Summary
Intracellular pathogens have devised mechanisms to exploit their host cells to ensure their survival and replication. The malaria parasite Plasmodium falciparum relies on an exchange of metabolites with the host for proliferation. We describe the first mass spectrometry-based metabolomic analysis of the parasite throughout its 48-hour intraerythrocytic developmental cycle. Our results reveal a general modulation of metabolite levels by the parasite, with numerous metabolites varying in phase with the developmental cycle. Others differed from uninfected cells irrespective of the developmental stage. Among these was extracellular arginine, which was specifically converted to ornithine by the parasite. To identify the biochemical basis for this effect, we disrupted the plasmodium arginase gene in the rodent malaria model P. berghei. These parasites were viable but did not convert arginine to ornithine. Our results suggest that systemic arginine depletion by the parasite may be a factor in human malarial hypoargininemia associated with cerebral malaria pathogenesis.
Introduction
Infection by the malaria parasite Plasmodium falciparum has severe and potentially lethal consequences for the metabolic state of the human host (Planche et al., 2005). The approximately 500 million cases of this devastating disease lead each year to 1–2 million deaths worldwide (Sachs and Malaney, 2002). In all parasitic infections there is a significant metabolic interaction between pathogen and host as the parasite diverts nutrients towards its own growth while the host struggles to maintain homeostasis and cope with waste products, toxins and tissue damage. In the case of P. falciparum-induced malaria, the massive metabolic demands of the rapidly proliferating parasite cells, coupled with the effects of massive erythrocyte lysis and ischemic damage arising from the sequestration of parasitized erythrocytes within the microvasculature, are responsible for the pathogenesis of the disease and its manifestations (Mackintosh et al., 2004). These clinical manifestations include hypoglycemia, lactic acidosis, hemolytic anemia, hemoglobinuria and hypoargininemia.
The metabolites required by cells for macromolecular synthesis and growth are acquired either directly, by uptake from the environment, or else biosynthesized from the pool of available nutrients. Free-living protozoa maintain a versatile and dynamically regulated metabolic network that can support optimal growth from a wide variety of nutrient sources. In contrast, obligate intracellular parasites of mammals are restricted to a much more homeostatic and nutrient-rich environment and might be expected to have adapted to this specific milieu by altering the architecture of their metabolic network to maximize growth at the cost of flexibility. Within the human host, P. falciparum is found mostly within mature erythrocytes in the nutritionally complex environment of the bloodstream, and a wealth of evidence suggests that the metabolism of these blood stage parasites has diverged significantly from well-studied model eukaryotes such as yeast.
Upon invading an erythrocyte the parasite induces major modifications to the host cell to permit the directed exchange of metabolites [reviewed in (Kirk et al., 2005)]. New permeability pathways are induced on the host cell surface to enhance the influx and efflux of specific compounds. The parasite also initiates a catabolic process whereby hemoglobin from the erythrocyte cytoplasm is ingested and proteolyzed into its constituent amino acids in an acidic vacuole (Krugliak et al., 2002). Extensive genomic and biochemical evidence indicates that many areas of parasite metabolism have been radically streamlined or modified. For example, Plasmodia have lost the ability to synthesize the purine ring de novo, rendering them auxotrophs dependent on the uptake of purine nucleotides and nucleobases from the host (Gardner et al., 2002). These parasites are also incapable of amino acid biosynthesis, relying instead on hemoglobin catabolism and uptake from the extracellular space (Gardner et al., 2002). Energy metabolism consists almost entirely of glucose fermentation by the parasite to lactate, calling into question the role of the parasite mitochondrion in energy generation (Sherman, 1998). The function of the apicoplast, a non-photosynthetic plastid-like organelle, remains poorly elucidated, although it is known to play a role in fatty acid, heme and isoprenoid biosynthesis (Ralph et al., 2004).
Due to the importance of plasmodial metabolism for malaria pathogenesis and as a target for most current and candidate antimalarial pharmaceuticals, it is critical that we understand both the structure and dynamics of the parasite metabolic network. Indirect approaches to reconstructing this network, such as inference from genomic data or in vitro biochemistry, are at best incomplete and present significant obstacles in the case of highly diverged organisms. Metabolomic technologies (Kell, 2004; Want et al., 2005), however, are beginning to enable systems level measurements of changes in metabolic activity in response to genetic (Fischer and Sauer, 2003) and nutrient (Brauer et al., 2006) perturbations, as well as drug treatments (Nicholson et al., 2002) and other viral infections (Munger et al., 2006).
One focus of recent metabolomic investigations has been the nature of host-parasite interactions. Parasite pathogens and their hosts are by definition metabolically intertwined, and the pathology of many parasitic diseases is linked to dysregulation of host metabolism. Several studies have examined the systemic effects of parasite infection in vivo by NMR analysis of host biofluids with the aim of elucidating metabolic modulation by the parasite and identifying reliable biomarkers to aid in the diagnosis of Schistosoma (Wang et al., 2004), Trichinella (Martin et al., 2006) and Plasmodium (Li et al., 2008) infection. These studies have uncovered a number of significant effects that likely would not have been found by classical biochemical methods. In particular, an analysis of urine and plasma from mice infected by Plasmodium berghei (Li et al., 2008) discovered evidence of a disturbance of the gut microbiota and high levels of the lysine catabolic product pipecolic acid in host urine, an effect which thus far appears to be specific to plasmodial infection. Directed metabolomic experiments in cell culture have also revealed critical aspects of parasite metabolism, such as an apparent partitioning of carbon metabolism in the protozoan Trypanosoma brucei into different regimes corresponding to the differing nutrient availabilities in the mammalian and insect host tissues (Coustou et al., 2008). Recently, NMR metabolomics has also been used to determine the intracellular concentrations of a range of metabolites in the P. falciparum trophozoite stage (Teng et al., 2008). Applying metabolomic approaches to study the dynamics of P. falciparum-infected erythrocyte metabolism promises to similarly unravel some of the more poorly understood aspects of parasite biology.
Understanding the plasticity of Plasmodium metabolism to physiologically relevant perturbations will ultimately have great clinical relevance in light of the possibility that parasites experience dramatically different metabolic states within the human host and the fact that certain front-line drugs, such as artemisinin and its derivatives, act via an entirely unknown mechanism. We have taken the initial step towards this goal by conducting the first metabolomic study of the complete intraerythrocytic developmental cycle (IDC) of the most virulent human malaria parasite, Plasmodium falciparum.
Results
P. falciparum–infected RBC metabolite analysis
We have quantitatively measured the levels of known metabolites in synchronized cultures of P. falciparum (3D7 strain) over the course of its 48-hour blood-stage developmental cycle using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) method that simultaneously assays for ~200 compounds of validated identity (Lu et al., 2006). We analyzed both parasite-infected and uninfected red blood cell (RBC) cultures at seven timepoints during development to determine the parasite-induced alterations to normal RBC metabolism. The RBCs (infected and uninfected) and the liquid culture medium were collected and analyzed separately in order to profile the parasite-induced changes to both the intra- and extracellular metabolite pools. Roughly 90 metabolites were detected with a signal sufficient to construct temporal profiles of their relative levels (Figure 1, Figure S1 and S2, Tables S1 and S2). These compounds span a wide range of metabolic pathways and include amino acids, nucleotides, biosynthetic precursors, and central carbon metabolism intermediates. For comparative purposes, we also measured global parasite mRNA levels in parallel by DNA microarray analysis for all timepoints (Figure S3, Table S3).
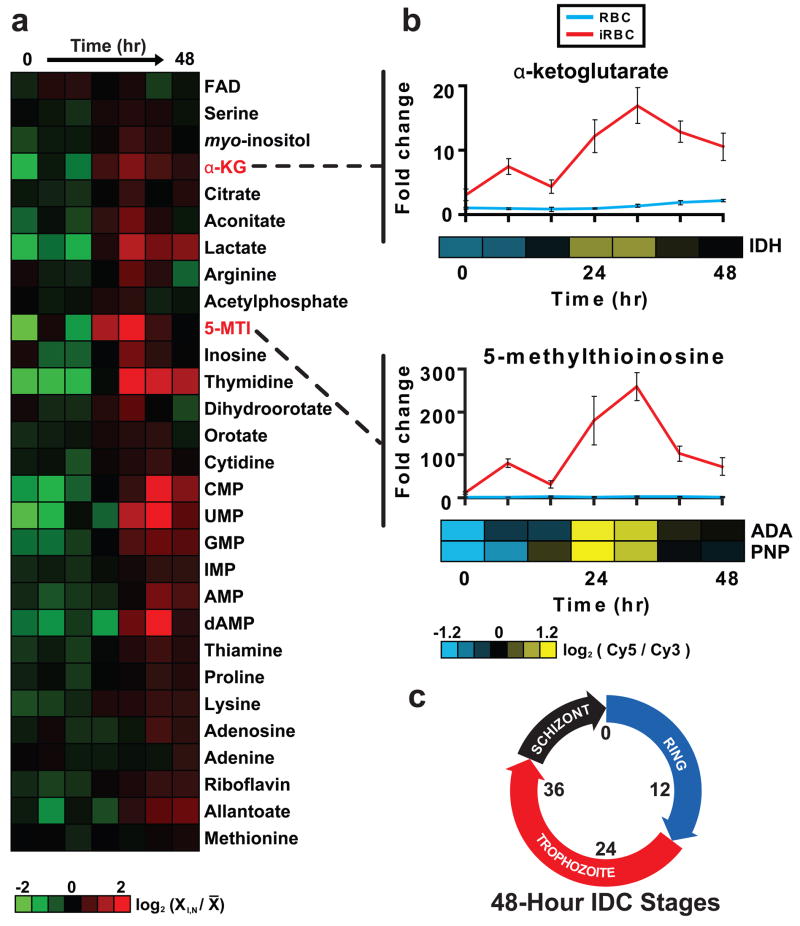
Figure 1. Periodic metabolite fluctuation during the intraerythrocytic developmental cycle (IDC).
A, Profiles of 29 intracellular metabolites with periodic changes in relative levels. Relative levels are expressed as the mean-centered log ratio of the normalized signal intensity in the infected RBC extract at each timepoint (X I, N / X̄). Profiles are ordered by the timing of their peak abundance level, from early (top) to late (bottom). B, Temporal profile of relative levels of α-ketoglutarate and 5-methylthioinosine (5-MTI) levels in RBC (blue lines) and infected RBC (red lines) extracts over the IDC. Shown for comparison are the gene expression profiles of the associated enzymes isocitrate dehydrogenase (IDH), adenosine deaminase (ADA) and purine nucleoside phosphorylase (PNP) as determined by DNA microarray analysis. The error is given as the standard deviation of n = 3 independent biological replicates. C, Schematic of the developmental stages during the 48-hour IDC.
Due to the highly periodic nature of gene expression observed during the IDC (Bozdech et al., 2003), we analyzed our metabolite profiles for periodic fluctuation to determine if metabolic requirements are also cyclical. Although many metabolite levels were either unchanged or else varied monotonically throughout the cycle, the subset shown in Figure 1 A exhibited detectable periodicity. Most of these metabolites show peak abundance during the late trophozoite/early schizont stages, a period of the IDC associated with high levels of energy metabolism and macromolecular biosynthesis.
The P. falciparum metabolome reflects the radical metabolic simplification that has occurred over its evolution towards a parasitic lifestyle. For example, we find no evidence for de novo amino acid biosynthesis, consistent with extensive genomic (Gardner et al., 2002) and biochemical (Krugliak et al., 2002) evidence that the Plasmodia have essentially lost this capacity entirely in favor of amino acid scavenging and hemoglobin catabolism. The metabolites that are detected, and which appear by this analysis to be cell cycle-regulated, are indicative of the major metabolic processes that are known to be essential for growth. For example, dihydroorotate and orotate, intermediates in the pyrimidine biosynthesis pathway, oscillate strongly in phase with the IDC (Figure 1 A). As the parasite’s sole source of pyrimidine bases, this pathway is essential for growth, and in fact has been proposed to be the effective target of the antimalarial drug atovaquone (Painter et al., 2007). Inosine-5′-phosphate (IMP) follows a similarly periodic profile (Figure 1 A). This metabolite is the product of an early step in the salvage pathway by which the Plasmodia, which are obligate purine auxotrophs, acquire purine bases (Downie et al., 2008). These metabolites reach their peak abundance coincident with a burst of nucleosides and nucleoside monophosphates occurring 32–40 hours post invasion (Figure 1 A), corresponding to the late trophozoite-early schizont transition in which the parasite successively replicates its genome 16–32 times and nucleotide demand is at its highest.
Energetically, it is well-established that the Plasmodia rely almost entirely on glycolysis and perform little or no oxidative phosphorylation (Lang-Unnasch and Murphy, 1998). While the parasites possess a mitochondrion, the mitochondrial genome is the smallest sequenced to date, their oxygen consumption is minimal and there has been no clear evidence of a functional tricarboxylic acid (TCA) cycle during the blood stages of growth (van Dooren et al., 2006). Strikingly, however, we observe coincident peaks in the TCA cycle intermediates citrate, aconitate and α-ketoglutarate during the trophozoite stage (Figure S4). Malate exhibited a periodicity score slightly below our significance threshold but is also most abundant during this stage. The P. falciparum genome encodes all the necessary enzymes to run a complete TCA cycle, and all are expressed during the IDC (Figure S4) (Bozdech et al., 2003); these data suggest that the enzymes are also active. In light of the recent observation that the sole pyruvate dehydrogenase encoded by the P. falciparum genome localizes to the apicoplast, where it presumably generates acetyl-CoA exclusively for fatty acid biosynthesis (Foth et al., 2005) instead of the TCA cycle, these data suggest TCA metabolism may be more complex and important than previously realized and a promising candidate for future study.
Within the set of non-periodic metabolites we find a substantial increase in the NAD+ content of infected RBCs, consistent with previous literature (Zerez et al., 1990). These high NAD+ levels are supported in part by biosynthesis from the precursor nicotinamide, which is significantly depleted from the culture medium during the late trophozoite/schizont stage of the life cycle (Figure S5). Elevated NAD+ levels have been suggested to be necessary to support the increased glycolytic flux in infected RBCs (Zerez et al., 1990). This flux up-regulation is evident in our data as an increase in the relative rates of glucose consumption and lactate generation (Figure S5). These results agree with genomic data that suggests that P. falciparum should be entirely dependent on exogenous nicotinamide or nicotinic acid for NAD+ biosynthesis (Gardner et al., 2002) and highlight either the uptake or synthesis pathway as a potential drug target.
Metabolite and mRNA Transcript Abundance During Development
To better understand the relationship between gene expression and metabolite levels, we calculated the correlations between each intracellular metabolite profile and the mRNA expression profile of the enzymes involved in its formation or degradation (Supplementary Table 4). Only a subset of metabolite-enzyme pairs are significantly correlated or anti-correlated. The strongest correlation was observed between 5-methylthioinosine (5-MTI), an intermediate in a Plasmodium-specific purine recycling pathway (Ting et al., 2005), and the upstream enzyme adenosine deaminase, although 5-MTI also correlates well with its downstream enzyme purine nucleoside phosphorylase (Figure 1 B). Interestingly, α-ketoglutarate falls into this set due to its high correlation with isocitrate dehydrogenase (Figure 1 B). No significant correlation is found between α-ketoglutarate and glutamate dehydrogenase, despite the presumptive major role of this enzyme in α-ketoglutarate formation. This points to the possibility that, despite the non-canonical nature of the plasmodium TCA cycle, intracellular α-ketoglutarate levels are strongly impacted by the activity of canonical TCA cycle enzymes.
Arginine is Rapidly and Specifically Depleted From the Culture Medium
P. falciparum digests up to 75% of its host cell hemoglobin (Hb) over its IDC to supply amino acids for protein synthesis and clear space within the host cell (Krugliak et al., 2002). Although the parasite lacks any capacity for de novo amino acid biosynthesis (Gardner et al., 2002), this process generates sufficient amounts of most amino acids to sustain growth even in the absence of external supplementation (Liu et al., 2006). We find that intracellular levels of the amino acids that are most abundant in hemoglobin (alanine, valine, and histidine) all rise significantly in the trophozoite stage, coincident with the initiation of hemoglobin digestion (Figure S1). These amino acids also accumulate in the extracellular medium, consistent with the observation that the parasite incorporates only a fraction of the Hb-derived amino acids into protein, excreting the excess as waste (Figure 2 A, Figure S2).
Figure 2. Amino acid levels in the culture medium.
A, Profiles of the 16 amino acids measured in culture medium samples. Relative levels are expressed as the background-subtracted log ratio of the normalized signal intensity in the infected RBC sample at each timepoint (XI, N) to the normalized signal intensity in the uninfected RBC time 0 sample (XU,0). Amino acids are ordered by the log ratio of the final timepoint, from highest (accumulating) (top) to lowest (depleting) (bottom). B, Arginine and ornithine concentrations in the culture medium of a 40-hour culture. A synchronized culture (6% initial parasitemia) was adjusted to 1% hematocrit in fresh RPMI 1640 culture medium at the trophozoite stage (approximately 24 hours post infection) and incubated 40 hours in parallel with an uninfected RBC control culture. Concentrations were determined by LC-MS/MS analysis using an internal standard solution containing known concentrations of isotopically labeled arginine and ornithine. The error is given as the standard deviation of n = 3 independent biological replicates.
The most dramatic relative change in extracellular amino acid levels occurs for the non-proteogenic amino acids ornithine and citrulline, which accumulate substantially in the medium during trophozoite and schizont stages (Figure 2 A). These amino acids are generally derived from arginine, which is significantly depleted over the same time interval (Figure 2 A). Standard RPMI 1640 medium used for Plasmodium culturing contains 1.15 mM arginine, which represents an 8–10 fold excess over normal human serum arginine levels (Lopansri et al., 2003). However, previous studies have concluded that arginine is unnecessary to support optimal parasite growth (Liu et al., 2006), which suggests that the parasite acquires sufficient quantities of arginine from hemoglobin digestion. This raises the question of the origin and nature of the observed effect on arginine and its downstream metabolites. Intriguingly, several clinical studies have demonstrated a strong correlation between reduced arginine levels in children and adults with malaria (Lopansri et al., 2003; Yeo et al., 2007).
Arginine Depletion is Due to Arginase
When we measured arginine, ornithine and citrulline levels in a timecourse initiated during the trophozoite stage, arginine levels were depleted to almost undetectable levels over 40 hours (>100-fold down) (Figure 2 B). In eukaryotic systems there are several enzymes that can interconvert between arginine, ornithine and citrulline, including arginase, nitric oxide synthase (NOS) and ornithine transcarbamylase. To determine which of these might be responsible for the observed effect, we cultured parasites in RPMI 1640 supplemented with equimolar amounts of unlabeled and uniformly 13C-15N-labeled L-arginine. After 40 hours, the only labeled form of ornithine detected was the fully 13C-15N-labeled form (Figure 3 A). Fully-labeled ornithine can be generated from fully-labeled arginine directly by arginase-catalyzed hydrolysis of arginine to ornithine and urea, or indirectly by the action of a citrullinase on fully-labeled citrulline to yield ornithine and ammonia. There is no recognizable citrullinase in the P. falciparum genome, but an arginase (PFI0320w) has been cloned and characterized (Muller et al., 2005). We therefore identify this enzyme as the likely source of extracellular ornithine. We also detected fully-labeled citrulline, which might derive from labeled arginine via the removal of the terminal nitrogen atom by either NOS (yielding nitric oxide) or arginine deiminase (yielding ammonia). Neither of these enzymes are predicted in the P. falciparum genome, but human RBCs contain NOS protein that has been shown to be active under some circumstances (Kleinbongard et al., 2006). However, ornithine accounts for the majority of the depleted arginine (Figure 2 B).
Figure 3. Arginine depletion is due to the P. falciparum arginase.
A, P. falciparum was cultured for 40 hours in RPMI 1640 medium supplemented with equimolar amounts (1.15 mM) of unlabeled and U-13C-15N-labeled arginine. Culture medium samples were analyzed by LC-MS/MS for labeling in metabolites downstream of arginine. Shown are the signal intensities for unlabeled (grey) and U-13C-15N-labeled (black) ornithine and citrulline. These were the only forms detected for either amino acid. B, Uninfected RBCs were uniformly lysed in distilled H2O, diluted to 1% hematocrit in RPMI 1640, and incubated alongside a P. falciparum culture (1% hematocrit, 6% initial parasitemia, trophozoite stage). After 40 hours medium samples were collected and analyzed by LC-MS/MS. Shown are concentrations of arginine and ornithine in untreated RPMI medium (white bars), RPMI medium mixed with the RBC lysate for 40 hour (grey bars), and the extracellular medium from an infected RBC (iRBC) culture grown for 40 hour in RPMI (black bars). The error is given as the standard deviation of n = 3 independent biological replicates.
The P. falciparum Arginase is Responsible for Arginine Turnover
The P. falciparum arginase I homologue (Muller et al., 2005) contains no recognizable targeting or export signal motifs, and is most highly expressed during ring and schizont stages (Bozdech et al., 2003). However, mature human erythrocytes also contain active arginase I (NP_000036) and we observe arginine depletion and ornithine formation in uninfected blood cultures, although to a much lower degree (Figure 2 B). One explanation for the enhanced arginase activity in parasite-infected cultures is the release of host arginase during RBC lysis, permitting continuous degradation of the extracellular arginine pool. This lysis-mediated effect has been proposed to explain the observed hypoargininemia in sickle-cell disease patients (Morris et al., 2005) due to the rupturing of sickled erythrocytes. To test the contribution of RBC lysis to in vitro arginine degradation, we incubated a culture consisting of uninfected RBCs that had been uniformly disrupted by hypotonic lysis in parallel with an infected RBC culture. The ornithine levels in the lysate culture were significantly lower than in the infected RBC culture, demonstrating that even 100% RBC lysis is insufficient to account for the high level of arginase activity (Figure 3 B). This implicates the parasite arginase as the major factor in arginine depletion.
Arginase Knockout Parasites Exhibit Background Arginine Degradation Rates
To further test this hypothesis we generated an arginase knockout line of the mouse parasite Plasmodium berghei (ANKA strain) via site-specific recombination (Figure S6). The knockout (argKO) line occasionally exhibited a slight delay in ascending parasitemia, but mice infected with WT and argKO consistently developed comparably high blood parasitemias (Figure S7) and exhibited no difference in the onset of morbidity, establishing that the parasite arginase is not essential for in vivo growth during the IDC. When cultured ex vivo, WT P. berghei exhibited high levels of ornithine generation, while ornithine generation by argKO parasites was indistinguishable from that of uninfected RBCs (Figure 4 A). Energy metabolism, as measured by lactate excretion, was highly similar in WT and argKO parasites (Figure 4 B); the slightly elevated lactate levels in the argKO 12-hour timepoint may reflect the added energetic demand of generating ornithine by an alternative pathway, such as proline degradation. Thus high levels of arginase activity are found in a murine malaria parasite as well, but the enzyme does not significantly contribute to in vivo growth and thus may serve a secondary purpose.
Figure 4. Phenotypic analysis of the P. berghei arginase knockout (argKO) strain.
A, Ornithine levels in the culture medium of a 24-hour ex vivo culture (10% parasitemia, 1% hematocrit). B, Lactate levels from the same experiment. The p-values give the significance of difference between the ANKA- and argKO-infected RBC samples (*, p < 0.01; **, p < 10−4; ***, p < 0.05). The error is given as the standard deviation of n = 3 independent biological replicates.
Discussion
Host-pathogen interactions rely heavily on an exchange of nutrients between the host cell and the infectious agent. Using a comprehensive mass spectrometry-based approach, we have demonstrated that development of Plasmodium falciparum within red blood cells modulates the levels of dozens of compounds from most major metabolic processes, including amino acid and nucleotide metabolism, energy generation, and cellular redox potential. These data suggest a complex interplay between the host red blood cell and the parasite for nutrient acquisition, turnover and waste removal.
Specifically, we have identified a previously unrealized rapid and specific degradation of arginine deriving from the plasmodial arginase. A major metabolic activity of arginases in protozoa is the generation of ornithine for polyamine biosynthesis (Kropf et al., 2005). However, this requirement seems unlikely to account for the high levels of observed activity for two reasons: first, the vast majority of the metabolized arginine is excreted into the extracellular environment, not selectively retained in the cell; second, the growth rate is not affected by the absence of arginine in the culture medium, suggesting that arginine is not rate-limiting for growth in vitro (Liu et al., 2006). The argKO parasites presumably acquire sufficient amounts of ornithine either from the plasma or the activity of the host cell arginase.
An alternative model is that the parasite depletes the host arginine pool in order to modulate the activity of the host enzyme, NOS. NOS is used by the mammalian immune system to generate antimicrobial nitric oxide (NO) radicals from arginine. Previous studies in rodent malaria models have found that upon Plasmodium infection the expression and activity of NOS is induced in activated macrophages (Tachado et al., 1996). Depletion of the plasma arginine pool may suppress this immune response by removing the substrate for NO production. In addition, the excess ornithine generated could exert a further suppressive effect by competing with arginine for transporter-mediated uptake into macrophages. A similar strategy is employed by the enterobacterium Helicobacter pylori, which uses its arginase to deplete the host arginine pool and ablate the NOS response (Gobert et al., 2001). However, while NO directly suppresses H. pylori infection by killing the bacteria, this does not seem to be the case with Plasmodium. Reactive nitrogen intermediates have been shown to have direct antiparasitic activity in vitro, but only at concentrations orders of magnitude above physiological levels [reviewed in (Sobolewski et al., 2005)]. In addition, a study using NOS-deficient mouse lines established that host NO production does not affect the proliferation of asexual stage parasites (Favre et al., 1999). However, NO does inactivate the more vulnerable sexual stage gametocytes which are generated during the IDC and taken up by the mosquito host (Naotunne et al., 1993). This pathway may have evolved as a means for the more abundant and metabolically active asexual parasites to protect gametocytes from NO-mediated inactivation, increasing infectivity and allowing for more efficient propagation. The need to protect gametocytes from NO may become more acute for P. falciparum because of the longer period (~14 days) necessary for their maturation, during which the parasite needs to remain sequestered from circulation. A further consequence of hypoargininemia and low host nitric oxide levels may be the up-regulation of host endothelial cell surface receptors such as ICAM-1 (intracellular adhesion molecule 1), which increases the ability of the parasitized RBCs to cytoadhere to the vascular endothelium and thus avoid passage through the spleen and clearance (Sherman et al., 2003).
There is substantial evidence linking hypoargininemia to malarial infection and progression to cerebral malaria (Gramaglia et al., 2006; Lopansri et al., 2003; Yeo et al., 2007), but the origin of this effect has not been established to date. Previously proposed explanations included dietary deficiencies, defects in arginine transport or biosynthesis, and the activity of the free host arginase (Gramaglia et al., 2006; Lopansri et al., 2003). Additionally, arginine levels are strongly regulated during the host inflammatory response, and hypoargininemia is a hallmark of both pathogenic inflammations, such as bacterial sepsis, and those due to traumatic insult (Satriano, 2004). Our results, however, demonstrate that the Plasmodium arginase plays a significant role in arginine depletion (at least in vitro) and that disruption of the parasite enzyme ablates this effect without compromising in vivo viability. It has recently been reported that diminished NO bioavailability plays a critical role in the onset of experimental cerebral malaria (ECM) leading to mortality (Gramaglia et al., 2006). The same study also found that hypoargininemia correlated with disease progression. This, coupled with a similar result from a clinical study of human cerebral malaria (Lopansri et al., 2003), suggests that Plasmodium arginase could be an important factor in cerebral malaria pathogenesis, although the observation that plasma arginine levels in patients with severe malaria (which includes a substantial parasite burden in the blood) are not significantly different than in those with moderately severe malaria (Yeo et al., 2007) highlights the likely multifactorial nature of arginine regulation. Arginine supplementation has recently been shown to improve endothelial function and increase exhaled NO levels in clinical malaria patients (Yeo et al., 2007). Our results suggest that a combination therapy incorporating a parasite arginase inhibitor as a promising target for future investigation.
Metabolomic analysis promises to significantly deepen our understanding of Plasmodium biology and the malaria disease state. It will be useful in determining the metabolic differences between phenotypically divergent strains and elucidating the mechanism of action for poorly characterized drugs, by mapping responses at the metabolic level that are undetectable by such methods as microarray analysis. The ability to generate concurrent metabolomic and transcriptomic profiles will allow investigation of the relationship between enzyme expression and metabolite level. A recent analysis of the transcriptional profiles of P. falciparum isolated from clinical malaria patients suggested that in vivo parasites may exist in distinct physiological regimes corresponding to stress or starvation states which have not been observed in ex vivo cultures (Daily et al., 2007). As metabolic enzymes represent a significant portion of the genes differentially regulated between these states, metabolomic investigations of patient samples might significantly advance our understanding of malarial pathogenesis within the human host. Furthermore, this experimental system will provide a powerful tool to further investigate clinically important biochemical pathways in greater detail and elucidate the metabolomic response to physiologically relevant perturbations such as nutrient limitation, heat shock and drug exposures.
Experimental Procedures
P. falciparum culturing and metabolite extraction
P. falciparum cultures were maintained and synchronized by standard methods with slight modifications outlined in the Supplemental Methods. RBCs (infected or uninfected) were pelleted and immediately serially extracted, first with four volumes of 100% methanol at −75° C for 15 minutes, then twice with one volume 80:20 methanol:water at 4° C. For the second and third extractions, the mixture was sonicated 15 minutes on ice in a water bath sonicator. The culture medium supernatants from each extraction were pooled and centrifuged free of cell debris and protein. Supernatants were diluted in 4 volumes of 100% methanol at −75° C and centrifuged. Samples were analyzed within 24 hours of their generation.
LC-MS/MS instrumentation
Liquid chromatography – tandem mass spectrometry (LC-MS/MS) was performed using an Shimadzu LC-10A HPLC system and a Phenomenex Luna aminopropyl column (250 mm × 2 mm with a 5-μm particle size) coupled to a Thermo Electron Corporation Finnigan TSQ Quantum Ultra triple-quadrupole mass spectrometer. The data presented are means ± standard deviation for N = 3 biological replicates. Data extraction, treatment, periodicity and microarray correlation analyses are described in the Supplemental Methods.
DNA microarray analysis
For each timepoint, 0.5 mL of packed RBC (10% parasitemia) was pelleted by centrifugation, washed once in PBS and flash-frozen in liquid nitrogen. Total RNA isolation and amino-allyl cDNA labeling were as previously described (Bozdech et al., 2003). A pool of 3D7 total RNA from all IDC stages was generated as the reference sample. Microarray data have been deposited at the NCBI Gene Expression Omnibus (GEO) under accession number XXXXX.
Parasite infections
Donor BALB/c mice were injected with P. berghei ANKA WT or argKO stabilates. Parasitemia was monitored by examining blood films. Donor mice infected with argKO parasites were treated with 10 mg/kg pyrimethamine once parasites were detectable.
Generation of argKO P. berghei
To generate the pL0001.1 knockout vector, DNA fragments flanking the predicted P. berghei ortholog of PFI0320w (PB000787.03.0) were cloned into the vector pL0001 flanking the pyrimethamine selectable marker Toxoplasma gondii DHFR-TS. Linearized plasmid DNA was transfected and pyrimethamine-resistant parasites recovered from BALB/c mice. Locus disruption and clonality of the population was verified by Southern Blotting (Figure S6).
Ex vivo short term cultures of P. berghei
Blood from infected BALB/c mice with 20–30% parasitemia was collected in Hepes-buffered saline containing 10 U/ml heparin. Cells were centrifuged and resuspended in RPMI 1640 medium containing 20% fetal calf serum and 0.5% Albumax. Parasitemia was adjusted to 10% by dilution with uninfected RBC. Cultures were set up in triplicate at 1% hematocrit in 24-well plates and incubated in chambers gassed with 90% N2, 5% O2, and 5% CO2. Samples were collected at 0, 12 and 24 hours for metabolomic analysis.
Supplementary Material
Acknowledgments
We thank B. Bennett, E. De Silva, D. Gresham, Y. Kwon, M. Szpara, and I. Zhu for critical discussions and reading of the manuscript; V. Schramm for 5-MTI and 5-MTA purified standards; A. Waters for plasmid pL0001; S. Bajad for assistance with initial experimental setup. M.L. is funded by the Burroughs Wellcome Fund and an NIH Director’s New Innovators award (1DP2OD001315-01). J.D.R. is funded by a Beckman Young Investigators award and an NSF CAREER award. M.L and J.D.R. receive support from the Center for Quantitative Biology (P50 GM071508). J.M.B. and A.B.V. are funded by NIH/NIAID. K.O. is funded by an NSF Graduate Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS biology. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Yuan J, Bennett BD, Lu W, Kimball E, Botstein D, Rabinowitz JD. Conservation of the metabolomic response to starvation across two divergent microbes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19302–19307. doi: 10.1073/pnas.0609508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Biran M, Breton M, Guegan F, Riviere L, Plazolles N, Nolan D, Barrett MP, Franconi JM, Bringaud F. Glucose-induced remodeling of intermediary and energy metabolism in procyclic Trypanosoma brucei. J Biol Chem. 2008;283:16342–16354. doi: 10.1074/jbc.M709592200. [DOI] [PubMed] [Google Scholar]
- Daily JP, Scanfeld D, Pochet N, Le Roch K, Plouffe D, Kamal M, Sarr O, Mboup S, Ndir O, Wypij D, et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007;450:1091–1095. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- Downie MJ, Kirk K, Mamoun CB. Purine salvage pathways in the intraerythrocytic malaria parasite Plasmodium falciparum. Eukaryot Cell. 2008;7:1231–1237. doi: 10.1128/EC.00159-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre N, Ryffel B, Rudin W. Parasite killing in murine malaria does not require nitric oxide production. Parasitology. 1999;118(Pt 2):139–143. doi: 10.1017/s0031182098003618. [DOI] [PubMed] [Google Scholar]
- Fischer E, Sauer U. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur J Biochem. 2003;270:880–891. doi: 10.1046/j.1432-1033.2003.03448.x. [DOI] [PubMed] [Google Scholar]
- Foth BJ, Stimmler LM, Handman E, Crabb BS, Hodder AN, McFadden GI. The malaria parasite Plasmodium falciparum has only one pyruvate dehydrogenase complex, which is located in the apicoplast. Molecular microbiology. 2005;55:39–53. doi: 10.1111/j.1365-2958.2004.04407.x. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, Cheng Y, Mobley HL, Wilson KT. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13844–13849. doi: 10.1073/pnas.241443798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramaglia I, Sobolewski P, Meays D, Contreras R, Nolan JP, Frangos JA, Intaglietta M, van der Heyde HC. Low nitric oxide bioavailability contributes to the genesis of experimental cerebral malaria. Nature medicine. 2006;12:1417–1422. doi: 10.1038/nm1499. [DOI] [PubMed] [Google Scholar]
- Kell DB. Metabolomics and systems biology: making sense of the soup. Current opinion in microbiology. 2004;7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Kirk K, Martin RE, Broer S, Howitt SM, Saliba KJ. Plasmodium permeomics: membrane transport proteins in the malaria parasite. Curr Top Microbiol Immunol. 2005;295:325–356. doi: 10.1007/3-540-29088-5_13. [DOI] [PubMed] [Google Scholar]
- Kleinbongard P, Schulz R, Rassaf T, Lauer T, Dejam A, Jax T, Kumara I, Gharini P, Kabanova S, Ozuyaman B, et al. Red blood cells express a functional endothelial nitric oxide synthase. Blood. 2006;107:2943–2951. doi: 10.1182/blood-2005-10-3992. [DOI] [PubMed] [Google Scholar]
- Kropf P, Fuentes JM, Fahnrich E, Arpa L, Herath S, Weber V, Soler G, Celada A, Modolell M, Muller I. Arginase and polyamine synthesis are key factors in the regulation of experimental leishmaniasis in vivo. FASEB J. 2005;19:1000–1002. doi: 10.1096/fj.04-3416fje. [DOI] [PubMed] [Google Scholar]
- Krugliak M, Zhang J, Ginsburg H. Intraerythrocytic Plasmodium falciparum utilizes only a fraction of the amino acids derived from the digestion of host cell cytosol for the biosynthesis of its proteins. Molecular and biochemical parasitology. 2002;119:249–256. doi: 10.1016/s0166-6851(01)00427-3. [DOI] [PubMed] [Google Scholar]
- Lang-Unnasch N, Murphy AD. Metabolic changes of the malaria parasite during the transition from the human to the mosquito host. Annu Rev Microbiol. 1998;52:561–590. doi: 10.1146/annurev.micro.52.1.561. [DOI] [PubMed] [Google Scholar]
- Li JV, Wang Y, Saric J, Nicholson JK, Dirnhofer S, Singer BH, Tanner M, Wittlin S, Holmes E, Utzinger J. Global metabolic responses of NMRI mice to an experimental Plasmodium berghei infection. Journal of proteome research. 2008;7:3948–3956. doi: 10.1021/pr800209d. [DOI] [PubMed] [Google Scholar]
- Liu J, Istvan ES, Gluzman IY, Gross J, Goldberg DE. Plasmodium falciparum ensures its amino acid supply with multiple acquisition pathways and redundant proteolytic enzyme systems. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8840–8845. doi: 10.1073/pnas.0601876103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopansri BK, Anstey NM, Weinberg JB, Stoddard GJ, Hobbs MR, Levesque MC, Mwaikambo ED, Granger DL. Low plasma arginine concentrations in children with cerebral malaria and decreased nitric oxide production. Lancet. 2003;361:676–678. doi: 10.1016/S0140-6736(03)12564-0. [DOI] [PubMed] [Google Scholar]
- Lu W, Kimball E, Rabinowitz JD. A high-performance liquid chromatography-tandem mass spectrometry method for quantitation of nitrogen-containing intracellular metabolites. J Am Soc Mass Spectrom. 2006;17:37–50. doi: 10.1016/j.jasms.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Mackintosh CL, Beeson JG, Marsh K. Clinical features and pathogenesis of severe malaria. Trends Parasitol. 2004;20:597–603. doi: 10.1016/j.pt.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Martin FP, Verdu EF, Wang Y, Dumas ME, Yap IK, Cloarec O, Bergonzelli GE, Corthesy-Theulaz I, Kochhar S, Holmes E, et al. Transgenomic metabolic interactions in a mouse disease model: interactions of Trichinella spiralis infection with dietary Lactobacillus paracasei supplementation. Journal of proteome research. 2006;5:2185–2193. doi: 10.1021/pr060157b. [DOI] [PubMed] [Google Scholar]
- Morris CR, Kato GJ, Poljakovic M, Wang X, Blackwelder WC, Sachdev V, Hazen SL, Vichinsky EP, Morris SM, Jr, Gladwin MT. Dysregulated arginine metabolism, hemolysis-associated pulmonary hypertension, and mortality in sickle cell disease. JAMA. 2005;294:81–90. doi: 10.1001/jama.294.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller IB, Walter RD, Wrenger C. Structural metal dependency of the arginase from the human malaria parasite Plasmodium falciparum. Biological chemistry. 2005;386:117–126. doi: 10.1515/BC.2005.015. [DOI] [PubMed] [Google Scholar]
- Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS pathogens. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naotunne TS, Karunaweera ND, Mendis KN, Carter R. Cytokine-mediated inactivation of malarial gametocytes is dependent on the presence of white blood cells and involves reactive nitrogen intermediates. Immunology. 1993;78:555–562. [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- Painter HJ, Morrisey JM, Mather MW, Vaidya AB. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature. 2007;446:88–91. doi: 10.1038/nature05572. [DOI] [PubMed] [Google Scholar]
- Planche T, Dzeing A, Ngou-Milama E, Kombila M, Stacpoole PW. Metabolic complications of severe malaria. Curr Top Microbiol Immunol. 2005;295:105–136. doi: 10.1007/3-540-29088-5_5. [DOI] [PubMed] [Google Scholar]
- Ralph SA, van Dooren GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, Tonkin CJ, Roos DS, McFadden GI. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nature reviews. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- Sachs J, Malaney P. The economic and social burden of malaria. Nature. 2002;415:680–685. doi: 10.1038/415680a. [DOI] [PubMed] [Google Scholar]
- Satriano J. Arginine pathways and the inflammatory response: interregulation of nitric oxide and polyamines: review article. Amino Acids. 2004;26:321–329. doi: 10.1007/s00726-004-0078-4. [DOI] [PubMed] [Google Scholar]
- Sherman IW. Carbohydrate Metabolism of Asexual Stages. In: Sherman IW, editor. Malaria, Parasite Biology, Pathogenesis and Protection. Washington, D.C.: ASM Press; 1998. pp. 135–143. [Google Scholar]
- Sherman IW, Eda S, Winograd E. Cytoadherence and sequestration in Plasmodium falciparum: defining the ties that bind. Microbes Infect. 2003;5:897–909. doi: 10.1016/s1286-4579(03)00162-x. [DOI] [PubMed] [Google Scholar]
- Sobolewski P, Gramaglia I, Frangos J, Intaglietta M, van der Heyde HC. Nitric oxide bioavailability in malaria. Trends Parasitol. 2005;21:415–422. doi: 10.1016/j.pt.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Tachado SD, Gerold P, McConville MJ, Baldwin T, Quilici D, Schwarz RT, Schofield L. Glycosylphosphatidylinositol toxin of Plasmodium induces nitric oxide synthase expression in macrophages and vascular endothelial cells by a protein tyrosine kinase-dependent and protein kinase C-dependent signaling pathway. J Immunol. 1996;156:1897–1907. [PubMed] [Google Scholar]
- Teng R, Junankar PR, Bubb WA, Rae C, Mercier P, Kirk K. Metabolite profiling of the intraerythrocytic malaria parasite Plasmodium falciparum by (1)H NMR spectroscopy. NMR Biomed. 2008 doi: 10.1002/nbm.1323. [DOI] [PubMed] [Google Scholar]
- Ting LM, Shi W, Lewandowicz A, Singh V, Mwakingwe A, Birck MR, Ringia EA, Bench G, Madrid DC, Tyler PC, et al. Targeting a novel Plasmodium falciparum purine recycling pathway with specific immucillins. J Biol Chem. 2005;280:9547–9554. doi: 10.1074/jbc.M412693200. [DOI] [PubMed] [Google Scholar]
- van Dooren GG, Stimmler LM, McFadden GI. Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol Rev. 2006;30:596–630. doi: 10.1111/j.1574-6976.2006.00027.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Holmes E, Nicholson JK, Cloarec O, Chollet J, Tanner M, Singer BH, Utzinger J. Metabonomic investigations in mice infected with Schistosoma mansoni: an approach for biomarker identification. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12676–12681. doi: 10.1073/pnas.0404878101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Want EJ, Cravatt BF, Siuzdak G. The expanding role of mass spectrometry in metabolite profiling and characterization. Chembiochem. 2005;6:1941–1951. doi: 10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- Yeo TW, Lampah DA, Gitawati R, Tjitra E, Kenangalem E, McNeil YR, Darcy CJ, Granger DL, Weinberg JB, Lopansri BK, et al. Impaired nitric oxide bioavailability and L-arginine reversible endothelial dysfunction in adults with falciparum malaria. J Exp Med. 2007;204:2693–2704. doi: 10.1084/jem.20070819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerez CR, Roth EF, Jr, Schulman S, Tanaka KR. Increased nicotinamide adenine dinucleotide content and synthesis in Plasmodium falciparum-infected human erythrocytes. Blood. 1990;75:1705–1710. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.