation constants obtained were 3.9 ± 0.6 × 10−4 s−1 (nNOSoxy) and 1.0 ± 0.1 × 10−4 s−1 (iNOSoxy), similar to previously reported values of 3.1 × 10−4 s−1 and 1.35 × 10−4 s−1 for nNOS [16] and iNOS [11], respectively. Because the NO dissociation rates are 3 to 4 orders of magnitude slower than the kox values reported for iNOSoxy and nNOSoxy at half-air saturation, (kox of 3.0 s−1 and 0.19 s−1 for iNOS and nNOS, respectively) [11;12] we conclude that an NO dissociation step is not involved in the oxidation mechanism of NOS heme FeII-NO complexes.
Figure 2.
Oxygen dependence of kox and possible buildup of reaction intermediates
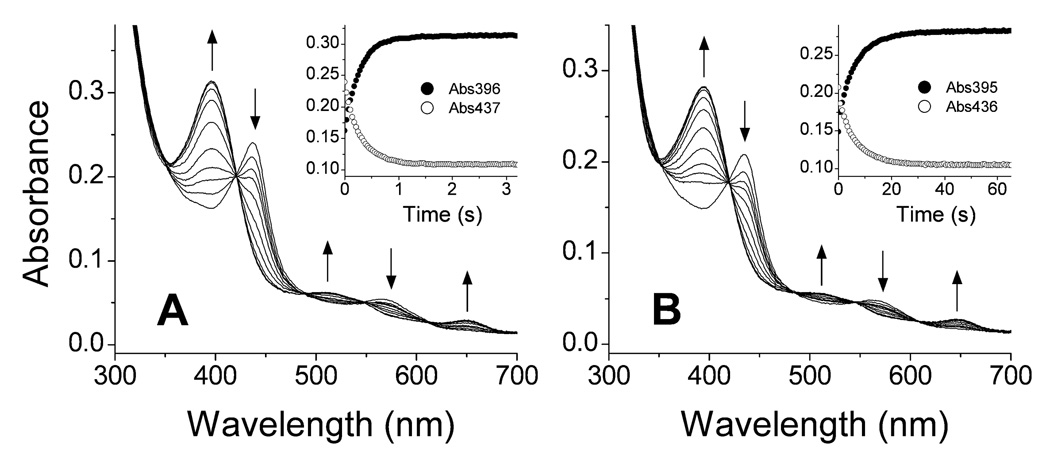
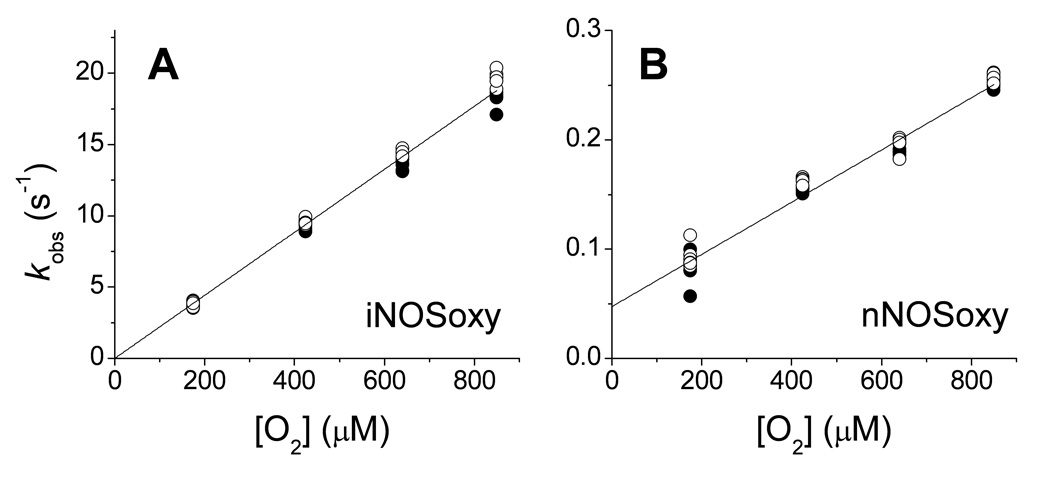
We utilized stopped-flow spectroscopy to study the oxidation reactions of pre-formed iNOSoxy and nNOSoxy heme FeII-NO complexes at different oxygen concentrations, to determine the oxygen dependence of kox and to observe if any heme-based reaction intermediates would build up. Figure 3A contains representative spectral data collected during reaction of the iNOSoxy FeII-NO complex with 170 µM oxygen in the presence of both H4B and L-Arg. The spectral changes are consistent with conversion of the FeII-NO complex into the FeIII high-spin form of iNOSoxy. The reaction here was well-described as a single step process, as indicated by the several isosbestic points in the spectra (Fig. 3A) and by the single exponential decay of the absorbance signal at different wavelengths (Fig. 3A, inset). The rate of FeII-NO complex disappearance (437 nm) was similar to the rate of ferric enzyme product formation (396 nm) (Fig. 3A, inset). The spectral and fitting results we obtained for replica reactions run at other O2 concentrations were highly similar. In all cases, the reactions proceeded as single step processes with no apparent buildup of enzyme reaction intermediates. The plot of the observed rates (kox values) versus oxygen concentration shows a linear dependency across the entire range of experimental O2 concentrations (Figure 4A), and did not display saturation kinetic behavior. From the plot, rate constants for the bimolecular reaction (k1 and k−1, see methods) were obtained. There was no substantial change in the accuracy of fit when the k−1 term was not included, and therefore we assume that it is too small to be accurately determined by our method. A k1 value of 26500 ± 340 M−1s−1 was calculated for the reaction of the iNOSoxy heme FeII-NO complex with O2 (Table 1). We used the same procedure to analyze the reaction of the nNOSoxy heme FeII-NO complex with O2. Fig. 3B contains representative spectra recorded for reaction of the complex with 170 µM O2 in the presence of both L-Arg and H4B. The overlapped spectral traces create isosbestic points at approximately the same wavelengths as we observed in the iNOSoxy reactions. The reactions of nNOSoxy were also single exponential and showed no detectable buildup of enzyme intermediates in any case. When the observed rates were plotted versus O2 concentration a linear dependency was observed (Fig. 4B) with no saturation across the entire experimental O2 concentration range. The calculated rate constants were k1 = 230 ± 8 M−1s−1 and k−1 = 0.047 ± 0.004 s−1 (Table 1). Thus, reaction of the nNOSoxy heme FeII-NO complex with O2 exhibited similar behavior to that of the iNOSoxy heme FeII-NO complex, but was about 100 times slower at any given O2 concentration (compare Fig. 3B (inset) and Fig. 4B).
Figure 3.
Figure 4.
Table 1.
Rates of Ferrous Heme-NO dissociation and oxidation for NOSoxy and other heme proteins.
| Protein | NO dissociation |
NO oxidation a | References | |
|---|---|---|---|---|
| iNOSoxy | 1.0 × 10−4 s−1 | k1 = 2.65 × 104 M−1s−1 | Present study | |
| nNOSoxy | 3.9 × 10−4 s−1 | k1 = 2.30 × 102 M−11s−1 | k−1 = 4.7 × 10−2 s−1 | Present study |
| Neuroglobin | 2.0 × 10−4 s−1 | k1 = 1.6 × 101 M−1s−1 | k2 = 5.0 × 10−4 s−1 | [36;56] |
| Hemopexin | 9.1 × 10−4 s−1 | k1 = 2.4× 101 M−1s−1 | k2 = 1.4 × 10−3 s−1 | [35] |
| Myoglobin | 0.9 × 10−5 s−1 | k1 = 1.3 × 10−4 s−1 | k2 = 2.6 × 10−4 s−1 | [26;57] |
| Hemoglobin | 3.2 × 10−4 s−1 b | k1 = 2.0 × 10−4 s−1 | k2 = 1.0 × 10−4 s−1 | [27] |
| Guanylate cyclase | 7.0 × 10−4 s−1 | [57] | ||
| Cytochrome c oxidase | 1.0 × 10−2 s−1 | [58] | ||
| Cytochrome cd1 | 4.35× 101 s−1 | [44] | ||
The second kinetic parameters are referred to as k−1 and k2 because in the case of nNOS it reflects an apparent equilibrium and in the other proteins refers to a subsequent reaction.
This is the fast rate; bi-exponential fit gave values of: k1=3.2 × 10−4 s−1; k2 = 0.7 × 10−4 s−1 [27].
Nitrate and nitrite product formation
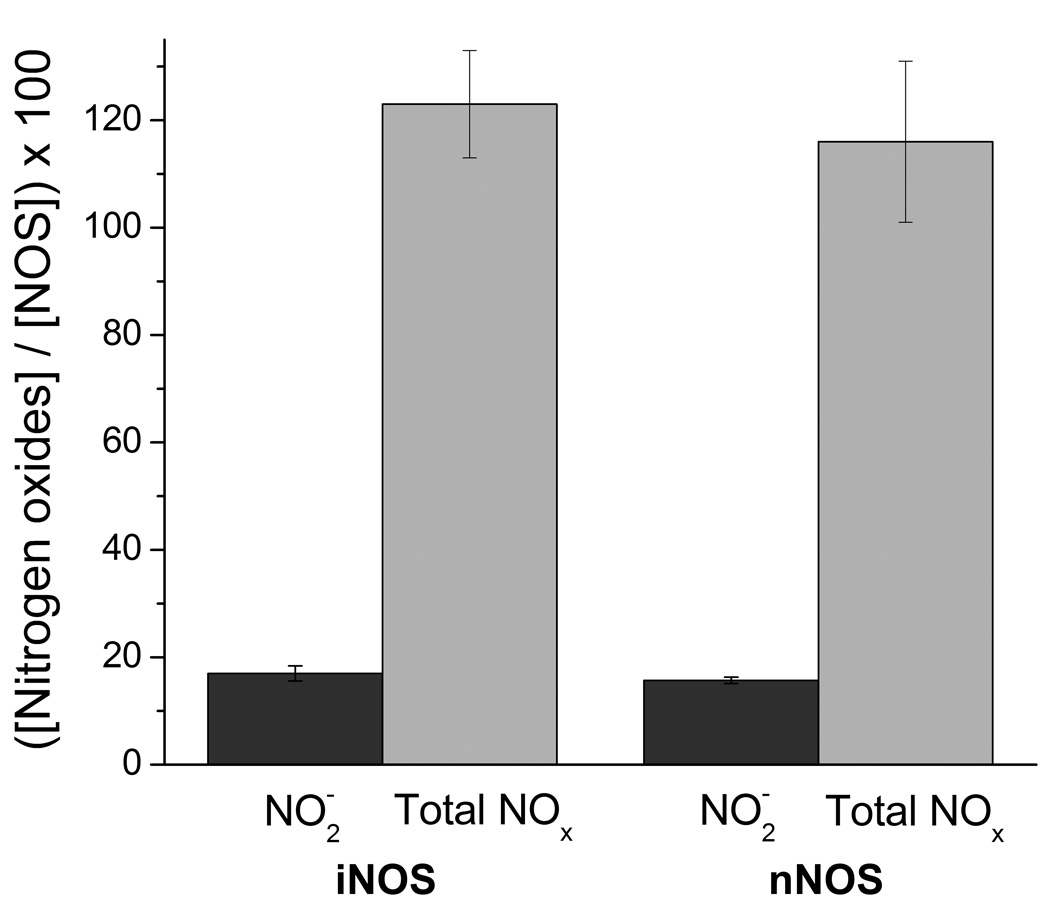
To quantify product formation, the iNOSoxy FeII-NO complex was formed and purified under anaerobic conditions (see Methods). Aliquots were mixed with air saturated buffer and the amounts of NO2 − and total NO2 − + NO3 − were quantified by chemiluminescence and photometric assays (see methods). Percent values were calculated dividing the concentration of the nitrite/nitrate species by the initial concentration of NOS in each sample and multiplying by 100 the resulting value. Thus, a 100% value is equivalent to 1 mole of nitrite/nitrate produced per mole of enzyme. The results are shown in Figure 5. The detected amounts of nitrite or total nitrite plus nitrate were: NO2 − = 17.0 ± 1.4 %; Total NO2 −/NO3 − = 123 ± 10 % (iNOS) and NO2 − 15.7 ± 0.6 %; Total NO2 −/NO3 − = 116 ± 15% (nNOS). Thus, the results with both enzymes were similar with approximately 1 mole of nitrate and 0.2 moles of nitrite produced per mole of FeII-NO complex. Some possible sources of the small amount of NO2 − can be the reaction of excess NO with O2 or reduction of NO3 − by excess dithionite. On the other hand, NO3 − can only be formed in the enzymatic reaction via the futile cycle (see Fig. 1). We conclude that the reaction of the NOS ferrous-NO complexes with oxygen leads quantitatively to nitrate.
Figure 5.
Discussion
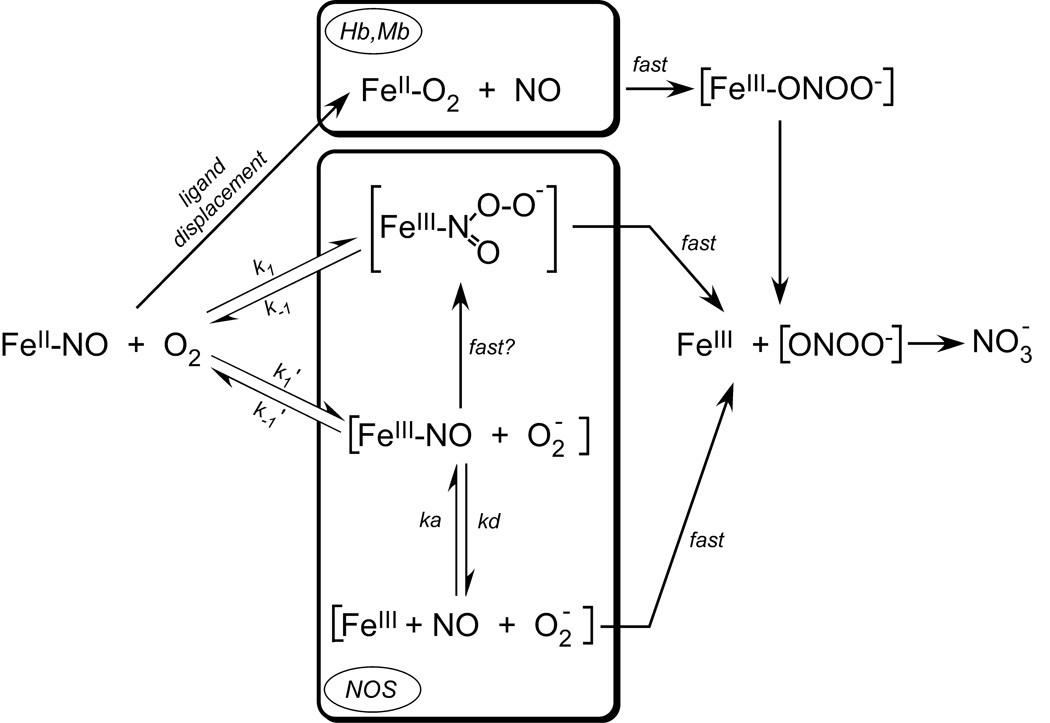
The reaction of O2 with the ferrous heme-NO complexes of hemoglobin and myoglobin have been closely studied and serve as a paradigm for our present study [24–26]. A central hallmark of these reactions is that they are relatively slow and proceed at rates that basically match their rates of heme FeII-NO dissociation (Table 1, [26]). This has led to a generally accepted mechanism illustrated in Fig. 6 (top portion), where NO dissociation from the ferrous heme is the initial and rate-limiting step for the overall oxidation. In these cases, NO dissociation allows O2 to bind to the ferrous heme and form a heme FeII-O2 complex, which then reacts rapidly with NO. This reaction appears to take place inside the heme pocket [26;27], as indicated by the rate being pH-independent and mono-molecular, and no nitrite being formed as product, which could otherwise result if any NO was released from the protein into solution.
Figure 6.
For NOS enzymes, the mechanism of heme FeII-NO oxidation by O2 does not involve an NO dissociation step, because the observed rates of heme FeIINO dissociation are two or three orders of magnitude slower than the observed rates of heme FeIINO oxidation. Our results suggest two alternative reaction mechanisms that are illustrated in Fig. 6. Both have O2 reacting directly with the NOS heme FeII-NO complex, in one case as an electrophile that attacks the nitrogen of the bound NO (k1/k−1) and in the other case as an oxidant in an outer-sphere electron transfer reaction (k1’/k−1’) that generates heme FeIII-NO and superoxide. In either case, the initial O2-dependent reaction appears to be the rate-limiting step, and conceivably both reactions would generate an N-bound heme-peroxynitrite complex as an immediate product (Fig. 6). Because we observed monophasic transitions for our iNOSoxy and nNOSoxy reactions with no apparent build-up of a heme-peroxynitrite product complex, this suggests that the immediate heme-product complex may dissociate (or rearrange) relatively quickly to form the observed ferric enzyme product (Fig. 6). This is consistent with the reaction rates not saturating even at the highest O2 concentrations that were experimentally available to us. Interestingly, buildup of an observable oxo-ferryl heme intermediate does occur when excess peroxynitrite is reacted with ferric iNOSoxy [28]. However, this reaction involves the initial formation of an O-linked heme-peroxynitrite complex, which is not likely to form in our reactions.
The outer-sphere electron transfer mechanism depicted in Fig. 6 (k1’/k−1’) is likely to be thermodynamically uphill based on a comparison of the midpoint potentials for the O2/O2 − couple (−330 mV) [29] versus the NOS FeIII-NO/FeII-NO couple (−2 mV) [30]. However, the equilibrium could be driven toward products if the subsequent reaction of superoxide and the heme FeIII-NO complex is fast. This pathway would form the same N-bound heme-peroxynitrite intermediate as in the mechanism proposed above. If an outer-sphere electron transfer occurs, it opens up the possibility that NO could dissociate from the heme FeIII-NO complex prior to reaction with superoxide (Fig. 6). However, the measured rates of NOS heme FeIII-NO dissociation (kd) range from 2 to 60 s−1 at similar temperatures [8;11;31]. Therefore, the probability of NO escape from the enzyme would depend on the initial O2 reaction proceeding by an outer-sphere electron transfer, and the resulting superoxide exhibiting a poor reactivity toward the heme FeIII-NO complex. We believe this combination of events is unlikely. Instead, the heme FeII-NO complex of iNOSoxy and nNOSoxy appears to undergo an efficient NO dioxygenase reaction to generate nitrate with little or no NO escaping from the enzyme during the reaction.
What makes the heme FeII-NO complexes of iNOSoxy and nNOSoxy more reactive toward O2 than the heme FeII-NO complexes of Hb or Mb? A difference in O2 access is not likely to be involved, because each of these heme proteins evolved to bind O2, and their ferrous forms all have fairly similar kon values for O2 and for other diatomic ligands [13;30;32]. Instead, the difference may be caused in part by the heme-thiolate bond causing the NOS heme FeIINO complex to have a lower midpoint potential. The midpoint potential of the FeIIINO/FeIINO couple in NOS (−2 mV) [30] is about 50 to 150 mV more negative than the same couple in histidine-ligated heme proteins [33]. Resonance Raman data are consistent with greater electron density in the NOS heme-NO complexes compared to globins, and suggest the bound NO exhibits some nitroxide-like character, which should increase its reactivity toward O2 if the mechanisms proposed in Fig. 6 are accurate. Conceivably, one might further increase the oxidation rate of a heme FeII-NO complex by O2 by further decreasing the heme midpoint potential. This effect may have been achieved for the W409F mutant of nNOS, whose more negative FeIII/FeII midpoint potential is associated with a 7-times faster kox than in wild-type nNOS [12]. In general, different heme proximal ligands (cysteine versus histidine) or related protein modifications near the heme may enable a range of heme midpoint potentials. At one extreme (more positive potential), this may diminish the O2 reactivity of a protein’s heme FeII-NO complex to such an extent that consecutive NO dissociation and O2 substitution steps dominate the oxidation mechanism (as for Hb and Mb). At the other extreme (more negative potential), it may increase the O2 reactivity of a protein’s heme FeII-NO complex enough to enable a direct and potentially faster oxidation reaction (as in NOS enzymes). Layered onto this “redox” regulation are likely to be effects from other aspects of protein structure that together will determine the mechanism and kinetics of heme FeII-NO oxidation for any given protein.
Recent evidence suggests that histidine-ligated heme proteins may exhibit a range of behaviors regarding the O2 oxidation of their heme FeII-NO complexes [34]. For example, the hexacoordinate heme proteins neuroglobin and hemopexin show interesting differences with the aforementioned Hb and Mb. The reactions of hemopexin or neuroglobin heme FeII-NO complexes with O2 proceed relatively fast and they form what appear to be detectable heme-peroxynitrite intermediates that have relatively slow conversion rates to ferric enzyme [34–36]. The reported rates are compared in Table 1. Interestingly, the heme FeIII/FeII midpoint potentials of hemopexin and neuroglobin are more negative than those of Hb and Mb, consistent with the concept of redox regulation as described above. Flavohemoglobins are another type of histidine-ligated heme protein that catalyze an NO dioxygenase reaction to form nitrate [37;38]. Their reaction is extremely fast with a kcat of 112–670 s−1 at 37°C [39;40]. The bulk of evidence suggests the their mechanism involves ligand substitution on the ferrous heme (O2 for NO) as described for reactions of the heme FeII-NO complexes of Mb and Hb in Fig. 6 [37;41–43]. This would imply an unusually fast NO dissociation from the flavohemoglobin heme FeII-NO complex. Recently, fast NO dissociation rates were observed (43.5 s−1 at 20 °C) [44] for the heme FeII-NO complex of cytochrome cd1, which appears to explain its fast steady-state activity [44;45]. Apparently, broad variations in NO kd and/or in heme midpoint potential may help to determine the mechanism and speed by which various heme protein FeII-NO complexes are oxidized by O2. These concepts merit further investigation.
Concluding remarks
NOS enzymes catalyze a more rapid oxidation of their heme FeII-NO complex than any other heme-containing protein that has been studied to date. The basis is likely related to their heme environment, and particularly to the characteristics of the heme-thiolate bond. Further studies are under way to test the importance of heme midpoint potenial, heme distortion, NOS quaternary structure (dimer versus monomer), heme pocket access, active site residues, and the influence of bound cofactor or substrate. Because NOS form a heme FeII-NO complex during their normal catalytic cycle it may have forced them to develop a way to efficiently oxidize these complexes, which otherwise can constitute a dead-end in catalysis. Other heme-thiolate proteins appear to show differences in their NO reactivity [46], and many cytochrome P450’s are inactivated by NO [47;48]. It will be interesting to see if the reaction described here for NOS is applicable to other heme-thiolate proteins.
There is a remarkable difference between iNOSoxy and nNOSoxy in their rates of their heme FeII-NO oxidation. The fast oxidation of the iNOS heme FeII-NO complex could be designed to synthesize peroxynitrite. This would be in accord with a physiological role of iNOS in host defense, and might help to explain why protein nitration is observed in some cells upon iNOS expression [28;49;50]. In this context, it is interesting to note that the bacterial NOS-like enzymes from Deinococcus radiodurans [51] and Streptomyces turgidiscabies [52] have both been found to catalyze amino-acid nitration. In comparison, the relatively slower oxidation of the nNOS heme FeII-NO complex has been suggested to enable its function in biological O2 sensing [53;54] or to serve as a pulse source of NO in signal transduction cascades [55]. Our current work provides a foundation to test these possibilities.
Materials and methods
Reagents
O2 and N2 (medipure grade) gases were obtained from Praxair. NO gas was purchased from Linde LLC. H4B was purchased from Schircks Laboratories (Jona, Switzerland). Glacial acetic acid was purchased from Mallinckrodt Baker. Other chemicals were obtained from either Fisher Scientific (Pittsburgh, PA) or Sigma (St. Louis, MO).
Protein purification
The oxygenase domains of rat nNOS and mouse iNOS were overexpressed in E. coli and purified as previously described [13;14]. Protein concentration was determined from the absorbance at 444 nm of the ferrous-CO complex using an extinction coefficient of 74 mM−1cm−1 [15].
Ferrous heme-NO dissociation rates
The dissociation rates of NO from ferrous heme-NO complexes were studied in the presence of CO and using sodium dithionite as an NO scavenger as described [11;16]. The rate for the reaction of dithionite with NO is 1.4 × 103 M−1s−1 at 20 °C in 50 mM phosphate buffer at pH 7.0 [17]. Although oxyhemoglobin is a more efficient NO scavenger, dithionite has been successfully used when NO dissociation is a slow process, such as for ferrous-NO complexes of hemoglobin or NOS [11;16;17]. Solutions of iNOSoxy or nNOSoxy (40–80 µM) were made anaerobic in the presence of 2.5 mM L-Arg, 20 µM H4B, 1 mM DTT in EPPS 40 mM pH 7.6 buffer were made anaerobic and then the ferrous heme-NO complexes were formed by adding successively sodium dithionite (final concentration 1.5 mM) and NO from a saturated NO-solution in EPPS buffer (final concentration 200 µM, NO saturated buffer concentration is 2.05 mM at 20 °C). The reaction was studied at 10 °C and was started by adding 100 µl of ferrous heme-NO solution to 900 µl of CO-saturated EPPS 40 mM, pH 7.6 buffer containing 2.5 mM L-Arg, 20 µM H4B, 1 mM DTT and 1.5 mM dithionite. The conversion of the heme FeII-NO complex to the FeII-CO complex was monitored by UV-Visible spectroscopy and changes at 445 nm were fit to single exponential equations using Origin Pro 7.5 software (OriginLab, Northampton, MA).
Ferrous heme-NO oxidation
Ferrous heme-NO complexes were prepared as follows: protein solutions containing 10 µM of ferric NOS were prepared in 40 mM EPPS buffer, pH 7.6, 10% Glycerol, 150 mM NaCl, 0.5 mM EDTA, 20 µM H4B and 2.5 mM L-Arg. Samples were made anaerobic in sealed cuvettes by several cycles of nitrogen/vacuum. Then, FeIII-heme was titrated with sodium dithionite to produce the ferrous enzyme. Reduction of the enzyme was monitored in either a Cary 100 or a Shimadzu UV-2401 PC spectrophotometer. Ferrous enzyme was then titrated with a NO-saturated buffer solution. Each FeII-NO complex was then transferred to a syringe housed in a temperature-controlled Hi-Tech SF-61 stopped-flow instrument (Hi-Tech Scientific, Salisbury, UK) equipped with a diode array detector. The NOSoxy FeII-NO complex was rapidly mixed with O2-containing buffer with the same composition as the protein buffer. Reactions were carried out at 10 °C. To assess the oxygen-dependency of the reaction, variable concentrations of oxygen were produced by mixing different amounts of O2-saturated and N2-saturated buffers. The concentration of oxygen in the O2 saturated buffer at 10°C was calculated to be 1.7 mM [18]. As the mixing ratio was 1:1 the final O2 concentration achieved varied from 0 to 850 µM.
Kinetic analysis
Reaction rates were calculated from the absorbance changes at the wavelength of the Soret peaks of FeIII-heme (≈ 396nm, ferric heme recovery) or heme FeII-NO (≈ 436nm, ferrous heme-NO decay). Data were fitted to a single-exponential equation. The oxygen dependency of the observed rates versus O2 concentration was fit to a linear equation in the form: kobs = k1 [O2] + k−1, were k1 and k−1 are the apparent rates of O2 association and dissociation, respectively. For iNOSoxy the value of k−1 is too small to be determined accurately and a linear fit through zero in the form kobs = k1 [O2] was used. Data analysis was carried out using Origin Pro 7.5 software (OriginLab, Northampton, MA).
Determination of end products
iNOSoxy and nNOSoxy samples (approx 80 µM) were prepared in 40 mM EPPS buffer, pH 7.6, 10% Glycerol, 150 mM NaCl, 0.5 mM EDTA, 200 µM H4B and 2.5 mM L-Arg. The ferrous-NO complex was formed by successive titrations with sodium dithionite (to form ferrous enzyme) and NO-saturated buffer as described above. In order to minimize generation or carryover of oxidized nitrogen species from the excess dithionite and NO, the cuvettes were next transferred to a glove-box (Belle Technology, Portesham, Dorset, UK) that was kept below 10 ppm O2 and the ferrous-NO proteins were run through a Sephadex G-25 column (PD-10, GE Healthcare) equilibrated in anaerobic EPPS buffer 40 mM, pH 7.6, 10% Glycerol, 150 mM NaCl. The eluted fractions were transferred again to an anaerobic cuvette to determine their final concentrations (20–30 µM) and to insure that NO had remained bound to ferrous heme. Then the ferrous-NO complexes were mixed with different amounts of air-saturated buffer (final protein concentrations 5–25 µM). Nitrite and total nitrite plus nitrate (NOx) concentrations in the samples were measured using chemiluminescence as previously described [19]. Before measurement, samples were pretreated with 10% w/v ZnSO4 and 0.5 N NaOH and centrifuged to remove protein. Nitrite was converted to NO in line during the measurement by a solution containing an excess of potassium iodide in glacial acetic acid. Total NOx was converted to NO by a saturated solution of VCl3 in 1 M HCl. NO generated was detected by the Sievers NOA 280i (GE Analytical Instruments, Boulder, CO). All samples were measured at least in triplicate and the nitrite and NOx concentrations were determined by interpolation using authentic standards of nitrite and nitrate, respectively. Additional measurements of nitrite and total nitrite and nitrate (NOx) concentrations were carried out photometrically using the Griess assay on the same samples. The protein was removed from the samples by centrifugation through Amicon Ultra centrifugal devices with a 10 kDa MW cut-off (Millipore). In this case, to estimate the total amount of nitrite plus nitrate the samples were reduced by a cadmium-copper catalyst (Nitralyzer II, World Precision Instruments, Sarasota, FL, USA) and assayed following the procedure provided by the supplier.
Acknowledgements
We would like to thank Allison Janocha and Serpil Erzurum for their assistance on the chemiluminescence assays. This work was supported by National Institutes of Health Grants CA53914, GM51491, and HL76491 to D.J.S. J.T. is supported by a postdoctoral fellowship (0625632B) from the American Heart Association.
Abbreviations
- EDTA
ethylene diamine tetraacetic acid
- EPPS
4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
- FeII-NO
ferrous heme-NO complex
- FeIII-NO
ferric heme-NO complex
- FAD
flavin dinucleotide
- FMN
flavin mononucleotide
- H4B
(6R)-5,6,7,8-tetrahydro-L-biopterin
- kr
reduction rate of the heme by the reductase (flavoprotein) domain of NOS
- kd
dissociation rate of NO from the ferric heme-NO complex of NOS
- kox
oxidation rate of the ferrous heme-NO complex of NOS
- NADPH
nicotinamide adenine dinucleotide phosphate, reduced form
- NOS
nitric oxide synthase
- nNOS
neuronal nitric-oxide synthase
- nNOSoxy
oxygenase domain of the neuronal nitric-oxide synthase
- iNOS
inducible nitric-oxide synthase
- iNOSoxy
oxygenase domain of the inducible nitric-oxide synthase
- WT
wild-type.
References
- 1.Pfeiffer S, Mayer B, Hemmens B. Nitric Oxide: Chemical Puzzles Posed by a Biological Messenger. Angew. Chem. Int. Ed. 1999;38:1714–1731. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1714::AID-ANIE1714>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. Biochem. J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stuehr DJ, Santolini J, Wang ZQ, Wei CC, Adak S. Update on mechanism and catalytic regulation in the NO synthases. J. Biol. Chem. 2004;279:36167–36170. doi: 10.1074/jbc.R400017200. [DOI] [PubMed] [Google Scholar]
- 4.Gorren ACF, Mayer B. Nitric-oxide synthase: A cytochrome P450 family foster child. Biochimica et Biophysica Acta-General Subjects. 2007;1770:432–445. doi: 10.1016/j.bbagen.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 5.Stuehr DJ, Kwon NS, Nathan CF, Griffith OW, Feldman PL, Wiseman J. N omega-hydroxy-L-arginine is an intermediate in the biosynthesis of nitric oxide from L-arginine. J. Biol. Chem. 1991;266:6259–6263. [PubMed] [Google Scholar]
- 6.Shatalin K, Gusarov I, Avetissova E, Shatalina Y, McQuade LE, Lippard SJ, Nudler E. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1009–1013. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crane BR. The enzymology of nitric oxide in bacterial pathogenesis and resistance. Biochem. Soc. Trans. 2008;36:1149–1154. doi: 10.1042/BST0361149. [DOI] [PubMed] [Google Scholar]
- 8.Negrerie M, Berka V, Vos MH, Liebl U, Lambry JC, Tsai AL, Martin JL. Geminate recombination of nitric oxide to endothelial nitric-oxide synthase and mechanistic implications. J. Biol. Chem. 1999;274:24694–24702. doi: 10.1074/jbc.274.35.24694. [DOI] [PubMed] [Google Scholar]
- 9.Abu-Soud HM, Wang J, Rousseau DL, Fukuto JM, Ignarro LJ, Stuehr DJ. Neuronal nitric oxide synthase self-inactivates by forming a ferrous-nitrosyl complex during aerobic catalysis. J. Biol. Chem. 1995;270:22997–23006. doi: 10.1074/jbc.270.39.22997. [DOI] [PubMed] [Google Scholar]
- 10.Santolini J, Adak S, Curran CM, Stuehr DJ. A kinetic simulation model that describes catalysis and regulation in nitric-oxide synthase. J. Biol. Chem. 2001;276:1233–1243. doi: 10.1074/jbc.M006858200. [DOI] [PubMed] [Google Scholar]
- 11.Santolini J, Meade AL, Stuehr DJ. Differences in three kinetic parameters underpin the unique catalytic profiles of nitric-oxide synthases I, II, and III. J. Biol. Chem. 2001;276:48887–48898. doi: 10.1074/jbc.M108666200. [DOI] [PubMed] [Google Scholar]
- 12.Adak S, Wang Q, Stuehr DJ. Molecular basis for hyperactivity in tryptophan 409 mutants of neuronal NO synthase. J. Biol. Chem. 2000;275:17434–17439. doi: 10.1074/jbc.M000846200. [DOI] [PubMed] [Google Scholar]
- 13.Abu-Soud HM, Gachhui R, Raushel FM, Stuehr DJ. The ferrous-dioxy complex of neuronal nitric oxide synthase. Divergent effects of L-arginine and tetrahydrobiopterin on its stability. J. Biol. Chem. 1997;272:17349–17353. doi: 10.1074/jbc.272.28.17349. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh DK, Wu C, Pitters E, Moloney M, Werner ER, Mayer B, Stuehr DJ. Characterization of the inducible nitric oxide synthase oxygenase domain identifies a 49 amino acid segment required for subunit dimerization and tetrahydrobiopterin interaction. Biochemistry. 1997;36:10609–10619. doi: 10.1021/bi9702290. [DOI] [PubMed] [Google Scholar]
- 15.Stuehr DJ, Ikeda-Saito M. Spectral characterization of brain and macrophage nitric oxide synthases. Cytochrome P-450-like hemeproteins that contain a flavin semiquinone radical. J. Biol. Chem. 1992;267:20547–20550. [PubMed] [Google Scholar]
- 16.Scheele JS, Bruner E, Kharitonov VG, Martasek P, Roman LJ, Masters BS, Sharma VS, Magde D. Kinetics of NO ligation with nitric-oxide synthase by flash photolysis and stopped-flow spectrophotometry. J. Biol. Chem. 1999;274:13105–13110. doi: 10.1074/jbc.274.19.13105. [DOI] [PubMed] [Google Scholar]
- 17.Moore EG, Gibson QH. Cooperativity in the dissociation of nitric oxide from hemoglobin. J. Biol. Chem. 1976;251:2788–2794. [PubMed] [Google Scholar]
- 18.Battino R. IUPAC Solubility Data Series, Vol.7, Oxygen and Ozone. Oxford, England: Pergamon Press; 1981. [Google Scholar]
- 19.Dweik RA, Laskowski D, bu-Soud HM, Kaneko F, Hutte R, Stuehr DJ, Erzurum SC. Nitric oxide synthesis in the lung. Regulation by oxygen through a kinetic mechanism. J. Clin. Invest. 1998;101:660–666. doi: 10.1172/JCI1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevenson TH, Gutierrez AF, Alderton WK, Lian L, Scrutton NS. Kinetics of CO binding to the haem domain of murine inducible nitric oxide synthase: differential effects of haem domain ligands. Biochem. J. 2001;358:201–208. doi: 10.1042/0264-6021:3580201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato H, Nomura S, Sagami I, Ito O, Daff S, Shimizu T. CO binding studies of nitric oxide synthase: effects of the substrate, inhibitors and tetrahydrobiopterin. FEBS Lett. 1998;430:377–380. doi: 10.1016/s0014-5793(98)00699-1. [DOI] [PubMed] [Google Scholar]
- 22.Bengea S, Araki Y, Ito O, Igarashi J, Sagami I, Shimizu T. Analysis of the kinetics of CO binding to neuronal nitric oxide synthase by flash photolysis: dual effects of substrates, inhibitors, and tetrahydrobiopterin. J. Inorg. Biochem. 2004;98:1210–1216. doi: 10.1016/j.jinorgbio.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Abu-Soud HM, Wu C, Ghosh DK, Stuehr DJ. Stopped-flow analysis of CO and NO binding to inducible nitric oxide synthase. Biochemistry. 1998;37:3777–3786. doi: 10.1021/bi972398q. [DOI] [PubMed] [Google Scholar]
- 24.Andersen HJ, Skibsted LH. Kinetics and mechanism of thermal oxidation and photooxidation of nitrosylmyoglobin in aqueous solution. J. Agric. Food Chem. 1992;40:1741–1750. [Google Scholar]
- 25.Arnold EV, Bohle DS. Isolation and oxygenation reactions of nitrosylmyoglobins. Methods Enzymol. 1996;269:41–55. doi: 10.1016/s0076-6879(96)69008-9. [DOI] [PubMed] [Google Scholar]
- 26.Moller JK, Skibsted LH. Mechanism of nitrosylmyoglobin autoxidation: temperature and oxygen pressure effects on the two consecutive reactions. Chemistry. 2004;10:2291–2300. doi: 10.1002/chem.200305368. [DOI] [PubMed] [Google Scholar]
- 27.Herold S, Rock G. Mechanistic studies of the oxygen-mediated oxidation of nitrosylhemoglobin. Biochemistry. 2005;44:6223–6231. doi: 10.1021/bi0475929. [DOI] [PubMed] [Google Scholar]
- 28.Marechal A, Mattioli TA, Stuehr DJ, Santolini J. Activation of peroxynitrite by inducible nitric-oxide synthase: a direct source of nitrative stress. J. Biol. Chem. 2007;282:14101–14112. doi: 10.1074/jbc.M609237200. [DOI] [PubMed] [Google Scholar]
- 29.Ilan YA, Czapski G, Meisel D. The one-electron transfer redox potentials of free radicals. I. The oxygen/superoxide system. Biochim. Biophys. Acta. 1976;430:209–224. doi: 10.1016/0005-2728(76)90080-3. [DOI] [PubMed] [Google Scholar]
- 30.Ost TW, Daff S. Thermodynamic and kinetic analysis of the nitrosyl, carbonyl, and dioxy heme complexes of neuronal nitric-oxide synthase. The roles of substrate and tetrahydrobiopterin in oxygen activation. J. Biol. Chem. 2005;280:965–973. doi: 10.1074/jbc.M411191200. [DOI] [PubMed] [Google Scholar]
- 31.Ray SS, Tejero J, Wang ZQ, Dutta T, Bhattacharjee A, Regulski M, Tully T, Ghosh S, Stuehr DJ. Oxygenase domain of Drosophila melanogaster nitric oxide synthase: unique kinetic parameters enable a more efficient NO release. Biochemistry. 2007;46:11857–11864. doi: 10.1021/bi700803p. [DOI] [PubMed] [Google Scholar]
- 32.Cooper CE. Nitric oxide and iron proteins. Biochim. Biophys. Acta. 1999;1411:290–309. doi: 10.1016/s0005-2728(99)00021-3. [DOI] [PubMed] [Google Scholar]
- 33.Andersen JF, Ding XD, Balfour C, Shokhireva TK, Champagne DE, Walker FA, Montfort WR. Kinetics and equilibria in ligand binding by nitrophorins 1–4: evidence for stabilization of a nitric oxide-ferriheme complex through a ligand-induced conformational trap. Biochemistry. 2000;39:10118–10131. doi: 10.1021/bi000766b. [DOI] [PubMed] [Google Scholar]
- 34.De Marinis E, Casella L, Ciaccio C, Coletta M, Visca P, Ascenzi P. Catalytic peroxidation of nitrogen monoxide and peroxynitrite by globins. IUBMB. Life. 2009;61:62–73. doi: 10.1002/iub.149. [DOI] [PubMed] [Google Scholar]
- 35.Fasano M, Antonini G, Ascenzi P. O2-mediated oxidation of hemopexin-heme(II)-NO. Biochem. Biophys. Res. Commun. 2006;345:704–712. doi: 10.1016/j.bbrc.2006.04.154. [DOI] [PubMed] [Google Scholar]
- 36.Herold S, Fago A, Weber RE, Dewilde S, Moens L. Reactivity studies of the Fe(III) and Fe(II)NO forms of human neuroglobin reveal a potential role against oxidative stress. J. Biol. Chem. 2004;279:22841–22847. doi: 10.1074/jbc.M313732200. [DOI] [PubMed] [Google Scholar]
- 37.Gardner PR. Nitric oxide dioxygenase function and mechanism of flavohemoglobin, hemoglobin, myoglobin and their associated reductases. J. Inorg. Biochem. 2005;99:247–266. doi: 10.1016/j.jinorgbio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Frey AD, Kallio PT. Bacterial hemoglobins and flavohemoglobins: versatile proteins and their impact on microbiology and biotechnology. FEMS Microbiol. Rev. 2003;27:525–545. doi: 10.1016/S0168-6445(03)00056-1. [DOI] [PubMed] [Google Scholar]
- 39.Gardner AM, Martin LA, Gardner PR, Dou Y, Olson JS. Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 tyrosine hydroxyl is essential for dioxygen binding and catalysis. J. Biol. Chem. 2000;275:12581–12589. doi: 10.1074/jbc.275.17.12581. [DOI] [PubMed] [Google Scholar]
- 40.Gardner PR, Gardner AM, Martin LA, Dou Y, Li T, Olson JS, Zhu H, Riggs AF. Nitric-oxide dioxygenase activity and function of flavohemoglobins. sensitivity to nitric oxide and carbon monoxide inhibition. J. Biol. Chem. 2000;275:31581–31587. doi: 10.1074/jbc.M004141200. [DOI] [PubMed] [Google Scholar]
- 41.Hausladen A, Gow A, Stamler JS. Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10108–10112. doi: 10.1073/pnas.181199698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardner PR, Gardner AM, Martin LA, Salzman AL. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. U. S. A. 1998;95:10378–10383. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hausladen A, Gow AJ, Stamler JS. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14100–14105. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rinaldo S, Arcovito A, Brunori M, Cutruzzola F. Fast dissociation of nitric oxide from ferrous Pseudomonas aeruginosa cd1 nitrite reductase. A novel outlook on the catalytic mechanism. J. Biol. Chem. 2007;282:14761–14767. doi: 10.1074/jbc.M700933200. [DOI] [PubMed] [Google Scholar]
- 45.Sam KA, Fairhurst SA, Thorneley RN, Allen JW, Ferguson SJ. Pseudoazurin dramatically enhances the reaction profile of nitrite reduction by Paracoccus pantotrophus cytochrome cd1 and facilitates release of product nitric oxide. J. Biol. Chem. 2008;283:12555–12563. doi: 10.1074/jbc.M800954200. [DOI] [PubMed] [Google Scholar]
- 46.Quaroni LG, Seward HE, McLean KJ, Girvan HM, Ost TW, Noble MA, Kelly SM, Price NC, Cheesman MR, Smith WE, Munro AW. Interaction of nitric oxide with cytochrome P450 BM3. Biochemistry. 2004;43:16416–16431. doi: 10.1021/bi049163g. [DOI] [PubMed] [Google Scholar]
- 47.Morgan ET, Ullrich V, Daiber A, Schmidt P, Takaya N, Shoun H, McGiff JC, Oyekan A, Hanke CJ, Campbell WB, Park CS, Kang JS, Yi HG, Cha YN, Mansuy D, Boucher JL. Cytochromes P450 and flavin monooxygenases--targets and sources of nitric oxide. Drug Metab Dispos. 2001;29:1366–1376. [PubMed] [Google Scholar]
- 48.Aguiar M, Masse R, Gibbs BF. Regulation of cytochrome P450 by posttranslational modification. Drug Metab Rev. 2005;37:379–404. doi: 10.1081/dmr-46136. [DOI] [PubMed] [Google Scholar]
- 49.Aulak KS, Miyagi M, Yan L, West KA, Massillon D, Crabb JW, Stuehr DJ. Proteomic method identifies proteins nitrated in vivo during inflammatory challenge. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12056–12061. doi: 10.1073/pnas.221269198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia Y, Zweier JL. Superoxide and peroxynitrite generation from inducible nitric oxide synthase in macrophages. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6954–6958. doi: 10.1073/pnas.94.13.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adak S, Bilwes AM, Panda K, Hosfield D, Aulak KS, McDonald JF, Tainer JA, Getzoff ED, Crane BR, Stuehr DJ. Cloning, expression, and characterization of a nitric oxide synthase protein from Deinococcus radiodurans. Proc. Natl. Acad. Sci. U. S. A. 2002;99:107–112. doi: 10.1073/pnas.012470099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kers JA, Wach MJ, Krasnoff SB, Widom J, Cameron KD, Bukhalid RA, Gibson DM, Crane BR, Loria R. Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature. 2004;429:79–82. doi: 10.1038/nature02504. [DOI] [PubMed] [Google Scholar]
- 53.Abu-Soud HM, Rousseau DL, Stuehr DJ. Nitric oxide binding to the heme of neuronal nitric-oxide synthase links its activity to changes in oxygen tension. J. Biol. Chem. 1996;271:32515–32518. doi: 10.1074/jbc.271.51.32515. [DOI] [PubMed] [Google Scholar]
- 54.Guo FH, De Raeve HR, Rice TW, Stuehr DJ, Thunnissen FB, Erzurum SC. Continuous nitric oxide synthesis by inducible nitric oxide synthase in normal human airway epithelium in vivo. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7809–7813. doi: 10.1073/pnas.92.17.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salerno JC. Neuronal nitric oxide synthase: prototype for pulsed enzymology. FEBS Lett. 2008;582:1395–1399. doi: 10.1016/j.febslet.2008.03.051. [DOI] [PubMed] [Google Scholar]
- 56.Van Doorslaer S, Dewilde S, Kiger L, Nistor SV, Goovaerts E, Marden MC, Moens L. Nitric oxide binding properties of neuroglobin - A characterization by EPR and flash photolysis. Journal of Biological Chemistry. 2003;278:4919–4925. doi: 10.1074/jbc.M210617200. [DOI] [PubMed] [Google Scholar]
- 57.Kharitonov VG, Sharma VS, Magde D, Koesling D. Kinetics of nitric oxide dissociation from five- and six-coordinate nitrosyl hemes and heme proteins, including soluble guanylate cyclase. Biochemistry. 1997;36:6814–6818. doi: 10.1021/bi970201o. [DOI] [PubMed] [Google Scholar]
- 58.Sarti P, Giuffre A, Forte E, Mastronicola D, Barone MC, Brunori M. Nitric oxide and cytochrome c oxidase: mechanisms of inhibition and NO degradation. Biochem. Biophys. Res. Commun. 2000;274:183–187. doi: 10.1006/bbrc.2000.3117. [DOI] [PubMed] [Google Scholar]