Abstract
Gap junction pharmacology is a nascent field. Previous studies have identified molecules that enhance intercellular communication, and may offer potential for innovative antiarrhythmic therapy. However, their specific molecular target(s) and mechanism(s) of action remain unknown. Previously, we identified a 34-amino acid peptide (RXP-E) that binds the carboxyl terminal domain of Cx43 (Cx43CT) and prevents cardiac gap junction closure and action potential propagation block. These results supported the feasibility of a peptide-based pharmacology to Cx43, but the structure of the core active element in RXP-E, an essential step for pharmacological development, remained undefined. Here, we used a combination of molecular modeling, surface plasmon resonance, nuclear magnetic resonance and patch clamp strategies to define, for the first time, a unique ensemble of pharmacophores that bind Cx43CT and prevent closure of Cx43 channels. Two particular molecules are best representatives of this family: a cyclized heptapeptide (called CyRP-71), and a linear octapeptide of sequence RRNYRRNY. These two small compounds offer the first structural platform for the design of Cx43-interacting gap junction openers. Moreover, the structure of these compounds offers an imprint of a region of Cx43CT that is fundamental to gap junction channel function.
Keywords: Gap junctions, Arrhythmias, Connexin 43
INTRODUCTION
Gap junctions are intercellular channels formed by oligomerization of connexin proteins. In the heart, the most abundant connexin is the 43kDa isotype connexin43 (Cx43). Cardiac gap junctions conduct electrical impulses between cells to maintain normal rhythm, and their closure can be a substrate for cardiac arrhythmias.1 As such, drugs that selectively open gap junctions may offer a novel strategy for antiarrhythmic therapy and/or treatment of cardiovascular disorders.2–5
Gap junction pharmacology is a nascent field (see6 for Review). Recently, hexa-peptides such as AAP10 and its stable analogue ZP123 (rotigaptide) together with a novel peptide, GAP134, have been found to modify gap junctional communication, and to show potential as anti-arrhythmic agents.7–10 This accumulated evidence supports the notion of gap junction modification as a suitable pharmacological target.10 Yet, further development of these molecules is limited by the fact that their precise molecular target remains undefined, thus reducing their potential as platforms for target-specific drug design.11
As an alternative strategy, we have applied knowledge on the mechanisms of Cx43 chemical gating to design molecules that bind the carboxyl terminal domain of Cx43 (Cx43CT) and modify its function. Gating of Cx43 relies on an intramolecular particle–receptor interaction between the CT domain and the cytoplasmic loop.12–15 Using phage display, we identified a series of peptides containing the sequence “RXP” (arginine, any amino acid, proline) as a consensus Cx43CT binding motif, and reported that a particular 34-amino acid peptide within this RXP series (dubbed RXP-E) binds to Cx43, prevents heptanol- and low pH-induced gap junction closure, and prevents action potential propagation block.16,17
While these studies have shown significant and promising results, further applications of RXP-E are hampered because of the molecular size and low membrane permeability of this peptide, as well as the metabolic instability and poor oral bioavailability of peptides in general. Peptide-mimetics, on the other hand, can be developed to retain the desired biological properties of a peptide. Steps in the design of mimetic molecules include identification of the essential active components (or amino acids) of the peptide sequence (the pharmacophores), determination of their structure/conformation in aqueous solution and finally, development of a corresponding pharmacophore model.18 Here, we have combined molecular modeling (based on structural analysis of the RXP series16) and experimental methods to identify the first group of pharmacophores (cyclized and linear peptides 6 to 8 amino acids long), that bind Cx43CT and prevent closure of Cx43 channels. This ensemble of pharmacophores represents a new platform for future development of small molecules with high efficacy and affinity that can prevent closure of gap junctions. Furthermore, we provide the first three-dimensional imprint of a potential site in Cx43CT that can be used for binding of exogenous molecules.
MATERIALS AND METHODS
Experimental methods for molecular modeling, electrophysiological experiments, surface plasmon resonance and GST-binding assays, as well as nuclear magnetic resonance experiments followed standard, previously published procedures.16,20 Details (including statistical analysis) are provided in the online supplement.
RESULTS
Cyclized hexapeptides based on analysis of the RXP series
Molecular modeling
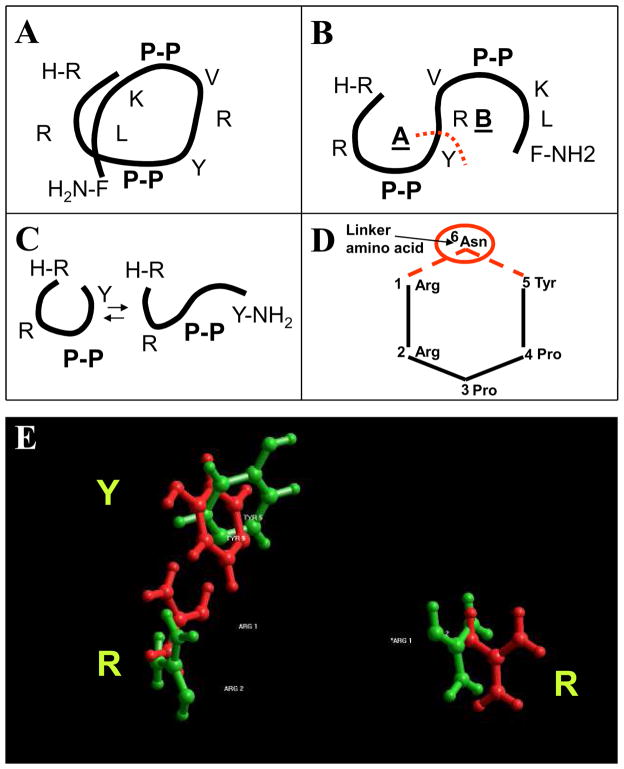
Our previous studies revealed that 12-mer peptides that bind Cx43CT share at least two common features: the presence of an RXP motif, and a predominance of basic amino acids16, their table 1 and Online Figure I). Thus, for identification of a core active element with Cx43CT binding activity, we modeled the structure of a peptide that fulfilled the following criteria: 1) was capable of binding Cx43CT, 2) showed an abundance of RXP sites, 3) was strongly basic, and 4) its primary sequence suggested a high-ordered secondary structure. A peptide labeled “RXP-4” by Shibayama et al16 best fulfilled these criteria. Analysis of its sequence (RRPPYRVPPKLF) showed 4 different RXP motifs (one of them in C-to-N direction, another one placed terminally), a balance of charge of +4 at physiological pH (7.4), and a combination of two proline-proline repeats likely to induce turns in the peptide, thus facilitating a helix-like conformation. We therefore used RXP-4 as a starting point to identify a novel Cx43CT-binding platform. Molecular modeling predicted that, in an alpha-helical conformation, the proline-proline repeats in RXP-4 would face opposite directions (Figure 1A). If extended, this alpha-helix yielded two horseshoe conformations (Figure 1B), both containing RXP motifs. Separating the two horseshoe-like sequences between tyrosine in position 5 and arginine in position 6 (red dotted line in Figure 1B) yielded two shorter peptides, both having terminal RXP motifs and a high content of basic residues (RRPPY and RVPPKLF; see Figure 1B). Given that RRP is a terminal RXP motif in the original RXP-4 peptide, the RRPPY sequence was chosen for further analysis.
Figure 1.
Strategy for identification of Cx43-binding pharmacophores based on RXP series.16 A: Position of amino acids in peptide RXP-4 when modeled as an alpha-helix. B: Representation of alpha-helix in A as two horseshoe-like structures. Red dotted line marks site where sequence was separated for further study. C: Representation of two likely configurations of peptide RRPPY, as a horseshoe, or as linearized molecule. D: Placement of asparagine between first arginine and last tyrosine fixed the R1-Y5 spacing into a cyclic conformation (compound CyRP-61). E: Molecular modeling results comparing position of arginines and tyrosine residues in RXP-4 with those in CyRP-61.
In a penta-peptide of sequence RRPPY, amino acids R1 and Y5 are placed at the opposite terminal ends, and the horseshoe conformation is anticipated to be at equilibrium with a corresponding “open” linear conformation (see Figure 1C). Thus, to stabilize the horseshoe conformation and keep the correct distance between R1 and Y5, the peptide was backbone-cyclized using an asparagine (N6) as a linker amino acid between R1 and Y5. The cyclic peptide (cyclo-RRPPYN) is illustrated in Figure 1D (small numbers mark the position given to each amino acid). This peptide was dubbed “CyRP-61” as it was the first Cyclized, RXP-derived hexaPeptide in this new series (see Table 1). Molecular modeling predicted an excellent correlation of the position in space of side chains R1R2Y5 in CyRP-61 (green in Figure 1E) with the equivalent R1R2Y5 side chains of RXP-4 (red in Figure 1E). Similar results were obtained by the conservative substitution N-Q at position 6 (CyRP-62; see Table 1). This design strategy, previously used to transform peptide AAP10 to a more potent tri-peptide analog,4 was predicted to generate more stable, smaller peptides with a core active structure capable of binding Cx43CT. This prediction was tested by surface plasmon resonance (SPR) experiments, as described below.
Table 1.
Sequence of Cyclic RXP-derived peptides.
| Name | Molecular Weight | Sequence |
|---|---|---|
| CyRP-61 | 783.89 | Cyclo-RRPPYN |
| CyRP-62 | 797.92 | Cyclo-RRPPYQ |
| CyRP-63 | 806.93 | Cyclo-RRPPWN |
| CyRP-71 | 954.11 | Cyclo-RRPPYRQ |
| CyRP-72 | 961.10 | Cyclo-RRPPYRN |
CyRP-62 binds to Cx43CT
SPR allows for assessment of ligand-analyte binding in real time, and was used for characterization of the RXP series.16 Cx43CT was covalently bound to the matrix of a sensor chip. Figure 2A shows plots of angle of incidence of resonance (in “response units”) as a function of time. Various concentrations of CyRP-62 were introduced in the microfluidics system at the time indicated by the upward arrow. Despite the small size of the analyte (798 Da), which approached the detection limits of the SPR instrument (~500–700 Da), a clear change in resonance was recorded, thus indicating direct binding of CyRP-62 to Cx43CT. Dissociation ensued upon washout (downward arrow). The rapid time course of association and dissociation precluded us from direct calculation of the dissociation constants. Yet, the results show that CyRP-62 binds Cx43CT. In previous studies, we demonstrated that peptide RXP-E prevents chemical gating of Cx43.16 As a next step, we tested whether this property was preserved in the CyRP peptides.
Figure 2.
CyRP peptides bind to Cx43CT and interfere with octanol-induced uncoupling of Cx43 channels. A: Surface plasmon resonance results. Change in angle of incidence of resonance (“resonance units”) plotted against time after onset of peptide exposure. Upward and downward arrows indicate onset and washout of CyRP-62 superfusion. B–D: Patch clamp results from N2A cells expressing Cx43. Peptides were diluted in patch pipettes (100 μM). Time “zero:” onset of octanol superfusion. Red and black symbols indicate, respectively, results from cells exposed to a given peptide, or kept as control. Significance values were p<0.001, p=0.148 and p=0.019 for data in panels B, C and D, respectively (each data set compared to results obtained without peptide).
Patch clamp results
Junctional conductance was measured in N2a cell pairs expressing Cx43. Patch pipettes were filled with an internal solution containing the test peptide. Time course and extent of octanol-induced uncoupling was compared to that observed in the absence of the peptide. Figure 2B shows average measurements of junctional conductance (Gj) recorded at various times after the onset of octanol superfusion. Red symbols and line correspond to data obtained when the cyclized hexapeptide CyRP-62 was dissolved in the internal pipette solution, and black symbols correspond to the average time course of octanol-induced uncoupling in control conditions. The data show that, in the presence of the cyclized peptide, the progression of octanol-induced uncoupling was delayed, and 100% of cells remained coupled after ten minutes after of octanol superfusion (see also Online Figure IA). The minimum Gj value recorded at the end of the 10-minute octanol exposure was significantly different from that obtained in the absence of the peptide (p<0.01).
The results in Figure 2B suggested that CyRP-62 contains an active core sequence capable of interfering with octanol-induced uncoupling. Other studies have demonstrated the importance of aromatic side chains in the preservation of pharmacophore activity.18 Consistent with this notion, substitution of Y5 with a larger aromatic residue (W) disrupted the activity of the peptide (Figure 2C; see also Table 1; peptide CyRP-63). Indeed, only one out of six pairs remained coupled at the end of octanol exposure (Online Figure IB) and the average Gj value was not statistically different from zero (p>0.05). The side chains of N and Q differ in having only one and two methylene groups, respectively. Consistent with this structural preservation, the activity of peptide CyRP-61 (N in position 6) was similar to that observed for CyRP-62 (Figure 2D and Online Figure IC). These data show that CyRP peptides represent a new class of core active molecules that interfere with Cx43 chemical gating. Additional improvement was achieved by increasing the balance of charge via addition of an arginine residue in the cyclized sequence.
Increased charge: Cyclized heptapeptides
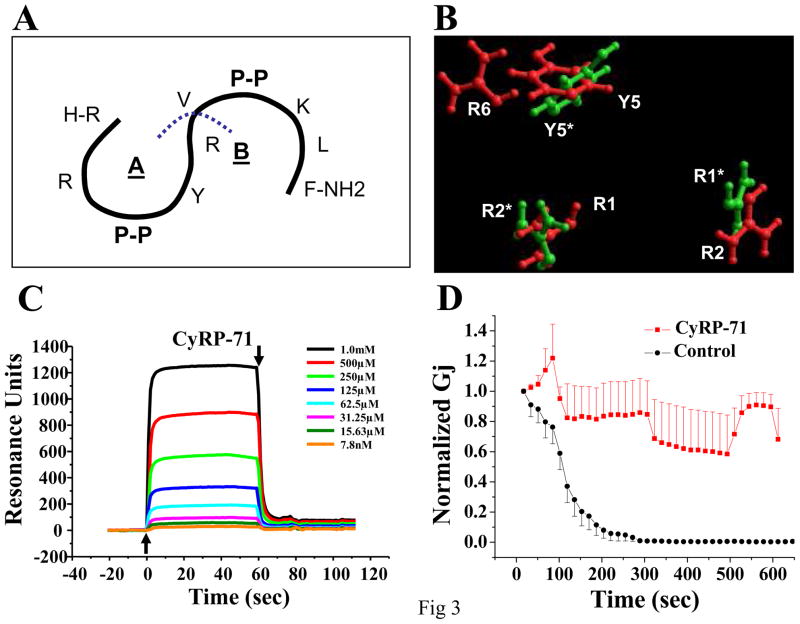
One of the characteristic features of RXP peptides was the high balance of positive charge in their sequence (see16). Thus, an alternative cyclized peptide (in this case, a heptapeptide) was generated by separating the RXP-4 sequence between amino acids R6 and V7 (blue line in Figure 3A), then forming a backbone-cyclic peptide using either asparagine or glutamine to complete the structure (see also Table 1; peptides labeled CyRP-71 and CyRP-72, respectively). These peptides, though a bit larger, had an increased electrostatic balance (+3 instead of +2). As shown in Figure 3B, an overlay of CyRP-71 with CyRP-61 reveals that both peptides share similar coordinates for the position of amino acids R1, R2 and Y5, while CyRP-71 presents an additional arginine (R6). This simple modification had a significant impact on the binding Cx43CT, as demonstrated by the SPR experiment shown in Figure 3C (compare data with those in Figure 2A; notice different scale in the ordinates). When normalized by molecular weight, the amplitude of the SPR response elicited by CyRP-62 (125 μM) was 3.71 (in arbitrary units; a.u.); in comparison, the response elicited by CyRP-71 was 9 times higher (33.92 a.u; immediately consecutive experiments conducted on the same sensor chip). In fact, the molecular weight-normalized response from CyRP-71 binding was larger than that obtained from RXP-E on the same sensor chip (9.91 a.u.), though there was a noticeable difference in the time course of the reaction (data not shown). Finally, patch clamp results showed that CyRP-71 also prevented octanol-induced uncoupling (Figure 3D). In fact, there was an improvement in the ability of this core active component to prevent uncoupling after insertion of the basic residue, further underlying the importance of the balance of charge in the activity of these compounds (see also Online Figure ID). Overall, this is the first demonstration of small cyclized peptides that can interfere with the octanol-induced closure of Cx43 channels.
Figure 3.
Addition of an arginine in CyRP sequence improved function. A: Diagram indicating RXP-4 sequence. Here, the peptide was separated between R6 and V7. B: Cyclized heptapeptide (CyRP-71; red) showed strong correlation in space with analog CyRP-61 (green and asterisks), though an arginine was exposed as potential site for additional interaction with a ligand. C: SPR results demonstrated binding of CyRP-71 to a sensor chip coated with recombinant Cx43CT. D: Time course of octanol-induced uncoupling in Cx43-expressing N2A cells. Cell pairs were recorded with pipettes containing CyRP-71 (100 μM; red symbols), or only the internal filling solution (black symbols). Difference between groups was significant (p<0.001).
Structural correlation between RXP-E and CyRP-71
In previous studies, we reported that RXP-E prevents chemically-induced uncoupling of Cx43 channels, and action potential propagation block.16 Here, we tested whether structural features thought to be of relevance in the cyclized peptides were also present in RXP-E. Figure 4 (top) shows the entire RXP-E sequence (in the one letter amino acid code), divided in three regions: The first RXP-containing domain (the amino side; colored red), the linker (green), and the second RXP-containing domain (the carboxyl side; colored blue). Panels A and C show the overlay of peptide CyRP-71 (green) with either the carboxyl terminus (amino acids 29–34) or the amino terminus (amino acids 1–11) of RXP-E (red), respectively. Clearly, the spatial coordinates of the basic residues R1, R2 and R6 of the cyclic peptide match with those of residues K29, R31 and R34 of RXP-E. The similar dimensions, spatial distribution and electrical charge of the core residues make it likely for the carboxyl terminal region of RXP-E and the cyclic peptides to share a common binding motif. The opposite conclusion can be drawn from the comparison of the cyclic peptide with the amino end (N-terminal) of RXP-E; in that case, the space occupied by R6 in the cyclized peptide would correspond to the location of two acidic residues (D2, D3) in RXPE. If binding between Cx43CT and the peptide is mediated, at least in part, by electrostatic forces, the presence of negative charges at a relevant position in space would prevent binding. Overall, these modeling results led us to predict that binding of RXP-E to Cx43CT occurs via the carboxyl end of RXP-E. The SPR results shown in Figures 4B and 4D were consistent with this hypothesis. Duplicates of either the carboxyl side, or the amino side of RXPE, separated by the linker region, were used (see sequences on top of the respective panels). As shown in panel 4B, a significant change in the angle of incidence for resonance was detected when a peptide containing the carboxyl end of RXPE was presented to Cx43CT (upward and downward arrows represent the addition of the peptide and its washout, respectively). In contrast, no binding was detected for the peptide formed by the amino end of RXP-E and its linker (panel 4D). Overall, these results show a convergence between modeling predictions and experimental results and suggest defined structural constraints for the binding of RXP-derived peptides to Cx43CT. This structural information led us to the prediction of a minimum Cx43CT binding motif.
Figure 4.
Structure-function comparison of CyRP-71 with RXP-E. Top: amino acid sequence (one-letter code) of RXP-E (see also16). A: Molecular overlay of predicted spatial position for amino acids R1, R2 and R6 of CyRP-71 (green) with positively charged amino acids K29, R31 and R34 in C-end of RXP-E (red). L Spatial correlation was consistent with the ability of peptide to bind Cx43CT by SPR (panel B; peptide sequence on top; peptide concentrations as noted). C: At amino end of RXP-E, overlay of positively charged residues R5 and H10 of RXP-E (red) over R1–R2 of CyRP-71 (green) predicts that two acidic residues (D2 and D3) occupy the position held by a basic amino acid (R6) in CyRP-71. The latter would be inconsistent with occupation of the same binding pocket. D: As predicted, a peptide of the amino end of RXP-E (sequence on top of panel) failed to bind Cx43CT by SPR.
Identification of a minimal active motif: RRNY
Cyclized compounds show longer bioavailability given their limited degradation in the intracellular space. An alternative path toward pharmacological development is the use of synthetic scaffolds that mimic peptide structure. Toward that aim, we sought to minimize the active sequence of the cyclized compounds by deleting the proline residues in CyRP-61 (Figure 5A). Indeed, in an alpha-helix, the proline-proline component would retain R1, R2 and Y5 at the appropriate spatial coordinates (see also Figure 1C). We therefore speculated that, as this spatial conformation was achieved by introduction of N6, the prolines would no longer be essential to hold the relative distances. Molecular modeling confirmed this expectation, as shown by the overlay of the side chains for R1, R2, Y4 in a short tetra-peptide RRNY (green) with R1 R2 and Y5 of the cyclized CyRP-61 hexa-peptide (red; Figure 5B). Functional and biochemical assays were therefore designed to assess the prediction that RRNY is a Cx43CT binding motif. As a first step, we tested whether RRNY-containing peptides could bind Cx43CT. Assessment of binding was carried out by SPR. Recombinant Cx43CT was used as ligand, covalently linked to the sensor chip, and peptides were presented as analytes. The mass of the analyte was increased by concatenating two “RRNY” motifs (i.e., RRNYRRNY). An example of sensograms obtained upon introduction of this peptide into the chip containing Cx43CT, is presented in Figure 5C. Clearly, angle of incidence for resonance changed rapidly upon onset of superfusion (time “zero”), and returned to baseline upon washout (see downward arrow). The rapid transitions signaled a fast on-off ligand-analyte interaction, and prevented us from determining dissociation constants for these peptides. However, the data showed that RRNYRRNY was capable of interacting with Cx43CT in a concentration-dependent manner. Similar results were obtained in 3 separate runs. Additional studies showed that RRNY-containing peptides can pulldown Cx43 from an adult rat heart lysate preparation (Online Figure II).
Figure 5.
Strategy for identification of sequence RRNY as potential core active molecule. A: Proline residues of hexapeptide CyRP-61 were removed, under the assumption that the structure formed by residues R1, R2 and Y5 represented the core Cx43-binding element. B: Overlay of predicted positions of amino acids R1, R2 and Y5 of CyRP-61 with those of RRNY. C: SPR traces obtained by presenting peptide RRNYRRNY to Cx43CT. Peptide concentrations as noted. D: Peptide RRNYRRNY in the pipette (red symbols) prevented octanol-induced uncoupling of Cx43 (p=0.005). Octanol uncoupling proceeded as control (black symbols) in the presence of a linearized version of CyRP-61 (RRPPYN; green symbols; p=0.63).
The binding results led us to assess whether the motif RRNY would be sufficient for preventing octanol-induced uncoupling. Cx43-expressing N2a cells were dialyzed with an internal pipette solution containing peptide RRNYRRNY. Time course of octanol-induced uncoupling is shown in Figure 5D. In the presence of the peptide, average Gj recorded 10 minutes after the onset of octanol was significantly different from that recorded in control at the same time point (p<0.05) and coincided with the preservation of electrical coupling in four of the six cells studied (see also Online Figure IIIA). These results indicate that RRNY is a minimum sequence capable of interfering with the chemical regulation of gap junctions by octanol. Moreover, since amino acids RRNY are contiguous in the cyclized CyRP-61 peptide (see Figure 1D and Table 1), we speculated that a linearized (non-cyclic) RRPPYN peptide would fail to affect uncoupling, given the loss of continuity of the RRNY motif. As expected, the time course and extent of octanol-induced uncoupling in the presence of the linear RRPPYN peptide was not different from control (Figure 5D; trace in green; also Online Figure IIIB), thus supporting the notion that pharmacophore activity is related not only to the presence of specific amino acids, but to the preservation of their molecular conformation in space.
CyRP-71 and the integrity of the Cx43CTdomain
Figures 1–5 described the various steps taken to identify potential leading compounds. Additional experiments focused on CyRP-71, given its binding and functional efficacy, and its potential as a more bio-stable compound. We hypothesized that binding of CyRP-71 to Cx43CT is linked to the ability of the peptide to prevent uncoupling. As shown in Figure 6A, this hypothesis was supported by experiments demonstrating that CyRP-71 failed to prevent octanol-induced uncoupling in Cx43 channels lacking the CT domain (mutant M257; [16]). In addition, this peptide did not modify the time course of octanol-induced uncoupling in N2a cells expressing a different connexin isoform, Cx40 (Figure 6B). Consistent with this observation, CyRP-71 caused only a minor SPR deflection when interacting with recombinant Cx40CT (Online Figure IV). The overall data suggest that there is a degree of structural specificity to the effect of CyRP-71, and that the Cx43CT domain is an essential component for peptide action.
Figure 6.
CyRP-71 did not prevent octanol-induced uncoupling in N2a cells expressing Cx43 mutant M257 (panel A) or an alternative connexin isotype, Cx40 (panel B). CyRP-71 prevented acidification-induced uncoupling of Cx43-expressing N2a cells (panel C). CyRP-71 (0.1 mmol/L) was diluted in internal pipette solution buffered to pH 6.2. In the absence of peptide, Gj decreased to 17.3 ± 1.7% of initial value. In the presence of CyRP-71, Gj decreased only to 49.0 ± 5.9% of maximum (p=0.002). Panel D: CyRP-71 prevented acidification-induced uncoupling of neonatal rat ventricular gap junctions (pH 6.2). Fifteen minutes after patch break, Gj in control and in the presence of linear peptide RRPPYR decreased to 8.4 ± 3.2% and 16.6 ± 6.5% of initial value, respectively (p value RRPPYR=0.48 when compared to control). In the presence of CyRP-71 (0.1 mmol/L), Gj decreased only to 47.0 ± 9.5% of maximum (p=0.006).
CyRP-71 and acidification-induced uncoupling
Octanol served as a screening tool to identify peptide activity. However, a more biologically-relevant question is whether, as in the case of RXP-E, the candidate compound can prevent low pH-induced block.16 Experiments were conducted both in Cx43-expressing N2a cells (Figure 6C) and in pairs of neonatal cardiac myocytes (Figure 6D). Patch pipettes were filled with an internal solution buffered to pH 6.2. In the absence of peptide (black symbols and traces) Gj decreased progressively, reaching minima of 17.3 ± 1.7% in N2a cells (Figure 6C) and 8.4 ± 3.2% (Figure 6D) in cardiac myocytes. In contrast, cells exposed to CyRP-71 remained coupled throughout the same time course, Gj decreasing only to 49.0 ± 5.9% (6C) and 47.0 ± 9.5% (6D) of control (red symbols). The green symbols in Figure 6D depict data obtained with a linearized peptide RRPPYR. Results were not different from control, indicating that the structure of CyRP-71, rather than only the net balance of charge, was important for its functional effect.
CyRP-71 and Cx43CT interaction resolved by NMR
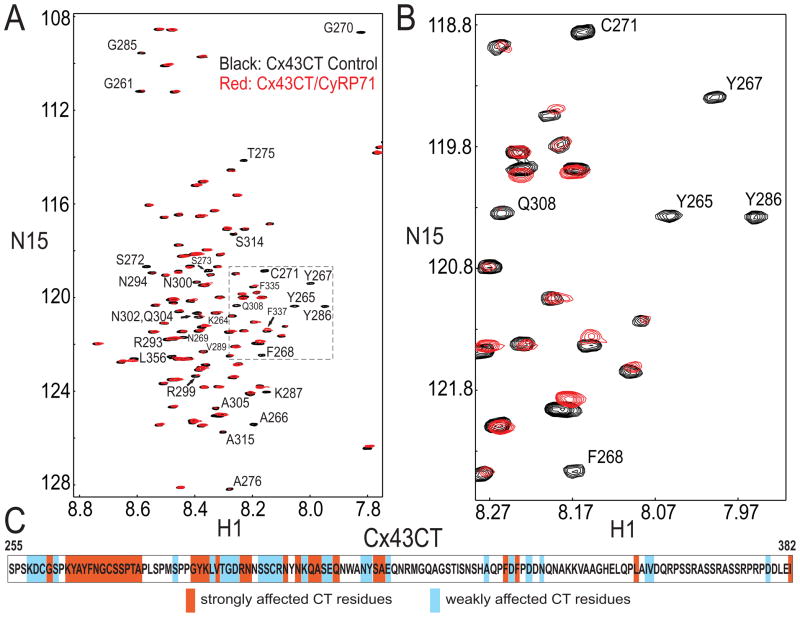
The SPR results demonstrated direct interaction between CyRP-71 and Cx43CT. Yet, further refinement of CyRP-71 will require a better understanding of the structural constraints imposed by its binding site in Cx43CT. As an initial step, we identified the amino acids in Cx43CT whose position in space change when presented to the peptide. Resonance assignments for Cx43CT have been published before (X). Figure 7, panels A and B, show an 15N-HSQC spectrum for Cx43CT alone (black) which has been overlaid with spectra obtained in the presence of CyRP71 (red). A summary of the Cx43CT residues affected by CyRP-71 is presented in panel C. The predominant regions of resonance shifts were: a) near or within the proline-rich region of Cx43CT (K258-A276), b) in the extreme end of the CT domain (I382), and c) in the region contained within amino acids G285 to E316 of Cx43. These results confirm the binding of Cx43CT to CyRP-71 and provide a first lead as to the amino acids in the CT domain that are involved in the interaction. Future studies will be needed to determine which residues form the actual binding site, and which shifts may result from secondary changes, perhaps some consequent changes in Cx43CT dimerization21.
Figure 7.
NMR analysis of structural modification of recombinant Cx43CT when in the presence of CyRP71. A: Cx43CT was titrated with CyRP71 to 1:500 and 1:900 molar ratio. 15N-HSQC spectrum for Cx43CT alone (black) was overlaid with spectra obtained in the presence of CyRP71 (1:500, red; 1:900, green). B: Close-up of resonance peaks from box in panel A. C: Summary of Cx43CT residues affected by CyRP71.
DISCUSSION
We have previously demonstrated that RXP-E binds to Cx43CT and prevents chemically-induced gap junction closure.16 As such, RXP-E represented a proof of principle, and the RXP series a starting point to develop a pharmacophore model for new compounds capable of modifying gap junctions. Structure-function analysis of the RXP series led us to a new ensemble of pharmacophores that bind Cx43CT and prevent chemically-induced uncoupling. To our knowledge, the two core active structures hereby reported (one cyclized; one linearized) represent the smallest known Cx43CT-binding molecules with regulatory activity over gap junctions. Moreover, CyRPs are the first known cyclized molecules capable of binding Cx43CT. Thus, our results represent a potential foundation for development of target-based gap junction pharmacology.
Our compounds represent a new a pharmacophore model for Cx43 regulation. Yet, major milestones need to be reached before considering them of practical application. First, our studies demonstrate that selected peptides prevent closure of Cx43 channels induced by octanol superfusion. CyRP-71 also prevented pH-induced uncoupling. Yet, other assays will be needed to obtain a wider view of the overall functional effects of this peptide on Cx43. Second, we have shown that our candidate compounds interact with Cx43, but a remaining question is whether these peptides bind other cellular components (including other ion channel proteins) and alter their function and/or regulation. Additional studies will be required to characterize the selectivity and specificity of these compounds, and their range of pharmacological applications. Yet, it is important to keep in mind that RXP-E, a leading molecule from the RXP series, prevents action potential propagation block without modifying other nonjunctional membrane channels17, and our studies show structural similarities between the pharmacophores and the active domain of RXP-E. A lack of increase of Gj during application of both RXPE17 and CyRP71 may also indicate that these pharmacophores act not by recruiting new channels, but by stabilizing the open state of those at the membrane. Third, we chose to explore the effect of CyRP-71 at one peptide concentration. Thus, we know that 100 μmol/l of CyRP-71 delivered through the patch pipette is enough to prevent octanol, and low-pH induced uncoupling (Figure 6). Yet, further structure-based refinement of the pharmacophore will be needed to improve its affinity for the target. Fourth, the linkage between Cx43CT binding in SPR and the patch clamp data may still seem correlative, although the hypothesis of a causative association is supported by results obtained with the mutant lacking Cx43CT(Figure 6A). These caveats notwithstanding, an important goal has been achieved: to find a compound of low molecular weight (<1 kDa) which binds the regulatory domain of Cx43, affects function, and shows selectivity for Cx43 over Cx40. Improving the affinity and selectivity of these compounds will make them valuable as potential scaffolds to act as carriers for cargoes with biological activity.
Our functional studies are complemented with NMR analysis of Cx43CT in the presence of CyRP-71. The data provide us with initial identification of those amino acids whose position in space is affected by the peptide (Figure 7). Differences were found with the resonance shifts caused by RXP-E.17 Of note, resonance shifts detected by HSQC may be consequent to direct binding, distant changes in conformation and/or modifications of the dimerized state of Cx43CT21. The larger, more complex structure of RXP-E (several “RXP” domains; likely more than one binding site) may cause a wider range of modifications, direct and indirect, that overlap (and may obscure) those occurring within the binding pocket. The cyclized peptide, on the other hand, may be less restricted for secondary interactions and therefore yield a different resonance shift map. Future identification of the actual binding pocket will be carried out by Nuclear Overhauser Effect Spectroscopy (NOESY). This analysis will be an important step toward optimizing pharmacophore binding and selectivity. Overall, these peptides represent a potential imprint of a region of Cx43CT that is amenable for binding to exogenous molecules that will affect the function of the Cx43 channel as a whole. Combined structural and biological studies may lead to a future generation of molecules of higher efficacy, selectivity and bio-stability, capable of crossing the cell membrane barrier to reach their target in a living cell. While the latter is only a goal, our data suggest that we are heading in the right direction. Identification of the CyRP group of compounds bears relevance, as cyclic peptides are more stable within the intracellular space and as such, have more potential for future pharmacological applications.
Peptide RRNYRRNY failed to prevent uncoupling in approximately 40% of the cell pairs studied. From that standpoint, its efficacy was less than that previously described for RXP-E. On the other hand, CyRP-71 showed an effect similar to that of RXP-E. Interestingly, CyRP-71 also shows homology with the Cx43CT-binding element of RXP-E. This cyclized molecule offers itself as an excellent platform for the next generation of compounds, utilizing peptide-mimetic substitutions on the core structure to minimize the size and maximize the activity, stability and bioavailability while preserving pharmacological effect.
In summary, we have described a new generation of Cx43-binding peptides. Our efforts focused on a series of in silico modeling steps, combined with biochemical and cellular experiments to identify the core active structure of the RXP series. We have identified new candidate molecules capable of binding Cx43CT and preventing chemically-induced uncoupling of Cx43 channels. This is the first demonstration of a small, cyclic core active structure that chemically and functionally interacts with Cx43 to prevent gap junction closure. Our data opens a new line of investigation for development of target-based gap junction pharmacology.
Acknowledgments
SOURCES OF FUNDING
Supported by grants NIH-HL39707, HL087226 and GM57691, grant GM072631 (PLS), and a grant from the Susan G Komen Foundation (SMT).
Footnotes
DISCLOSURES
Compounds provided by Zealand Pharma; BDL holds modest ownership interest.
LITERATURE CITED
- 1.Herve JC, Dhein S. Pharmacology of cardiovascular gap junctions. Adv Cardiol. 2006;42:107–31. doi: 10.1159/000092565. [DOI] [PubMed] [Google Scholar]
- 2.Dhein S. Cardiac ischemia and uncoupling: gap junctions in ischemia and infarction. Adv Cardiol. 2006;42:198–212. doi: 10.1159/000092570. [DOI] [PubMed] [Google Scholar]
- 3.Srinivas M, Duffy HS, Delmar M, Spray DC. Prospects for pharmacological targeting of gap junction channels. In: Zipes DZ, Jalife J, editors. Cardiac Electrophysiology: From Cell to Bedside. 4. Philadelphia: Saunders; 2004. pp. 158–167. [Google Scholar]
- 4.Eloff BC, Gilat E, Wan X, Rosenbaum DS. Pharmacological modulation of cardiac gap junctions to enhance cardiac conduction: evidence supporting a novel target for antiarrhythmic therapy. Circulation. 2003;108:3157–3163. doi: 10.1161/01.CIR.0000101926.43759.10. [DOI] [PubMed] [Google Scholar]
- 5.Haugan K, Petersen JS. Gap junction modifying antiarrhythmic peptides: therapeutic potential in atrial fibrillation. Drugs Future. 2007;32:245–260. [Google Scholar]
- 6.Lewandowski R, Petersen JS, Delmar M. Connexins as potential targets for cardiovascular pharmacology. In: Zipes DP, Jalife J, editors. Cardiac Electrophysiology: From Cell to Bedside. 5. Philadelphia: Saunders; In Press. [Google Scholar]
- 7.Muller A, Schaefer T, Linke W, Tudyka T, Gottwald M, Klaus W, Dhein S. Actions of the antiarrhythmic peptide AAP10 on intercellular coupling. Naunyn Schmiedebergs Arch Pharmacol. 1997;356(1):76–82. doi: 10.1007/pl00005031. [DOI] [PubMed] [Google Scholar]
- 8.Kjolbye AL, Haugan K, Hennan JK, Petersen JS. Pharmacological modulation of gap junction function with the novel compound rotigaptide: a promising new principle for prevention of arrhythmias. Basic Clin Pharmacol Toxicol. 2007;101:215–30. doi: 10.1111/j.1742-7843.2007.00123.x. [DOI] [PubMed] [Google Scholar]
- 9.Rossman EI, Liu K, Morgan GA, Swillo RE, Krueger JA, Butera J, Gruver M, Kantrowitz J, Feldman HS, Petersen JS, Haugan K, Gardell SJ, Hennan JK. The Gap Junction Modifier, GAP-134, Improves Conduction and Reduces Atrial Fibrillation/Flutter in the Canine Sterile Pericarditis Model. J Pharmacol Exp Ther. 2009;329:1127–1133. doi: 10.1124/jpet.108.150102. [DOI] [PubMed] [Google Scholar]
- 10.Axelsen LN, Haugan K, Stahlhut M, Kjolbye AL, Hennan JK, Holstein-Rathlou NH, Petersen JS, Nielsen MS. Increasing gap junctional coupling: a tool for dissecting the role of gap junctions. J Membr Biol. 2007;1:23–35. doi: 10.1007/s00232-007-9026-z. [DOI] [PubMed] [Google Scholar]
- 11.Dhein S, Polontchouk L, Salameh A, Haefliger JA. Pharmacological modulation and differential regulation of the cardiac gap junction proteins connexin 43 and connexin 40. Biol Cell. 2002;94:409–422. doi: 10.1016/s0248-4900(02)00018-7. [DOI] [PubMed] [Google Scholar]
- 12.Delmar M, Coombs W, Sorgen P, Duffy HS, Taffet SM. Structural basis for the chemical regulation of Connexin43 channels. Cardiovasc Res. 2004;62:268–275. doi: 10.1016/j.cardiores.2003.12.030. [DOI] [PubMed] [Google Scholar]
- 13.Morley GE, Taffet SM, Delmar M. Intramolecular interactions mediate pH regulation of connexin43 channels. Biophys J. 1996;70:1294–1302. doi: 10.1016/S0006-3495(96)79686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ek-Vitorin JF, Calero G, Morley GE, Coombs W, Taffet SM, Delmar M. pH regulation of connexin43: molecular analysis of the gating particle. Biophys J. 1996;71:1273–1284. doi: 10.1016/S0006-3495(96)79328-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seki A, Coombs W, Taffet SM, Delmar M. Loss of electrical communication, but not plaque formation, after mutations in the cytoplasmic loop of connexin43. Heart Rhythm. 2004;1:227–233. doi: 10.1016/j.hrthm.2004.03.066. [DOI] [PubMed] [Google Scholar]
- 16.Shibayama J, Lewandowski R, Kieken F, Coombs W, Shah S, Sorgen PL, Taffet SM, Delmar M. Identification of a novel peptide that interferes with the chemical regulation of connexin 43. Circ Res. 2006;98:1365–72. doi: 10.1161/01.RES.0000225911.24228.9c. [DOI] [PubMed] [Google Scholar]
- 17.Lewandowski R, Procida K, Vaidyanathan R, Coombs W, Jalife J, Nielsen MS, Taffet SM, Delmar M. RXP-E: a connexin43-binding peptide that prevents action potential propagation block. Circ Res. 2008;103:519–26. doi: 10.1161/CIRCRESAHA.108.179069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chea Y, Brooks RB, Marshall RG. Development of small molecules designed to modulate protein protein Interactions. J Comput Aided Mol Des. 2006;20:109–130. doi: 10.1007/s10822-006-9040-8. [DOI] [PubMed] [Google Scholar]
- 19.Oxford EM, Musa H, Maass K, Coombs W, Taffet SM, Delmar M. Connexin43 remodeling caused by inhibition of plakophilin-2 expression in cardiac cells. Circ Res. 2007;101:703–11. doi: 10.1161/CIRCRESAHA.107.154252. [DOI] [PubMed] [Google Scholar]
- 20.Lang BD, Delmar M, Coombs W. Surface plasmon resonance as a method to study the kinetics and amplitude of protein-protein binding. In: Dhein S, Mohr FW, Delmar M, editors. Practical Methods in Cardiovascular Research. Heidelberg, Germany: Springer; 2005. pp. 936–947. [Google Scholar]
- 21.Sorgen PL, Duffy HS, Spray DC, Delmar M. pH-dependent dimerization of the carboxyl terminal domain of Cx43. Biophys J. 2004;87:574–81. doi: 10.1529/biophysj.103.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno AP. Biophysical properties of homomeric and heteromultimeric channels formed by cardiac connexins. Cardiovasc Res. 2004;62:276–286. doi: 10.1016/j.cardiores.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Delmar M, Coombs W, Sorgen P, Duffy HS, Taffet SM. Structural bases for the chemical regulation of Connexin 43 channels. Cardiovasc Res. 2004;62:268–75. doi: 10.1016/j.cardiores.2003.12.030. [DOI] [PubMed] [Google Scholar]
- 24.Hirst-Jensen BJ, Sahoo P, Kieken F, Delmar M, Sorgen PL. Characterization of the pH-dependent interaction between the gap junction protein connexin43 carboxyl terminus and cytoplasmic loop domains. J Biol Chem. 2007;282:5801–5813. doi: 10.1074/jbc.M605233200. [DOI] [PubMed] [Google Scholar]