Abstract
Background and Purpose
The aim was to identify quantitative trait loci (QTL) for carotid intima-media thickness (CIMT) a risk factor for stroke and cardiovascular disease.
Methods
Probands were selected from Caribbean Hispanic subjects of the population-based Northern Manhattan Study. CIMT was measured by high resolution B-mode ultrasound and expressed as the mean (IMTx) and mean of the maximum (IMTm). Variance components methodology was used to detect linkage using SOLAR and calculate locus-specific heritability. Ordered-subset Analysis was done based on history of hypertension and total cholesterol levels.
Results
Among 100 Dominican families, 1390 subjects had CIMT measured (848 females; mean age 46.2 years). CIMT had a heritability of 0.65 after adjusting for age, ageˆ2, sex, cigarette pack-years, waist hip ratio, and BMI. Adjusted maximum multipoint LOD scores > 2 were found on chromosomes 14q (D14S606) and 7p (D7S817). Linkage to chromosome 14q was significantly increased in a subset of families with the greatest history of hypertension (MLOD=4.12). The QTL on Ch14q accounted for 0.21 of the heritability of IMTm, and on Ch7p 0.27 of the heritability of BIFm.
Conclusions
Several QTLs for CIMT were found on chromosomes 7p and 14q. The QTL on 14q replicates a suggestive linkage peak delimited in the Framingham Heart Study. These QTLs accounted for a substantial amount of trait heritability and warrant further fine mapping.
Keywords: Carotid Disease, Genetics, Linkage, Quantitative Traits, Risk Factors
The identification of the underlying genetic risk factors of stroke and cardiovascular diseases continue to elude us, particularly among the rapidly growing Hispanic population. Given the extreme complexity of genetic and environmental contributions to stroke, evaluation of risk factor phenotypes may reduce heterogeneity and facilitate gene discovery. Subclinical measurements of carotid intima-media thickness (IMT) offer the opportunity to evaluate the genetic determinants of these quantitative phenotypes. An AHA Scientific Statement advised that “Identification of important intermediate phenotypes (ie, phenotypes that mediate disease as opposed to phenotypes that represent the ultimate manifestation of a disease) may prove to be more amenable to genetic analysis.”1 The Genetics Panel of the NINDS Stroke Progress Review Group noted that “Quantitative intermediate phenotypes, such as IMT…offer significant potential for future stroke genetic studies.”2
Subclinical carotid disease is a well-documented risk marker for vascular disease. Ultrasound measures of carotid IMT have been demonstrated to be valid measures of pathologically-defined atherosclerosis, highly reproducible, associated with vascular risk factors, and predictive of stroke and myocardial infarction.3 4 5 6 7 Although prior studies have used measurements that have included carotid IMT, as well as plaque, there have differential relationships to risk factors and vascular outcomes.8 Carotid IMT is considered a separate phenotype from carotid plaque and likely to have distinct genetic determinants.9 10 The heritability of carotid IMT ranges from 0.30 to 0.60 depending on the study population.11 12 13 14
The overall goal of the Family Study of Stroke Risk and Carotid Atherosclerosis is to identify genetic determinants of specific cerebrovascular risk phenotypes which are precursors to stroke. The aim of the work presented here was to detect quantitative trait loci (QTL) for carotid IMT by performing a linkage analysis among high-risk Caribbean Hispanic families.
Methods
The details of the design of the family study have been described.15 In brief, a family study was derived from selected members of the Northern Manhattan Study (NOMAS). Probands were selected from high-risk Caribbean Hispanic members of NOMAS defined by the following criteria: (1) reporting a sibling with a history of myocardial infarction or stroke; or (2) having 2 of 3 quantitative risk phenotypes (maximal carotid plaque thickness, left ventricular mass, or homocysteine level above the 75th percentile in the NOMAS cohort). The majority of the probands (80%) were recruited based on the first criteria. Our family study was designed to enroll 1500 subjects to enable the detection of QTLs for traits with reasonable heritability. With our 100 families, we had over 80% power to obtain LOD scores over 2.0 for traits with a heritability greater than 0.18.
Families were enrolled if the proband was able to provide a family history, obtain the family member's permission for the research staff to contact them, and had at least 3 first-degree relatives able to participate. Although the proband was identified in Northern Manhattan, we enrolled family members in New York at Columbia University and in the Dominican Republic at the Clinicas Corazones Unidos in Santo Domingo. All subjects provided informed consent and the study was approved by the Institutional Review Boards of Columbia University, University of Miami, the National Bioethics Committee, and the Independent Ethics Committee of Instituto Oncologico Regional del Cibao in the Dominican Republic (DR).
Demographic, socioeconomic and risk factor data were collected through direct interview based on NOMAS instruments.16 Extensive questionnaires regarding hypertension, diabetes, smoking, alcohol use, and physical activity were completed. Measurements of height, weight, hip and waist circumference, and skin-fold thickness were obtained, as were serial blood pressures. Fasting blood was collected and processed for lipids (total cholesterol, LDL, triglyceride, HDL) and glucose. Extraction of DNA was done by the Columbia University Genome Center.
Phenotype Data: Carotid Measurements
All family members had high resolution B-mode ultrasound to measure carotid IMT. Carotid ultrasound was performed according to standard scanning and reading protocols by a trained and certified sonologist as detailed previously. 11 12 17 In brief, the carotid IMT protocols yield measurements of the distance between lumen-intima and media-adventitia ultrasound echoes, from which the IMT and arterial diameter are derived for the 3 carotid segments. The carotid segments were defined as follows: (1) near and far wall of the segment extending from 10 to 20 mm proximal to the tip of the flow divider into the common carotid artery (CCA); (2) near and far wall of the carotid bifurcation beginning at the tip of the flow divider and extending 10 mm proximal to the flow divider tip (BIF); and (3) near and far wall of the proximal 10 mm of the internal carotid artery (ICA).
IMT measurements were performed outside the areas of plaque as recommended by consensus documents.18 Plaque was defined as a focal thickening greater than 50% of the surrounding wall thickness. IMT was measured using an automated computerized edge tracking software M'Ath (Intelligence in Medical Technologies, Inc., Paris, France) from the recorded ultrasound clips which improves precision and reduces variance of the measurements. 19 Total IMT is calculated as a mean composite measure of the means of the near and the far wall IMT of all carotid sites (IMTx), and the maximum of the near and the far wall IMT of all carotid sites (IMTm). We also examined carotid segment-specific IMT phenotypes (BIFx, BIFm, CCAx, CCAm, ICAx, ICAm). Our carotid IMT reliability statistics demonstrated excellent results.20 Among 88 subjects, inter-reader reliability between 2 readers was demonstrated with a mean absolute difference in IMT of 0.11±0.09 mm, variation coefficient 5.5%, correlation coefficient 0.87, and the percent error 6.7%. Intra-reader mean absolute IMT difference was 0.07±0.04 mm (CCA near wall 0.06±0.05 mm and CCA far wall 0.04±0.04 mm), variation coefficient 5.4%, correlation coefficient 0.94, and the percent error 5.6%. In our laboratory, we have found that the measurement between near and far wall is reliable with comparable inter-reader reliability. The proportions of obtainable IMT measurements per carotid segment were: CCA near wall 95.5%, CCA far wall 95.7%; BIF near wall 87.9%, BIF far wall 91.6%; ICA near wall 70.6%, and ICA far wall 79.6%. Over 85% of subjects had measurements obtainable from 9 or more of the 12 carotid IMT sites.
Genotype Data
DNA was sent to the Center for Inherited Disease Research (CIDR) for genotyping. At CIDR, a 10 cM screen of 405 STR markers was performed after quality checks. STR genotypes were used to verify and adjust family structure using the programs Relpair and PREST.21 22 Mendelian error checking was performed on the final family structure using Pedcheck.23
Statistics
Variance components methodology in SOLAR was used to calculate two-point and multipoint LOD scores and identify QTLs.24 25 26 Heritability was evaluated using a pedigree-based maximum-likelihood method implemented.27 Since SOLAR requires that quantitative traits be normally distributed, traits were natural-log transformed and multiplied or shifted. Observations beyond 3 to 4 SD from the mean were dropped to ensure normality. An initial polygenic model for each trait was used to estimate significant covariates (p<0.10) that were used in all final analyses. The standard parameterization in SOLAR that we used represents a proportion of the total variance after the effect of all covariates has been removed. Thus, the residuals of the trait are used for analysis and checked for normality (kurtosis < 0.8) before proceeding.
Covariates that were tested included age, sex, waist hip ratio, body mass index, and history of hypertension, hypercholesterolemia, diabetes, and smoking. Most covariates were used as continuous variables while standard definitions were used for categorical covariates. Hypertension was defined as reported history of high blood pressure, systolic blood pressure ≥ 140 mmHg, or diastolic blood pressure ≥ 90 mmHg. Diabetes was defined by history or fasting blood sugar ≥ 126 mg/dL. Hypercholesterolemia was defined by history or total cholesterol > 240 mg/dL. Smoking was defined as never versus ever, and pack years were calculated as number of cigarette packs per day × years smoked.
Allele-sharing models were obtained by estimating identity by descent (IBD) for each marker. LOD scores were calculated using a log (base 10) ratio of the likelihoods of the polygenic and marker-specific models. Empirical p-values were calculated for each trait based on 10,000 replicates in which a fully-informative marker, unlinked to a given trait, was simulated and used to compute possible LOD scores. Locus-specific heritability, h2q (heritability attributed to the QTL), was calculated for specific loci after adjusting for the significant covariates.
Additionally, we performed ordered subsets linkage analysis (OSA).28 We ranked families separately by the percent of hypertension (percent with SBP > 140) and mean total cholesterol level in a family. For each ranking trait and ordering strategy, we added family-specific LOD scores in trait rank order until all families were included to evaluate the linkage results for chromosomes 14 and 7. For each ordering strategy and for each carotid trait, a permutation procedure was implemented to generate empirical p-values to test the hypothesis that ordering by family phenotype gave stronger linkage than random ordering. Specifically, 10,000 random family orderings were permuted and the maximum LOD scores from each random ordering were compared with the OSA results to derive the empirical p-values.
Results
Overall, 1508 subjects from 110 families (Dominican Republic 100, Puerto Rico 4, Cuba 2, Ecuador 2, Nicaragua 1, and Colombia 1) were enrolled. The mean family size was 13.9 ± 7.7; median 12.5, and range 3–90. Thirty percent (456) of the subjects were enrolled in the Dominican Republic. To reduce heterogeneity we restricted our analyses to the 100 Dominican families. The characteristics of these Dominican families were: 2184 individuals, 1460 sib pairs, 452 half-sib pairs, and 2273 avuncular pairs; 1390 subjects had carotid phenotypes measured (542 males; 848 females). Clinical characteristics are shown in Table 1. Our family study consisted of younger individuals (mean age 46.2 years) and the prevalence of small carotid plaques was only 22.8%.
Table 1.
Sociodemographic, vascular risk factors, and phenotype measurements among the 1390 subjects from 100 Dominican Families
| Demographics | % (n) | Phenotypes | |
| Sex (Female) | 61.3 (852) | Carotid plaque (All Subjects) | 22.8% (313) |
| ≥ High School Education | 49.0 (681) | Carotid plaque (Over Age 55) | 53% (224) |
| Enrolled in the DR | 32.2 (447) | Mean ± SD | |
| Mean Age ± SD | 46.2 ± 17.4 | Carotid IMT (mm) | 0.66 ± 0.08 |
| Carotid Max IMT (mm) | 0.84 ± 0.10 | ||
| Vascular Risk Factors | % | Carotid Bifurcation IMT (mm) | 0.68 ± 0.10 |
| Hypertension# | 40.2 (559) | Internal Carotid IMT (mm) | 0.61 ± 0.07 |
| Diabetes | 14.0 (194) | Common Carotid IMT (mm) | 0.62 ± 0.10 |
| Coronary artery diseaseˆ | 22.2 (308) | ||
| No physical activity | 56.6 (761) | ||
| Moderate alcohol intake | 48.1 (669) | ||
| Current smoker | 14.0 (195) | ||
| Mean ± SD | |||
| Body Mass Index (kg/m2) | 28.71 ± 5.8 | ||
| Waist Hip Ratio (WHR) | 0.90 ± 0.1 | ||
| Triceps skinfold thickness (mm) | 26.9 ± 11.7 | ||
| Fast Glucose (mg/dl) | 91.8 ± 37.7 | ||
| Total cholesterol (mg/dl)* | 185.4 ± 42.3 | ||
| LDL (mg/dl)* | 110.3 ± 35.1 | ||
| HDL (mg/dl) | 50.1 ± 13.5 | ||
| TG (mg/dl) | 126.4 ± 93.2 | ||
| SBP (mmHg)# | 122.0 ± 19.9 | ||
| DBP (mmHg)# | 77.2 ± 10.9 |
defined by self-report of a history of MI, angina, or prior interventional cardiac procedures
14% on statins;
28% on anti-hypertensives
Final heritability estimates of our carotid IMT phenotypes are shown in Table 2. Total mean carotid IMT had a heritability of 0.65 after adjusting for age, ageˆ2, sex, cigarette pack-years, body mass index, and waist hip ratio. Additionally, hypertension and diabetes met our covariate inclusion criteria (p < .10) for CCAx. Both age and ageˆ2 were retained as significant covariates in our heritability estimates. Heritabilities ranged from 0.41 to 0.65 across the different carotid segments and were significant for all segments.
Table 2.
Adjusted total heritability estimates of mean and max carotid IMT overall and stratified by specific carotid segments and the covariates associated with the trait and added to the adjusted models
| Trait | h2 ± SE | p-value | Variance* | Covariates |
|---|---|---|---|---|
| Mean Total IMT (IMTx) | 0.65 ± 0.05 | < 0.0001 | 0.51 | Age, ageˆ2,sex, PackYears, WHR, BMI |
| Mean Bifurcation IMT (BIFx) | 0.58 ± 0.05 | < 0.0001 | 0.44 | Age,ageˆ2, sex, PackYears, WHR, BMI |
| Mean Internal Carotid IMT (ICAx) | 0.47 ± 0.05 | < 0.0001 | 0.21 | Age, ageˆ2,sex, hypercholesterolemia, WHR, BMI |
| Mean Common Carotid IMT (CCAx) | 0.56 ± 0.05 | < 0.0001 | 0.54 | Age, sex, hypertension, diabetes, PackYears, WHR BMI |
| Max Total IMT (IMTm) | 0.62 ± 0.05 | < 0.0001 | 0.52 | Age, ageˆ2,sex, PackYears, WHR, BMI |
| Max Bifurcation IMT (BIFm) | 0.51 ± 0.05 | < 0.0001 | 0.47 | Age, ageˆ2, sex, age*sex, PackYears, WHR, BMI |
| Max Internal Carotid IMT (ICAm) | 0.41 ± 0.06 | < 0.0001 | 0.2 | Age, ageˆ2, sex, PackYears, WHR, BMI |
| Max Common Carotid IMT (CCAm) | 0.48 ± 0.05 | < 0.0001 | 0.53 | Age, sex, hypertension, hypercholesterolemia, diabetes, PackYears, WHR, BMI |
WHR – waist hip ratio
BMI – body mass index
Proportion of variance due to all final covariates
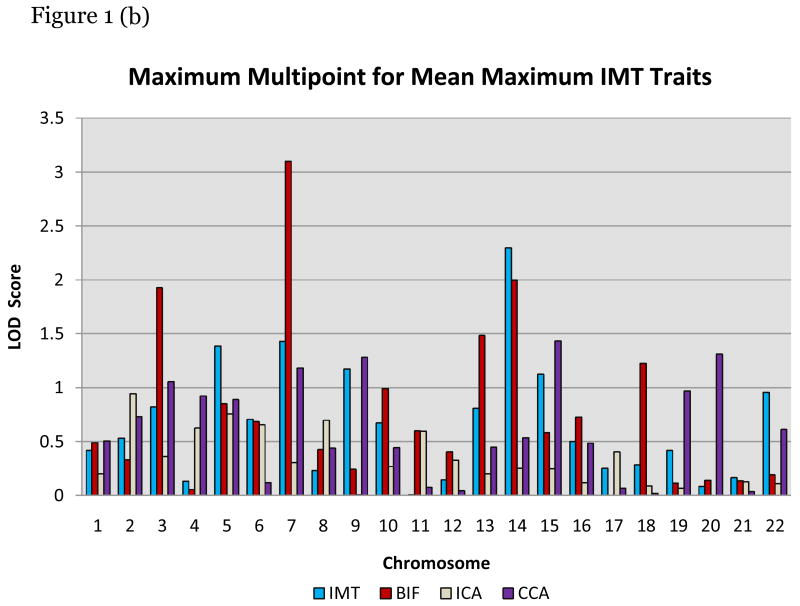
Maximum multipoint LOD scores > 1.5 are listed in Table 3. LOD scores >2 suggestive of linkage were found on chromosomes 7p and 14q (Figure 1). The maximum LOD score on 7p was at D7S817 for BIFm and BIFx. The maximum LOD score on chromosome 14q was at D14S606 for IMTm and BIFm and BIFx also map to this point. (Figure 2) Among the 69 families with the highest percentage of hypertension, our LOD score significantly increased (p = 0.0002) to 4.12 for IMTm on chromosome 14q and to 3.3 (p=0.0164) among the 80 families with lower total mean cholesterol levels.
Table 3.
SOLAR Multipoint LOD Scores > 1.5 by Trait and Ordered Subset Analyses ranking families by various systolic blood pressure and total cholesterol
| Trait | Location | CM | Marker | LOD Score | p-value | h2q | OSA % SBP > 140 (H to L) Max LOD (p-value, # family) |
OSA % SBP > 140 (L to H) Max LOD (p-value, # family) |
OSA Cholesterol (H to L) Max LOD (p-value, # family) |
OSA Cholesterol (L to H) Max LOD (p-value, # family) |
|---|---|---|---|---|---|---|---|---|---|---|
| IMTm | 14q31.1 | 95 | D14S606 | 2.3 | 0.0018 | 0.21 |
4.12 (0.0002, 69) |
2.30 (1, 100) |
2.54 (0.4055, 83) |
3.30 (0.0164, 80) |
| IMTx | 14q31.1 | 93 | D14S606 | 1.95 | 0.18 |
2.86 (0.0192, 69) |
1.95 (1, 100) |
2.21 (0.3866, 71) |
2.67 (0.056, 80) |
|
| BIFm | 3q11.2 | 119 | D3S2459 | 1.93 | 0.19 | |||||
| BIFm | 7p14.3 | 51 | D7S817 | 3.10 | 0.27 | 3.32 (0.4201, 74) |
3.13 (0.7167, 94) |
3.10 (1, 100) |
3.44 (0.2726, 97) |
|
| BIFm | 14q31.1 | 92 | D14S606 | 2.00 | 0.20 | |||||
| BIFx | 7p14.3 | 51 | D7S817 | 2.88 | 0.25 | 3.36 (0.1475, 74) |
3.09 (0.3916, 96) |
2.88 (1, 100) |
3.26 (0.221, 97) |
|
| BIFx | 14q31.1 | 95 | D14S606 | 1.63 | 0.17 | |||||
| CCAx | 5p13.3 | 45 | D5S1470 | 1.54 | 0.18 |
LOD SCORE=SOLAR multipoint LOD score
h2q=locus-specific heritability
p-value=empirical p-value based on 10,000 replicates
H to L = High to Low ranking
L to H = Low to High ranking
Figure 1.
(a) Maximum multipoint linkage results for mean overall IMT (IMTx) and at individual carotid sites (common carotid CCA, internal carotid ICA, and carotid bifurcations (BIF). (b) Maximum multipoint linkage results for mean of the maximum measurements for overall IMTm and the specific segments.
Figure 2.
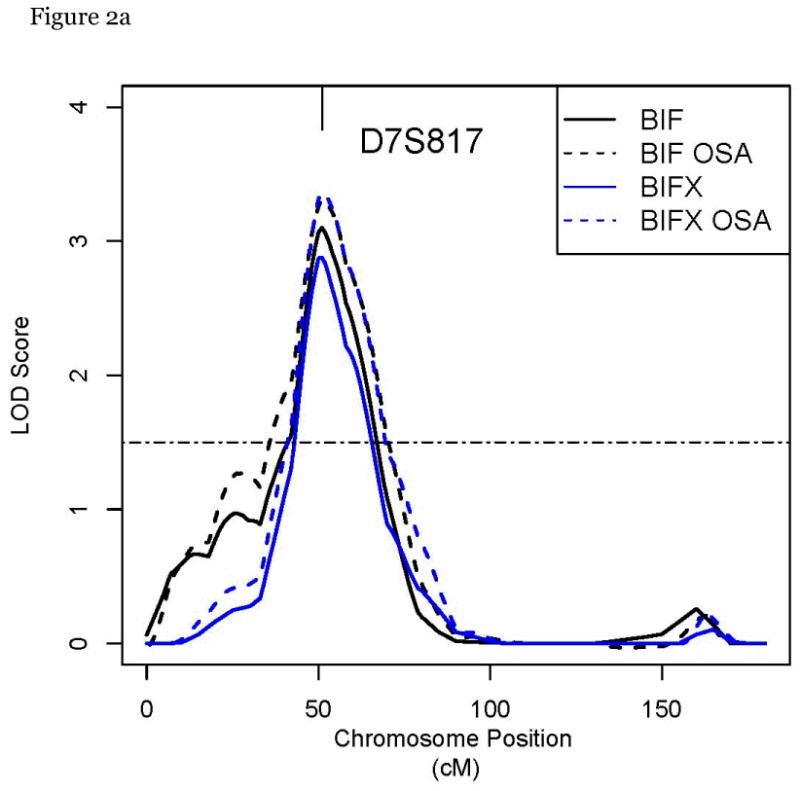

a, b: Multipoint linkage plots for chromosome 7p and 14q for our IMT traits including OSA by the prevalence of elevated systolic BP > 140
Discussion
The main goal of our Family Study is to identify susceptibility genes for cerebrovascular risk phenotypes, such as subclinical carotid disease, and thereby facilitate the search for genetic mechanisms of stroke and cardiovascular disease. We have established the high heritability of carotid IMT within our Dominican families and completed a linkage analysis as a hypothesis-generating method to identify regions of interest that might contain genes influencing our quantitative traits. We have identified peaks on chromosomes 7p and 14q to be pursued with fine mapping. Moreover, these QTLs appear to explain a reasonable amount of the overall heritability in our trait. The results from our QTL analysis of our Dominican Family Study are the first stage in identifying new genes for carotid IMT.
Several studies have evaluated candidate genes for carotid IMT, however, the results have been inconsistent.29 Of the many candidate gene studies reviewed, only one variant, the 5A/6A polymorphism of the matrix metalloproteinase (MMP3) gene, showed consistently positive associations with carotid IMT in a small number of studies. Studies provide evidence for modest associations for variants in APOE, Angiotensin Converting Enzyme (ACE) and NOS3 candidate gene with carotid IMT. In our former candidate gene study, carotid IMT was associated with allelic variants of stromelysin-1 (MMP3), interleukin 6, and hepatic lipase.17 Of these candidate genes, three are located on chromosome 7 (PON1, NOS3, and interleukin-6), but not near our linkage peak, and one is on chromosome 15 (hepatic lipase).
Multiple genes are likely to influence carotid IMT which could account for some of the variation in findings among genetic studies. Other factors include population characteristics, genetic heterogeneity, small sample sizes and sampling errors, possible confounding, gene–environment interactions, and differences in IMT measurement methods. Findings from white populations may not be the same when evaluated in Hispanic populations. Moreover, candidate gene studies are limited to the apriori candidates chosen, while family study designs allow for gene discovery.
Other family studies have investigated carotid IMT. In the NHLBI Offspring Cohort of the Framingham Study, there was no association for carotid IMT meeting criteria for genome-wide significance.30 However, 11 SNPs with p < 10-5 were identified for maximum internal carotid IMT and 5 SNPs with p < 10-5 by family based association testing (FBAT) for mean common carotid IMT. In addition, several regions of linkage to internal carotid IMT were identified on chromosome 12, confirming previous results from the same population.31
Suggestive evidence for linkage with carotid IMT was reported at our peak at D14S606 (two point LOD=1.75) in Framingham Heart Study Offspring cohort.27 Replication of linkage evidence in our study at D14S606 substantially corroborates this QTL. This marker has also been associated with arterial stiffness in African Americans in the Hypertension Genetic Epidemiology Network study.32 Candidate genes near D14S606 include secreted modular calcium binding protein (SMOC-1), a secreted glycoprotein that mediates cell-matrix interactions and fibulin-5 precursor (FBLN5), a secreted protein that promotes cell-cell adhesion. Both of them have been implicated in vascular remodeling and therefore could contribute to inter-individual IMT variations.
Linkage peak on chromosomes 7p has never been previously reported in relation to carotid IMT. Several candidate genes are located within the chromosomal 7p linkage region, including neuropeptide Y (main effect in increased food intake and decreased physical activity), lamin A/C (implicated in familial partial lipodystrophy, a syndrome of monogenic insulin resistance and diabetes), and secreted frizzled-related protein 4 precursor (SFRP4, an antagonist of the WNT/β-Catenin signaling pathway). Mounting evidence has suggested that the WNT/β-Catenin signaling pathway regulates vascular smooth muscle proliferation and apoptosis. It is conceivable that perturbation of this important pathway would lead to phenotypic variations of IMT.
Our Family Study is uniquely designed to evaluate the genetic precursors of stroke and cardiovascular disease with a focus on subclinical disease in Dominicans. A number of family studies have investigated the contribution of genetics to cardiovascular disease, but primarily among white non-Hispanic populations. The San Antonio Family Heart Study includes Mexican Americans while the Multi-ethnic Study of Atherosclerosis (MESA) now includes a family cohort of 450 Hispanic families, mainly sib trios.33 34 Only MESA includes some Caribbean Hispanics and is capable of evaluating the genetics of cerebrovascular risk phenotypes, such as carotid disease. Carotid IMT was also evaluated in the Mexican American Coronary Artery Disease family study.35 The multivariate adjusted heritability was 0.40 and the strongest evidence of linkage was on chromosome 2 at D2S2944 with smaller LOD scores on chromosomes 6 and 13. Although these results may help generate hypotheses for several genes that may be evaluated for association with subclinical atherosclerosis, the validation in different race-ethnic populations is lacking.
Several studies have shown that OSA offers promise in dissecting the genetic basis for complex traits by reducing heterogeneity and increasing statistical power.28 We have found that Dominican families with a higher prevalence of hypertension significantly contributed to the linkage signal on chromosome 14q for carotid traits. This suggests that loci in chromosome 14q may interact with hypertension to determine carotid variations in Dominican populations. To avoid potential confounding on phenotype-genotype correlation, this interaction should be considered in future fine mapping or association studies.
The strengths of our study are the well characterized families, large family sizes, and detailed systematic measurements of our quantitative phenotype. By focusing on one ethnic group using a family study design we have minimized the effects of heterogeneity. Approaches to mapping quantitative phenotypes also offer efficient statistical advantages over discrete traits. Some of the limitations of our study include the focus on one ethnic group in whom findings may not be applicable to other ethnic groups. Even within one ethnic group there may still be genetic heterogeneity which could confound our results. Our findings require fine mapping to identify the genes and variations underlying carotid IMT traits.
The American Heart Association has called for research that prioritizes studies that “characterize genes and genetic variants that are associated with cardiovascular disease across individuals, communities, and populations” and “develop new technologies in cardiovascular disease characterization, risk assessment, and outcome prediction.”1 Finding susceptibility genes for carotid IMT may lead to the discovery of new pathogenetic pathways for the development of carotid disease, and ultimately stroke, provide insights into disease modifying treatments, and aid in the development of earlier risk prediction and prevention. Our Family Study provides essential data on Caribbean Hispanics not available from other studies and helps fill gaps in our knowledge of the genetics of cerebrovascular risk phenotypes in minority populations.
Acknowledgments
The authors are grateful to all the families and research staff who participated in the study. We thank Drs. Katihurca Almonte and Carlos Garcia Lithgow for their support in the Dominican Republic. We also thank Dr. Luis Cuello Mainardi, Director of the Clinicas Corazones Unidos, where subjects were enrolled in the DR, and Drs. Rafael Lantigua and Andres Peralta for their guidance with regulatory environment in the DR.
This research was supported by grants from the National Institute of Neurological Disorders and Stroke R01 NS NS40807 and RO1 NS047655.
Footnotes
Disclosures: The authors have no conflicts of interest to disclose regarding this work.
References
- 1.Arnett DK, Baird AE, Barkley RA, Basson CT, Boerwinkle E, Ganesh SK, Herrington DM, Hong Y, Jaquish C, McDermott DA, O'Donnell CJ. Relevance of genetics and genomics for prevention and treatment of cardiovascular disease. Circulation. 2007;115:2878–901. doi: 10.1161/CIRCULATIONAHA.107.183679. [DOI] [PubMed] [Google Scholar]
- 2.http://www.ninds.nih.gov/find_people/groups/stroke_prg/index.htm.
- 3.Chambless LE, Folsom AR, Clegg LX, Sharrett AR, Shahar E, Nieto FJ, Rosamond WD, Evans G. Carotid wall thickness is predictive of incident clinical stroke: the Atherosclerosis Risk in Communities (ARIC) study. Am J Epidemiol. 2000;151:478–487. doi: 10.1093/oxfordjournals.aje.a010233. [DOI] [PubMed] [Google Scholar]
- 4.Zureik M, Ducimetiere P, Touboul PJ, Courbon D, Bonithon-Kopp C, Berr C, Magne C. Common carotid intima-media thickness predicts occurrence of carotid atherosclerotic plaques: longitudinal results from the Aging Vascular Study (EVA) study. Arterioscler Thromb Vasc Biol. 2000;20:1622–1629. doi: 10.1161/01.atv.20.6.1622. [DOI] [PubMed] [Google Scholar]
- 5.O'Leary DH, Polak JF, Kronmal RA, et al. Carotid-artery intima and media thickness as a risk factor for myocardial infarction and stroke in older adults. N Engl J Med. 1999;340:14–24. doi: 10.1056/NEJM199901073400103. [DOI] [PubMed] [Google Scholar]
- 6.Sharrett AR, Ding J, Criqui MH, Saad MF, Liu K, Polak JF, Folsom AR, Tsai MY, Burke GL, Szklo M. Smoking, diabetes, and blood cholesterol differ in their associations with subclinical atherosclerosis: The Multiethnic Study of Atherosclerosis (MESA) Atherosclerosis. 2005;186:441–447. doi: 10.1016/j.atherosclerosis.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Touboul PJ, Labreuche J, Vicaut E, Amarenco P, GENIC Investigators Carotid intima-media thickness, plaques, and Framingham risk score as independent determinants of stroke risk. Stroke. 2005;36:1741–1745. doi: 10.1161/01.STR.0000174490.23495.57. [DOI] [PubMed] [Google Scholar]
- 8.Johnsen SH, Mathiesen EB. Carotid plaque compared with intima-media thickness as a predictor of coronary and cerebrovascular disease. Curr Cardiol Rep. 2009;11(1):21–7. doi: 10.1007/s11886-009-0004-1. [DOI] [PubMed] [Google Scholar]
- 9.Pollex RL, Hegele R. Genetic determinants of carotid ultrasound traits. Curr Atheroscler Rep. 2006 May;8(3):206–15. doi: 10.1007/s11883-006-0075-z. [DOI] [PubMed] [Google Scholar]
- 10.Spence JD, Hegele RA. Noninvasive phenotypes of atherosclerosis. Arterioscler Thromb Vasc Biol. 2004 Nov;24(11):e188. doi: 10.1161/01.ATV.0000146160.22637.33. [DOI] [PubMed] [Google Scholar]
- 11.Lange LA, Bowden DW, Langefeld CD, Wagenknecht LE, Carr JJ, Rich SS, et al. Heritability of carotid artery intima-medial thickness in type 2 diabetes. Stroke. 2002;33:1876–81. doi: 10.1161/01.str.0000019909.71547.aa. [DOI] [PubMed] [Google Scholar]
- 12.Xiang AH, Azen SP, Buchanan TA, Raffel LJ, Tan S, Cheng LS, et al. Heritability of subclinical atherosclerosis in Latino families ascertained through a hypertensive parent. Arterioscler Thromb Vasc Biol. 2002;22:843–8. doi: 10.1161/01.atv.0000015329.15481.e8. [DOI] [PubMed] [Google Scholar]
- 13.Fox CS, Polak JP, Chazaro I, Cupples A, Wolf PA, D'Agostino RA, O'Donnell CJ. Genetic and Environmental Contributions to Atherosclerosis Phenotypes in Men and Women. Heritability of Carotid Intima-Media Thickness in the Framingham Heart Study. Stroke. 2003;34:397–401. doi: 10.1161/01.str.0000048214.56981.6f. [DOI] [PubMed] [Google Scholar]
- 14.Juo SH, Lin HF, Rundek T, Sabala EA, Boden-Albala B, Park N, Lan MY, Sacco RL. Genetic and environmental contributions to carotid intima-media thickness and obesity phenotypes in the Northern Manhattan Family Study. Stroke. 2004;35:2243–7. doi: 10.1161/01.STR.0000142132.20442.d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sacco RL, Sabala EA, Rundek T, Juo SH, Huang JS, DiTullio M, Homma S, Almonte K, Lithgow CG, Boden-Albala B. Design of a family study among high-risk Caribbean Hispanics: the Northern Manhattan Family Study. Ethn Dis. 2007;17:351–7. [PMC free article] [PubMed] [Google Scholar]
- 16.Elkind MSV, Sciacca RR, Boden-Albala B, Rundek T, Paik MC, Sacco RL. Moderate Alcohol Consumption Reduces Risk of Ischemic Stroke: The Northern Manhattan Study. Stroke. 2006;37:13–19. doi: 10.1161/01.STR.0000195048.86810.5b. [DOI] [PubMed] [Google Scholar]
- 17.O'Leary DH, Polak JF, Wolfson SK, Jr, Bond MG, Bommer W, Sheth S, Psaty BM, Sharrett AR, Manolio TA, CHS Collaborative Research Group Use of sonography to evaluate carotid atherosclerosis in the elderly: the Cardiovascular Health Study. Stroke. 1991;22:1155–1163. doi: 10.1161/01.str.22.9.1155. [DOI] [PubMed] [Google Scholar]
- 18.Touboul PJ, Hennerici MG, Meairs S, Adams H, Amarenco P, Bornstein N, Csiba L, Desvarieux M, Ebrahim S, Fatar M, Hernandez Hernandez R, Jaff M, Kownator S, Prati P, Rundek T, Sitzer M, Schminke U, Tardif JC, Taylor A, Vicaut E, Woo KS, Zannad F, Zureik M. Mannheim carotid intima-media thickness consensus (2004-2006) Cerebrovasc Dis. 2007;23:75–80. doi: 10.1159/000097034. [DOI] [PubMed] [Google Scholar]
- 19.Touboul PJ, Vicaut E, Labreuche J, Belliard JP, Cohen S, Kownator S, Pithois-Merli I, Paroi Artérielle et Risque Cardiovasculaire Study Investigators Design, baseline characteristics and carotid intima-media thickness reproducibility in the PARC study. Cerebrovasc Dis. 2005;19:57–63. doi: 10.1159/000081913. [DOI] [PubMed] [Google Scholar]
- 20.Rundek T, Elkind MS, Pittman J, Boden-Albala B, Martin S, Humphries SE, Hank Juo SH, Sacco RL. Carotid Intima-Media Thickness is Associated with Allelic Variants of Stromelysin-1, Interleukin-6 and Hepatic Lipase Genes: The Northern Manhattan Prospective Cohort Study. Stroke. 2002;333:1420–1423. doi: 10.1161/01.STR.0000015558.63492.B6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epstein MP, Duren WL, Boehnke M. Improved inference of relationships for pairs of individuals. American Journal of Human Genetics. 2000;67:1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McPeek, Sun Statistical Tests for Detection of Mis-specified Relationships by Use of Genome-Screen Data. American Journal of Human Genetics. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Connell JR, Weeks DE. PedCheck: A program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet. 63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amos CI, Krushkal J, Thiel TJ, Young A, Zhu DK, Boerwinkle E, de Andrade M. Comparison of model-free linkage mapping strategies for the study of a complex trait. Genet Epidemiol. 1997;14:743–748. doi: 10.1002/(SICI)1098-2272(1997)14:6<743::AID-GEPI30>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 25.Schork NJ. Extended multipoint identity-by-descent analysis of human quantitative traits: efficiency, power, and modeling considerations. Am J Hum Genet. 1993;53:1306–1319. [PMC free article] [PubMed] [Google Scholar]
- 26.Schork NJ. Extended pedigree patterned covariance matrix mixed models for quantitative phenotype analysis. Genet Epidemiol. 1992;9:73–86. doi: 10.1002/gepi.1370090202. [DOI] [PubMed] [Google Scholar]
- 27.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meigs JB, Manning AK, Dupuis J, Liu C, Florez JC, Cupples LA. Ordered Stratification to Reduce Heterogeneity in Linkage to Diabetes-related Quantitative Traits. Obesity. 2008;16:2314–22. doi: 10.1038/oby.2008.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manolio TA, Boerwinkle E, O'Donnell CJ, Wilson AF. Genetics of ultrasonographic carotid atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:1567–77. doi: 10.1161/01.ATV.0000138789.11433.c1. [DOI] [PubMed] [Google Scholar]
- 30.O'Donnell CJ, Cupples LA, D'Agostino RB, Fox CS, Hoffmann U, Hwang SJ, Ingellson E, Liu C, Murabito JM, Polak JF, Wolf PA, Demissie S. Genome-wide association study for subclinical atherosclerosis in major arterial territories in the NHLBI's Framingham Heart Study. BMC Med Genet. 2007;8 1:S4. doi: 10.1186/1471-2350-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fox CS, Cupples LA, Chazaro I, Polak JF, Wolf PA, D'Agostino RB, Ordovas JM, O'Donnell CJ. Genome wide linkage analysis for internal carotid artery intimal medial thickness: evidence for linkage to chromosome 12. Am J Hum Genet. 2004;74:253–61. doi: 10.1086/381559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherva R, Miller MB, Lynch AI, Devereux RB, Rao DC, Oberman A, Hopkins PN, Kitzman DW, Atwood LD, Arnett DK. A whole genome scan for pulse pressure/stroke volume ratio in African Americans: the HyperGEN study. Am J Hypertens. 2007;20:398–402. doi: 10.1016/j.amjhyper.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell BD, Kammerer CM, Blangero J. Genetic and Environmental Contributions to Cardiovascular Risk Factors in Mexican Americans The San Antonio Heart Study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- 34.http://www.mesa-nhlbi.org/MesaFamily/StudyOverview.aspx
- 35.Wang D, Yang H, Quiñones MJ, Bulnes-Enriquez I, Jimenez X, De La Rosa R, Modilevsky T, Yu K, Li Y, Taylor KD, Hsueh WA, Hodis HN, Rotter JI. A genome-wide scan for carotid artery intima-media thickness: the Mexican-American Coronary Artery Disease family study. Stroke. 2005;36:540–5. doi: 10.1161/01.STR.0000155746.65185.4e. [DOI] [PubMed] [Google Scholar]