Abstract
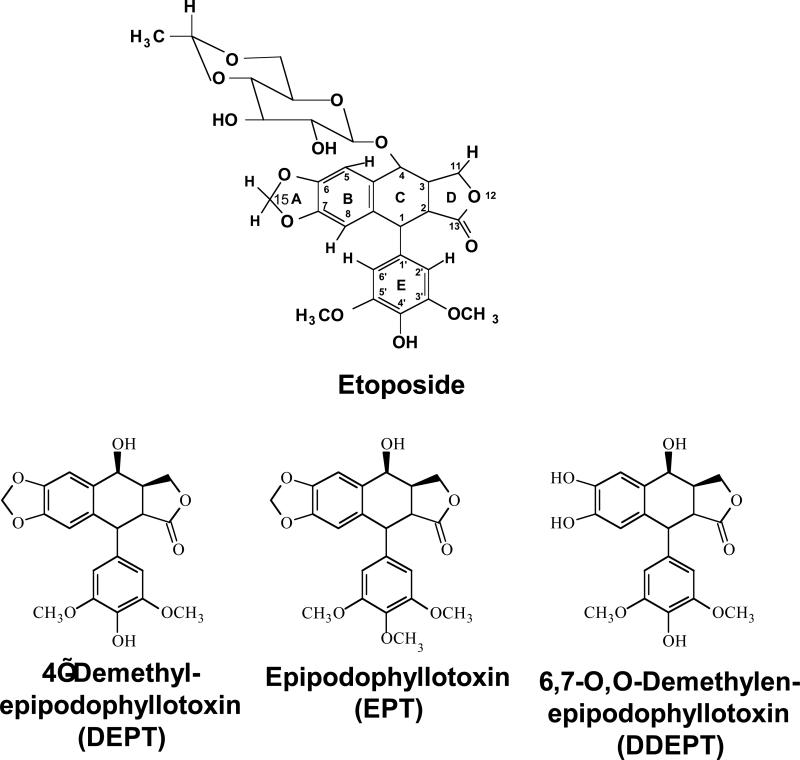
Etoposide is a widely prescribed anticancer agent that stabilizes topoisomerase II-mediated DNA strand breaks. The drug contains a polycyclic ring system (rings A–D), a glycosidic moiety at C4, and a pendant ring (E–ring) at C1. A recent study that focused on yeast topoisomerase II demonstrated that the H15 geminal protons of the etoposide A–ring, the H5 and H8 protons of the B–ring, and the H2’, H6’, 3’–methoxyl, and 5’–methoxyl protons of the E–ring contact topoisomerase II in the binary enzyme-drug complex [Wilstermann et al. (2007) Biochemistry 46, 8217−8225]. No interactions with the C4 sugar were observed. The present study used DNA cleavage assays, saturation transfer difference [1H]-NMR spectroscopy, and enzyme-drug binding studies to further define interactions between etoposide and human topoisomerase IIα. Etoposide and three derivatives that lacked the C4 sugar were analyzed. Except for the sugar, 4’-demethyl epipodophyllotoxin is identical to etoposide, epipodophyllotoxin contains a 4’-methoxyl group on the E–ring, and 6,7-O,O-demethylenepipodophyllotoxin replaces the A–ring with a diol. Results suggest that etoposide–topoisomerase IIα binding is driven by interactions with the A– and B–rings, and potentially by stacking interactions with the E–ring. We propose that the E–ring pocket on the enzyme is confined, as the addition of bulk to this ring adversely affects drug function. The A– and E–rings do not appear to contact DNA in the enzyme-drug-DNA complex. Conversely, the sugar moiety subtly alters DNA interactions. The identification of etoposide substituents that contact topoisomerase IIα in the binary complex has predictive value for drug behavior in the enzyme-etoposide-DNA complex.
Etoposide is a highly successful anticancer agent that has been used to treat a variety of human malignancies since the early 1980's (1-4). The drug is a semi-synthetic derivative of podophyllotoxin, a naturally occurring antimitotic agent from may apples that has been used as an herbal remedy for more than a millennium (1, 5).
The primary cellular target for etoposide is topoisomerase II (1-4, 6). This essential enzyme plays critical roles in a number of growth related processes in eukaryotic cells, including DNA replication and chromosome segregation (7-11). Topoisomerase II regulates levels of DNA supercoiling (i.e., under- and overwinding), and removes knots and tangles from the genetic material by passing a double helix through a transient double-stranded break that it generates in a separate DNA segment (7-13). To maintain genomic integrity during the cleavage event, the enzyme forms covalent bonds between active site tyrosyl residues and the 5’-DNA termini created by scission of the double helix (14-16). This covalent enzyme-cleaved DNA reaction intermediate is known as the cleavage complex (6).
Although lower eukaryotes such as yeast and Drosophila encode only a single form of topoisomerase II, vertebrates express two distinct isoforms of the enzyme, topoisomerase IIα and IIβ (9, 11, 17, 18). These isoforms display a high degree (∼70%) of amino acid sequence identity and similar enzymological characteristics, but are encoded by separate genes (9-11, 17-23). Topoisomerase IIα and topoisomerase IIβ are both nuclear enzymes, but have distinct patterns of expression and cellular functions. Topoisomerase IIα is essential for the survival of actively growing cells and its concentration increases dramatically during periods of proliferation (24-27). It is believed to be the isoform that functions in growth-dependent processes, such as DNA replication and chromosome segregation (7, 10). In contrast to the α isoform, topoisomerase IIβ is dispensable at the cellular level (28) and its expression appears to be constitutive, regardless of proliferative status (7, 23, 26, 29). Topoisomerase IIβ cannot compensate for the loss of topoisomerase IIα in mammalian cells, suggesting that these two isoforms do not play redundant roles, at least in replicative processes (23, 27, 30, 31).
Etoposide kills cells by inhibiting the ability of topoisomerase II to ligate cleaved DNA molecules (32, 33). This drug action leads to the accumulation of topoisomerase II-DNA cleavage complexes (4, 6, 32, 33). When DNA tracking systems such as the DNA replication or transcription machinery attempt to traverse these complexes, they convert them to permanent enzyme-linked double-stranded breaks in the genetic material (4, 13, 34, 35). The resulting breaks destabilize the genome and when present at sufficient concentrations, induce cell death pathways (13, 22, 34-39). The individual contributions of topoisomerase IIα and IIβ to the clinical efficacy of etoposide have yet to be determined. However, because the concentration of the α isoform is generally high in malignant tissues, most studies of etoposide action have focused on topoisomerase IIα (40-42).
Multiple lines of evidence, including mutagenesis, binding, and kinetic studies, indicate that interactions between topoisomerase II and etoposide, as opposed to drug-DNA interactions, are critical for drug activity and mediate the entry of etoposide into the ternary enzyme-drug-DNA complex (4, 13, 43-53). Therefore, a recent study utilized saturation transfer difference [1H]-nuclear magnetic resonance (STD [1H]-NMR) spectroscopy to identify the substituents on etoposide that contact topoisomerase II in the binary enzyme-drug complex (54). Work focused primarily on yeast topoisomerase II and included analysis of etoposide as well as a few related compounds. A brief NMR study with etoposide and human topoisomerase IIα also was included in the work. Results suggest that substituents on the A–, B–, and E–rings of etoposide (see Figure 1) interact with topoisomerase II, while the sugar moiety at C-4 and the D-ring do not (54).
Figure 1.
Structures of etoposide and etoposide derivatives that were employed in the present study.
To extend these findings and more fully define interactions between etoposide and the human enzyme, the present study assessed the ability of etoposide derivatives with an altered A– ring, E–ring, or C4 sugar moiety to induce enzyme-mediated DNA cleavage, interact with topoisomerase IIα, and compete with the parent drug for binding to the enzyme. Results suggest that the binding of etoposide to human topoisomerase IIα is driven by interactions with the A– and B–rings, and potentially by stacking interactions with the E–ring. Furthermore, drug contacts in the binary complex defined by STD [1H]-NMR have predictive value for the actions of etoposide within the ternary drug-enzyme-DNA complex.
EXPERIMENTAL PROCEDURES
Materials
Negatively supercoiled pBR322 plasmid DNA was prepared using a Plasmid Mega Kit (Qiagen) as described by the manufacturer. Etoposide and podophyllotoxin were purchased from Sigma. 4’-Demethyl epipodophyllotoxin (DEPT), epipodophyllotoxin (EPT), and 6,7-O,O-demethylenepipodophyllotoxin (DDEPT) were synthesized from podophyllotoxin as described (55-57). All drugs were stored at 4 °C as 20 mM stock solutions in 100% DMSO. Drugs used for NMR experiments were stored in 100% d-DMSO. [3H]etoposide was obtained from Moravek Biochemicals as a 1.5 mM stock in 100% ethanol. D2O (99.9%) was purchased from Aldrich. All other chemicals were analytical reagent grade.
Purification of Human Topoisomerase IIα
Human topoisomerase IIα was expressed in Saccharomyces cerevisiae and purified as described previously (48, 54, 58, 59). However, in the final step of the purification, the type II topoisomerase was eluted from the phosphocellulose column (P81, Whatman) with buffer containing 10 mM sodium phosphate, pH 7.7, 750 mM KCl, 1 mM EDTA, 1 mM EGTA, and 0.5 mM dithiothreitol.
Cleavage of Plasmid DNA by Human Topoisomerase IIα
DNA cleavage reactions were carried out using the procedure of Fortune and Osheroff (60). Assay mixtures contained 135 nM topoisomerase IIα and 10 nM negatively supercoiled pBR322 DNA in a total of 20 μL of cleavage buffer (10 mM Tris-HCl, pH 7.9, 100 mM KCl, 5 mM MgCl2, 0.1 mM NaEDTA, and 2.5% glycerol) that contained 0 to 200 μM etoposide, DEPT, EPT or DDEPT. DNA cleavage was initiated by the addition of enzyme and mixtures were incubated for 6 min at 37 °C to establish DNA cleavage-religation equilibria. Enzyme-DNA cleavage intermediates were trapped by adding 2 μL of 5% SDS and 1 μL of 375 mM EDTA, pH 8.0. Proteinase K was added (2 μL of 0.8 mg/mL) and reactions were incubated for 30 min at 45 °C to digest the topoisomerase IIα. Samples were mixed with 2 μL of 60% sucrose in 10 mM Tris-HCl, pH 7.9, 0.5% bromophenol blue, and 0.5% xylene cyanol FF, heated for 15 min at 45 °C, and subjected to electrophoresis in 1% agarose gels in 40 mM Tris-acetate, pH 8.3, 2 mM EDTA that contained 0.5 μg/mL ethidium bromide. DNA cleavage was monitored by the conversion of negatively supercoiled plasmids to linear molecules. DNA bands were visualized by ultraviolet light and quantified using an Alpha Innotech digital imaging system.
Drug-induced DNA Cleavage Mediated by Topoisomerase IIα in Cultured Human CEM Cells
Human CEM acute lymphoblastic leukemia cells (ATCC) were cultured under 5% CO2 at 37 °C in RPMI 1640 medium (Cellgro by Mediatech, Inc.) containing 10% heat-inactivated fetal calf serum (Hyclone) and 2 mM glutamine (Cellgro by Mediatech, Inc.). The In vivo Complex of Enzyme (ICE) bioassay (61, 62) (as modified on the TopoGEN, Inc. website) was employed to determine the ability of etoposide, DEPT, EPT, or DDEPT to induce topoisomerase IIα-mediated DNA breaks in CEM cells. Exponentially growing cultures were treated with 10 μM etoposide 10 μM DEPT, 50 μM EPT or 50 μM DDEPT for 2 h. Cells (∼5 × 106) were harvested by centrifugation and lysed by the immediate addition of 3 mL of 1% sarkosyl. Following gentle Dounce homogenization, cell lysates were layered onto a 2 mL cushion of CsCl (1.5 g/mL) and centrifuged in a Beckman NVT 90 rotor at 80,000 rpm (∼500,000 × g) for 5.5 h at 20 °C. DNA pellets were isolated, resuspended in 5 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, and blotted onto nitrocellulose membranes using a Schleicher and Schuell slot blot apparatus. Covalent cleavage complexes formed between topoisomerase IIα and chromosomal DNA were detected using a polyclonal antibody directed against human topoisomerase IIα (Kiamaya Biochemical Co.) at a 1:1000 dilution.
DNA Religation
DNA religation mediated by topoisomerase IIα was monitored according to the procedure of Byl et al. (63). Topoisomerase IIα DNA cleavage/religation equilibria were established as described above in the absence of compound, or in the presence of 100 etoposide, DEPT, EPT, or DDEPT. Religation was initiated by shifting reaction mixtures from 37 °C to 0 °C, and reactions were stopped at time points up to 40 s by the addition of 2 μL of 5% SDS followed by 1 μL of 375 mM NaEDTA, pH 8.0. Samples were processed and analyzed as described above for topoisomerase IIα plasmid DNA cleavage reactions.
STD [1H]-NMR Spectroscopy
All NMR experiments are performed at 283 K using a Bruker Avance DRX 400-MHz spectrometer equipped with a 5-mm BBI probe with z-gradients. NMR buffers contained 10 mM sodium phosphate, pH 7.7, 250 mM KCl, 0.1 mM Na2EDTA, and 5 mM MgCl2. NMR samples (500 μL) contained 5 μM human topoisomerase IIα and 250 μM etoposide, DEPT, EPT or DDEPT and were maintained at 4 °C until data were obtained. STD [1H]-NMR experiments employed a pulse scheme similar to that reported by Mayer and Meyer (64). A 2 s saturation pulse was used. The gradient pulse that was applied was 1 ms at 30% with a 500 μs recovery delay. The water signal was suppressed by tailoring a watergate pulse sequence to the beginning of the f2 presaturation STD-pulse program. For each experiment (on and off resonance irradiation), a total of 2000 scans were collected with a 3 s relaxation delay between each scan. On and off-resonance irradiations were performed at 0.5 and 17 ppm, respectively. Difference spectra were prepared by subtracting the on-resonance spectrum from the off-resonance spectrum. Signals resulting in the difference spectrum represent the NOE difference signals generated by the transfer of irradiation energy from the enzyme to the bound ligand. Ligand protons in close spatial proximity with the enzyme displayed larger NOE signals. Mapping of the NOE signals with their proton assignments on the ligand revealed the ligand binding epitope to the target topoisomerase II. Spectra were processed using Bruker Topspin software.
Topoisomerase II-Drug Binding
Competition binding studies were performed using a nitrocellulose filter binding technique (54). Nitrocellulose membranes (0.45 μm HA; Millipore) were soaked in binding buffer (10 mM sodium phosphate, pH 7.7, 250 mM KCl, 0.1 mM NaEDTA, and 5 mM MgCl2) for 10 min. Reaction mixtures contained 1.6 μM human topoisomerase IIα and 20 μM [3H]etoposide, as well as 0−100 μM non-labeled etoposide, DEPT, EPT or DDEPT in a total of 60 μL of binding buffer. Samples were incubated for 6 min at 30 °C and applied to the nitrocellulose membranes in vacuo. Filters were immediately washed three times with 1 mL of ice-cold binding buffer, dried, and submerged in 8 mL of scintillation fluid (Econo-Safe; Research Products International). Radioactivity remaining on membranes was quantified using a Beckman LS 5000 TD scintillation counter. The amount of radioactive etoposide remaining on the filter in the absence of enzyme was subtracted prior to binding calculations.
Site-specific DNA Cleavage
DNA sites cleaved by human topoisomerase IIα were determined by a modification (65) of the procedure of O'Reilly and Kreuzer (66). A linear 4330 bp fragment (HindIII/EcoRI) of pBR322 plasmid DNA singly labeled with 32P on the 5’-terminus of the HindIII site was used as the cleavage substrate. Reaction mixtures contained 0.35 nM DNA substrate and 60 nM topoisomerase IIα in 50 μL of cleavage buffer. Assays were carried out in the absence of compound, or in the presence of 25 μM etoposide, 25 μM DEPT, 250 μM EPT or 250 μM DDEPT. Reactions were initiated by the addition of the enzyme and were incubated for 10 min at 37 °C. Cleavage intermediates were trapped by adding 5 μL of 10% SDS followed by 5 μL of 250 mM NaEDTA, pH 8.0. Topoisomerase IIα was digested with proteinase K (5 μL of 0.8 mg/mL) for 30 min at 45 °C. Reaction products were precipitated twice in ethanol, dried, and resuspended in 40% formamide, 8.4 mM EDTA, 0.02% bromophenol blue, and 0.02% xylene cyanole FF. Samples were subjected to electrophoresis in a 6% sequencing gel. The gel was then fixed in 10% methanol/10% acetic acid for 5 min, dried, and DNA cleavage products were analyzed on a Bio-Rad Molecular Imager FX.
RESULTS AND DISCUSSION
Contribution of Etoposide Substitutents to Drug Activity Against Topoisomerase IIα
Etoposide, a widely prescribed topoisomerase II-targeted anticancer agent, is composed of a polycyclic ring system (rings A–D), a glycosidic moiety at the C4 position, and a pendant ring (E–ring) at the C1 position (Figure 1) (1-4). Because of its importance in cancer chemotherapy, numerous etoposide derivatives have been synthesized and analyzed (67-76). Despite the fact that these derivatives display a wide ability to induce topoisomerase II-associated DNA strand breaks, virtually no data is available that can assign a specific function to any substituent on the drug molecule.
A recent STD [1H]-NMR spectroscopy study, however, was able to identify the regions on etoposide that interact with yeast topoisomerase II and human topoisomerase IIα (54). Results demonstrated that the H15 geminal protons of the A–ring, the H5 and H8 protons of the B–ring, and the H2’ and H6’ protons and the 3’– and 5’–methoxyl protons of the pendant E–ring contact both enzymes in the binary protein-ligand complex (Figure 1). In contrast, no significant nuclear Overhauser enhancement (NOE) signals arising from the C–ring, the D–ring, or the C4 glycosidic moiety were observed, suggesting that there is (at best) limited interaction between these portions of etoposide and topoisomerase II in the binary complex. It should be noted that it was not possible to observe NMR signals from hydroxyl groups in this study, as they were obscured by the water peak.
To further define interactions between etoposide and human type II topoisomerases and to relate the structural NMR data obtained with the binary complex to drug function within the ternary enzyme-drug-DNA complex, the ability of etoposide and three derivatives to induce DNA cleavage mediated by human topoisomerase IIα was determined. The derivatives all lack the C4 sugar moiety, which does not contact the enzyme in the binary complex (Figure 1). The three derivatives are 4’-demethyl epipodophyllotoxin (DEPT)1, which aside from the C4 moiety is identical to etoposide; epipodophyllotoxin (EPT), which contains a methoxyl group in place of the 4’-OH on the E–ring; and 6,7-O,O-demethylenepipodophyllotoxin (DDEPT), which lacks C15 and its associated geminal protons and replaces the A–ring with a diol.
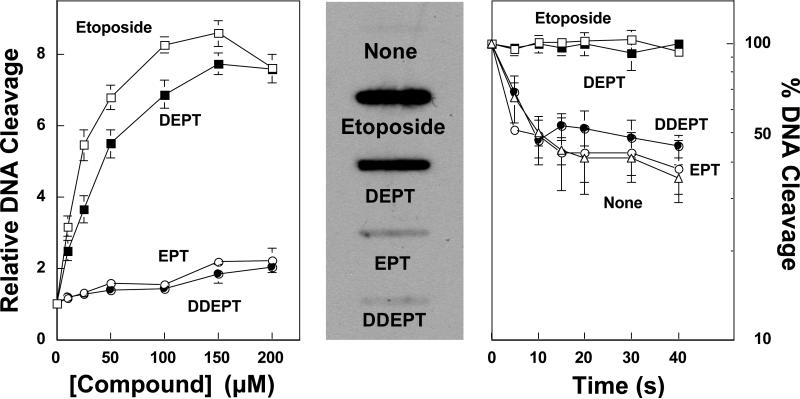
As seen in Figure 2 (left), DNA cleavage results with DEPT were similar to those obtained with etoposide. Thus, removal of the C4 glycosidic moiety had little effect on the ability of the drug to enhance DNA scission mediated by human topoisomerase IIα. In contrast, the addition of bulk to the E-ring in the form of a 4’-methoxyl group (EPT) or the loss of the H15 geminal protons (DDEPT) resulted in etoposide derivatives with little activity against the enzyme.
Figure 2.
Effects of etoposide derivatives on DNA cleavage and religation mediated by human topoisomerase IIα. Left: Levels of DNA cleavage were expressed as a fold–enhancement over reactions that were carried out in the absence of drug. Assay mixtures contained 0−200 μM etoposide (open squares), DEPT (closed squares), EPT (open circles), or DDEPT (closed circles). Error bars represent the standard deviation of three independent experiments. Center: The ICE bioassay was used to monitor the level of cleavage complexes in human CEM leukemia cells treated with etoposide derivatives. DNA (10 μg) from cultures treated with no compound (None), 10 μM etoposide, 10 μM DEPT, 50 μM EPT or 50 μM DDEPT for 2 h was blotted onto a nitrocellulose membrane and probed with a polyclonal antibody directed against human topoisomerase IIα. Results are representative of three independent experiments. Right: DNA religation was examined in the absence of compound (none, open triangles) or in the presence of 100 μM etoposide (open squares), DEPT (closed squares) EPT (open circles), or DDEPT (closed circles). Error bars represent the standard deviation of three independent experiments.
To further characterize drug activity, the ability of DEPT, EPT, and DDEPT to induce DNA cleavage by topoisomerase IIα in cultured human CEM leukemia cells was compared to etoposide (Figure 2, center). Consistent with the in vitro data, treatment of cells with etoposide or DEPT generated high levels of enzyme-linked DNA strand breaks, while treatment with EPT or DDEPT had little effect.
Finally, since etoposide increases levels of topoisomerase II-associated DNA breaks primarily by inhibiting the ability of the enzyme to religate cleaved nucleic acids, the effects of etoposide derivatives on DNA strand closure was determined (Figure 2, right). Once again, DEPT yielded results that were comparable to those with etoposide and strongly inhibited DNA religation mediated by topoisomerase IIα. In contrast, rates of religation in the presence of EPT or DDEPT were similar to reactions that contained no drug.
Taken together, these results indicate that removal of the sugar moiety, which does not contact topoisomerase IIα in the binary complex, has little effect on the actions of etoposide against the human enzyme. However, alterations in the A– or E–rings of etoposide, which are intimately associated with topoisomerase IIα in the binary complex, dramatically impair drug function. Thus, at least for the C4 glycosydic moiety, the A–ring, and the E–ring, data obtained from STD [1H]-NMR spectroscopy in the binary enzyme-drug complex have strong predictive value for etoposide-induced DNA scission in the ternary topoisomerase IIα-drug-DNA complex.
Interaction of Etoposide Derivatives with Topoisomerase IIα
In order to further assess the mechanistic basis for alterations in the activity of the etoposide derivatives employed, the interaction of these compounds with topoisomerase IIα was characterized by STD [1H]-NMR spectroscopy (54, 64, 77-80).
In the STD [1H]-NMR technique, a sample containing topoisomerase IIα and etoposide (or drug derivative) is selectively saturated with magnetization by irradiation at a frequency at which protein methyl groups, but no etoposide protons resonate (on-resonance frequency). Magnetization is spread rapidly throughout the protein by intramolecular spin diffusion. Substituents on the drug that interact with topoisomerase IIα are progressively saturated with magnetization via intermolecular, through-space, dipole-dipole interactions. In addition to this on-resonance spectrum, an off-resonance (reference) spectrum is generated by saturating the sample with a magnetization frequency that is different from the resonance frequencies of either topoisomerase IIα or the drug. The difference spectrum generated by subtracting the on-resonance spectrum from the off-resonance spectrum contains only the signals of the drug that are saturated through the intermolecular transfer of magnetization from the protein substituents indicating interaction with the enzyme (64, 77-80).
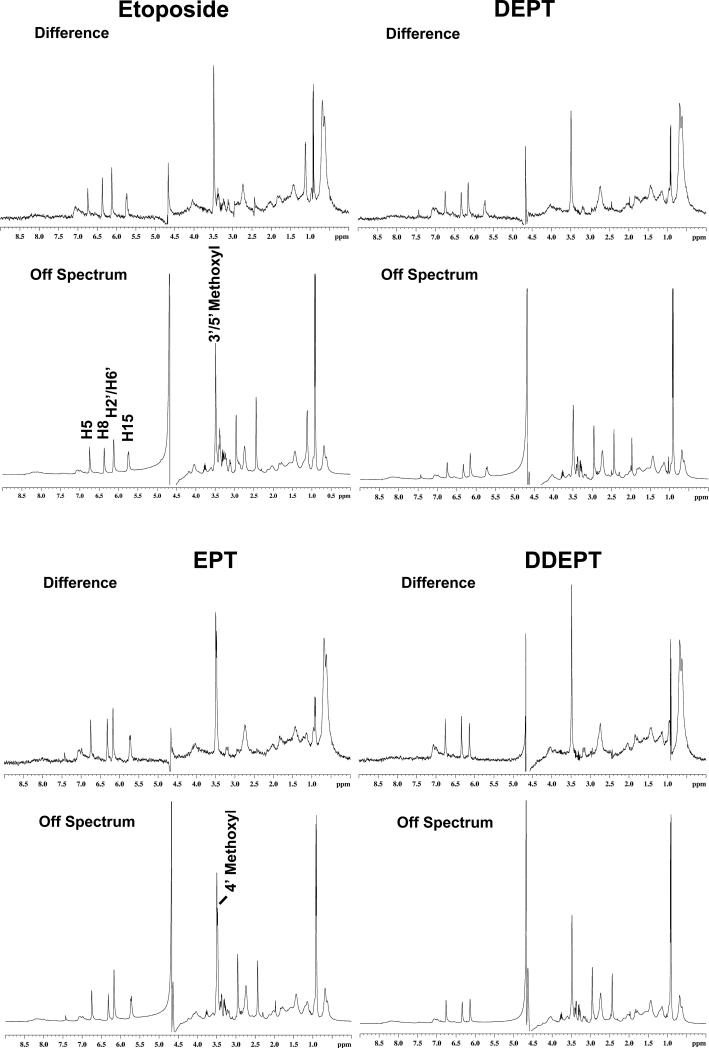
As a prelude to STD [1H]-NMR experiments, proton resonances of etoposide were assigned by 1-D NMR analysis (54, 81). As a control, a representative STD [1H]-NMR experiment that analyzed the binding of etoposide to human topoisomerase IIα is shown in Figure 3. As reported previously (54), the NOE signals from the bound drug seen in the difference spectrum indicate that the H15 geminal protons of the A–ring (5.75 ppm), the H5 and H8 protons of the B–ring (6.75 and 6.37 ppm, respectively), and the H2’ and H6’ protons (6.13 ppm) and the 3’– and 5’–methoxyl protons of the pendant E–ring (3.49 ppm) of etoposide interact with the human enzyme in the binary complex. Once again, no significant NOE signals were observed from the C–ring, the D–ring, or the C4 glycosidic moiety. Unfortunately, resonances for hydroxyl groups, including the 4’–OH of the E–ring, were obscured by the water peak and were not visualized in any of the NMR spectra.
Figure 3.
Interaction of etoposide (top left), DEPT (top right), EPT (bottom left) or DDEPT (bottom right) with human topoisomerase IIα as determined by STD [1H]-NMR spectroscopy. Difference and off resonance (reference) spectra are shown. Spectra are representative of at least two independent experiments.
Off-resonance and difference spectra for samples containing topoisomerase IIα and DEPT, EPT, or DDEPT are shown in Figure 3. Despite the loss of the C4 sugar moiety, the difference spectrum of DEPT was similar to that seen with etoposide. In addition, difference spectra generated for EPT and DDEPT were similar to that of etoposide, with the exception that a new NOE signal was observed for the 4’-methoxyl protons of EPT (3.48 ppm) and DDEPT lacked the H15 geminal protons. Furthermore, the area under common NOE peaks for all of the compounds differed by less than two–fold. These findings suggest that DEPT, EPT, and DDEPT bind to topoisomerase IIα with an overall geometry that is similar to that of the parent drug.
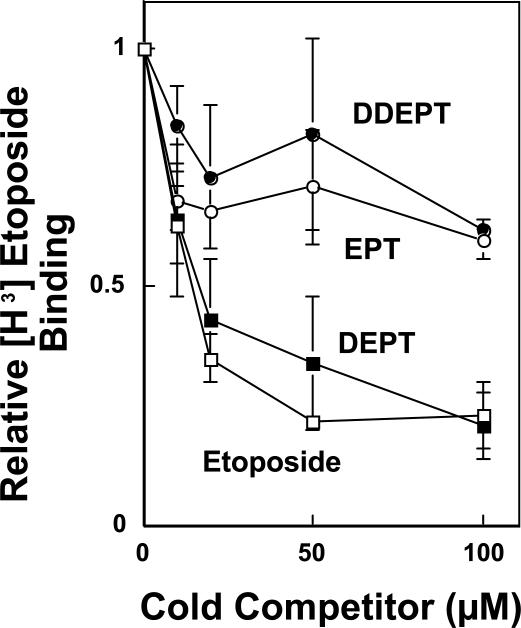
Because the STD [1H]-NMR experiments were carried out at saturating drug:topoisomerase IIα ratios, and because the area under the peak can reflect multiple factors, the spectra do not provide information on the affinities of etoposide derivatives for the enzyme. Therefore, the relative affinities of these etoposide derivatives for topoisomerase IIα were characterized by nitrocellulose filter binding competition assays. In these experiments, the ability of non-radioactive etoposide, DEPT, EPT, or DDEPT to compete with [3H]etoposide for binding to the human enzyme was determined. As seen in Figure 4, the concentrations of DEPT that were required to displace the bound [3H]etoposide were similar to those seen with unlabeled etoposide. This finding strongly suggests that the C4 sugar moiety does not contribute significantly to etoposide-topoisomerase IIα binding in the binary complex.
Figure 4.
Binding of etoposide and derivatives to human topoisomerase IIα. Reaction mixtures contained 20 μM [3H]etoposide and 0−100 μM non-labeled etoposide (open squares), DEPT (closed squares), EPT (open circles), or DDEPT (closed circles). Levels of [3H]etoposide binding to topoisomerase IIα in the absence of competitor drug were set to 1. Error bars represent the standard deviation of at least three independent experiments.
In contrast to DEPT, EPT and DDEPT were considerably less effective at competing with [3H]etoposide. These results suggest that substituents on the A–ring of etoposide play a critical role in mediating drug-enzyme binding interactions. They also indicate that the addition of bulk to the pendant E–ring greatly decreases the affinity of the drug for the human enzyme.
Although substituents on the E–ring of etoposide are necessary for drug function against topoisomerase II, they do not contribute significantly to drug-enzyme binding (54). In fact, when the 4’-OH and the 3’- and 5’-methoxyl groups were replaced with hydrogen atoms, NOE signals were observed by STD [1H]-NMR for all of the resulting protons on the E–ring, although with reduced signal (54). Based on these findings, it was proposed that protein associations with the E–ring are mediated by stacking interactions rather than by any specific group on the ring. Because the presence of the 4’-OH group has little effect on drug-enzyme binding, the substitution of a 4’-methoxyl group on the E–ring cannot be impairing drug interactions due to the loss of a critical binding moiety. Rather, the decreased drug affinity caused by presence of the 4’-methoxyl group in EPT most likely results from the introduction of steric bulk. If this is the case, it implies that the E–ring sits within a confined pocket in topoisomerase IIα.
Site-Specificity of DNA Cleavage Mediated by Human Topoisomerase IIα in the Presence of Etoposide Derivatives
Since etoposide imparts a distinctive DNA cleavage specificity to topoisomerase II, it is believed that some portion of the drug must interact with the double helix within the enzyme-DNA cleavage complex. Therefore, to determine whether modification of etoposide alters cleavage specificity, a singly end-labeled linear plasmid substrate was used to map sites of DNA scission by human topoisomerase IIα in the presence of etoposide, DEPT, EPT, or DDEPT (Figure 5). Due to the poor efficacy of EPT and DDEPT, the concentrations of these compounds were ten–fold higher (250 μM) than those employed for either etoposide or DEPT (25 μM).
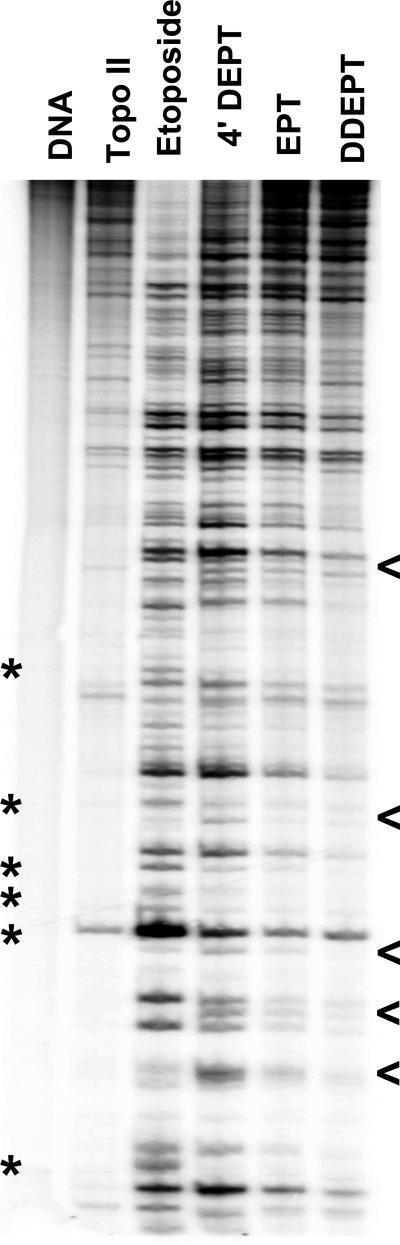
Figure 5.
DNA cleavage site utilization by human topoisomerase IIα in the presence of etoposide derivatives. A singly end-labeled linear 4330 bp fragment of pBR322 was used as the cleavage substrate. An autoradiogram of a polyacrylamide gel is shown. DNA cleavage reactions contained no compound (Topo II), 25 μM etoposide, 25 μM DEPT, 250 μM EPT or 250 μM DDEPT. A DNA control is shown in the far left lane. Bands that were present in etoposide-containing reactions that were weak or absent in reactions that contained DEPT are indicated by asterisks and bands that were present in DEPT-containing reactions that were weak or absent in reactions that contained etoposide are indicated by arrowheads. Data are representative of at least three independent experiments.
The DNA cleavage patterns generated in the presence of etoposide and DEPT were similar, but subtle differences were observed. Some bands that were present in reactions that contained etoposide were either weak or absent in reactions that contained DEPT (indicated by *) and vice versa (indicated by <). It was originally suggested (54) that the sugar moiety of etoposide did not contact DNA in the ternary complex based on the following: 1) the substitution of a thiophene for the 8”–methyl in teniposide or a C4 amino alkyl chain in TOP-53 did not greatly affect the DNA cleavage specificity of etoposide, and 2) every proton of the amino alkyl side chain of TOP-53 interacts strongly with topoisomerase II in the binary complex (6, 54, 62, 82). However, the differences observed in the DNA cleavage patterns induced by etoposide versus DEPT indicate that the presence of the glycosydic moiety of etoposide influences the selection of DNA cleavage sites by topoisomerase IIα. At the present time, it is not known whether this influence is due to a direct interaction between the C4 sugar and DNA or to an effect on the overall geometry of the cleavage complex.
Since DEPT, EPT, and DDEPT all lack the sugar moiety, their DNA cleavage patterns were compared to each other. Although levels of scission induced by EPT or DDEPT were lower than observed with DEPT, the site specificity of these three etoposide derivatives was essentially the same (Figure 5). These results strongly suggest that neither the A–ring nor the E–ring contact the DNA within the cleavage complex.
Conclusions
Although etoposide is one of the most widely prescribed drugs used for the treatment of human cancers (1-4), little information is available that identifies the specific substituents on the drug that mediate its interactions with topoisomerase II. However, based on results of STD [1H]-NMR and enzyme-drug binding in the binary topoisomerase II-etoposide complex, and DNA cleavage experiments in the ternary topoisomerase II-etoposide-DNA complex, a model is beginning to emerge (Figure 6). In this model, the binding of etoposide to human topoisomerase IIα is driven by interactions with the A–ring, B–ring, and potentially by stacking interactions with the E–ring. While the E–ring methoxyl groups and the 4’-OH do not contribute substantially to binding, they appear to be very important for drug function. The sugar moiety of etoposide does not contact the enzyme in the binary compslex, but subtly alters interactions with DNA. Given the lack of effect of A– and E–ring substituents on the specificity of topoisomerase II-mediated DNA cleavage, we propose that the D–ring of etoposide contacts the double helix within the cleavage complex. We currently are testing this hypothesis by examining a series of etoposide D-ring derivatives and establishing STD [1H]-NMR conditions for investigating interactions between etoposide, human topoisomerase IIα, and DNA in the ternary complex.
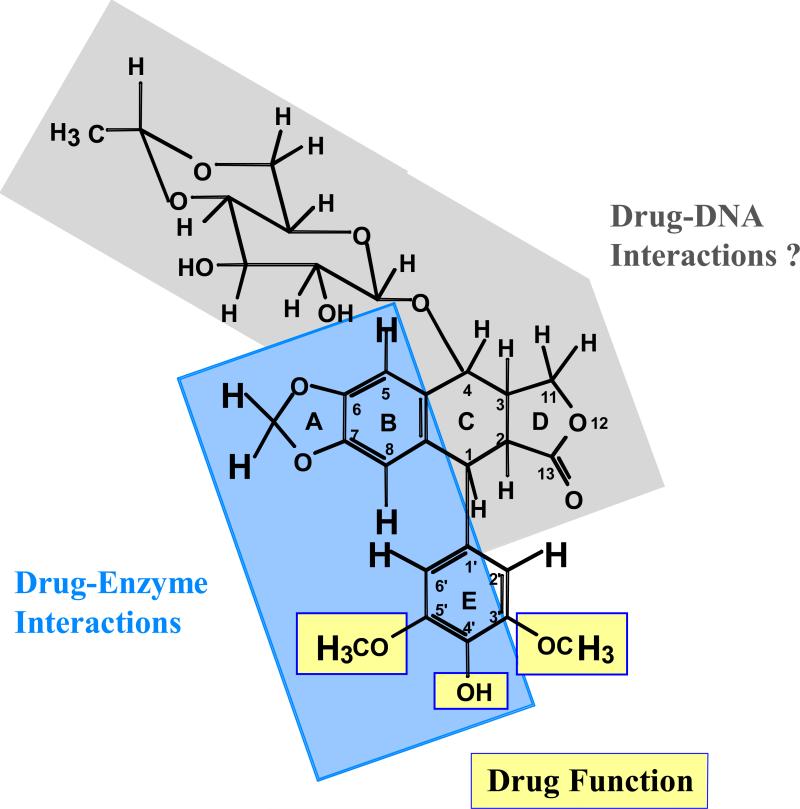
Figure 6.
Summary of etoposide substituents that interact with human topoisomerase IIα. Protons that interact with the enzyme are shown in large bold print, those that do not are shown in small print. Hydroxyl protons were obscured by the water peak and could not be visualized. The blue region on etoposide, including portions of the A–, B– and E–rings, is proposed to interact with topoisomerase IIα in the binary drug-enzyme complex. E–ring substituents highlighted with yellow boxes are important for drug function and interact with the enzyme, but do not appear to contribute significantly to binding (54). It is proposed that portions of the D–ring and/or sugar moiety of etoposide, which are shaded in gray, may interact with DNA in the drug-stabilized topoisomerase IIα-DNA cleavage complex.
Finally, the identification of groups on etoposide that interact with human topoisomerase IIα in the binary complex by STD [1H]-NMR appears to have predictive value for the behavior of the drug in the ternary enzyme-etoposide-DNA complex. Therefore, this technique may contribute to the future development of etoposide derivatives with enhanced activity against the human type II enzyme.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health grant GM33944 (NO) and National Science Foundation grant MCB-0334785 (DEG). RPB was a trainee under National Institutes of Health grant T32 CA09582.
Abbreviations: DEPT, 4’-demethyl epipodophyllotoxin; EPT, epipodophyllotoxin; and DDEPT, 6,7-O,O-demethylenepipodophyllotoxin.
REFERENCES
- 1.Hande KR. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur. J. Cancer. 1998;34:1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 2.Hande KR. Clinical applications of anticancer drugs targeted to topoisomerase II. Biochim. Biophys. Acta. 1998;1400:173–184. doi: 10.1016/s0167-4781(98)00134-1. [DOI] [PubMed] [Google Scholar]
- 3.Holden JA. DNA topoisomerases as anticancer drug targets: from the laboratory to the clinic. Curr. Med. Chem. Anticancer Agents. 2001;1:1–25. doi: 10.2174/1568011013354859. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr. Med. Chem. Anti-Canc. Agents. 2005;5:363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- 5.Nitiss JL, Liu YX, Hsiung Y. A temperature sensitive topoisomerase II allele confers temperature dependent drug resistance on amsacrine and etoposide: a genetic system for determining the targets of topoisomerase II inhibitors. Cancer Research. 1993;53:89–93. [PubMed] [Google Scholar]
- 6.Ross W, Rowe T, Glisson B, Yalowich J, Liu L. Role of topoisomerase II in mediating epipodophyllotoxin-induced DNA cleavage. Cancer Res. 1984;44:5857–5860. [PubMed] [Google Scholar]
- 7.Nitiss JL. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta. 1998;1400:63–81. doi: 10.1016/s0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- 8.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Quart. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 9.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 10.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 11.Velez-Cruz R, Osheroff N. DNA Topoisomerases: Type II. In: Lennarz W, Lane MD, editors. Encyclopedia of Molecular Biology. Elsevier Science; San Diego: 2004. pp. 806–811. [Google Scholar]
- 12.Wang JC. DNA Topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 13.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid. Res. Mol. Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 14.Sander M, Hsieh T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem. 1983;258:8421–8428. [PubMed] [Google Scholar]
- 15.Hsieh T. Knotting of the circular duplex DNA by type II DNA topoisomerase from Drosophila melanogaster. Journal of Biological Chemistry. 1983;258:8413–8420. [PubMed] [Google Scholar]
- 16.Zechiedrich EL, Christiansen K, Andersen AH, Westergaard O, Osheroff N. Double-stranded DNA cleavage/religation reaction of eukaryotic topoisomerase II: evidence for a nicked DNA intermediate. Biochemistry. 1989;28:6229–6236. doi: 10.1021/bi00441a014. [DOI] [PubMed] [Google Scholar]
- 17.Drake FH, Zimmerman JP, McCabe FL, Bartus HF, Per SR, Sullivan DM, Ross WE, Mattern MR, Johnson RK, Crooke ST, Mirabelli CK. Purification of topoisomerase II from amsacrine-resistant P388 leukemia cells. Evidence for two forms of the enzyme. J. Biol. Chem. 1987;262:16739–16747. [PubMed] [Google Scholar]
- 18.Drake FH, Hofmann GA, Bartus HF, Mattern MR, Crooke ST, Mirabelli CK. Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II. Biochemistry. 1989;28:8154–8160. doi: 10.1021/bi00446a029. [DOI] [PubMed] [Google Scholar]
- 19.Tsai-Pflugfelder M, Liu LF, Liu AA, Tewey KM, Whang-Peng J, Knutsen T, Huebner K, Croce CM, Wang JC. Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21−22. Proc. Natl. Acad. Sci. U.S.A. 1988;85:7177–7181. doi: 10.1073/pnas.85.19.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkins JR, Ayton P, Jones T, Davies SL, Simmons DL, Harris AL, Sheer D, Hickson ID. Isolation of cDNA clones encoding the beta isozyme of human DNA topoisomerase II and localisation of the gene to chromosome 3p24. Nucleic Acids Res. 1992;20:5587–5592. doi: 10.1093/nar/20.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan KB, Dorman TE, Falls KM, Chung TD, Mirabelli CK, Crooke ST, Mao J. Topoisomerase II alpha and topoisomerase II beta genes: characterization and mapping to human chromosomes 17 and 3, respectively. Cancer Res. 1992;52:231–234. [PubMed] [Google Scholar]
- 22.Wilstermann AM, Osheroff N. Stabilization of eukaryotic topoisomerase IIDNA cleavage complexes. Curr. Top. Med. Chem. 2003;3:321–338. doi: 10.2174/1568026033452519. [DOI] [PubMed] [Google Scholar]
- 23.Austin CA, Marsh KL. Eukaryotic DNA topoisomerase IIbeta. BioEssays. 1998;20:215–226. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 24.Heck MM, Earnshaw WC. Topoisomerase II: A specific marker for cell proliferation. J. Cell Biol. 1986;103:2569–2581. doi: 10.1083/jcb.103.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsiang YH, Wu HY, Liu LF. Proliferation-dependent regulation of DNA topoisomerase II in cultured human cells. Cancer Res. 1988;48:3230–3235. [PubMed] [Google Scholar]
- 26.Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth & Differentiation. 1991;2:209–214. [PubMed] [Google Scholar]
- 27.Grue P, Grasser A, Sehested M, Jensen PB, Uhse A, Straub T, Ness W, Boege F. Essential mitotic functions of DNA topoisomerase IIα are not adopted by topoisomerase IIβ in human H69 cells. J. Biol. Chem. 1998;273:33660–33666. doi: 10.1074/jbc.273.50.33660. [DOI] [PubMed] [Google Scholar]
- 28.Chen M, Beck WT. DNA topoisomerase II expression, stability, and phosphorylation in two VM-26-resistant human leukemic CEM sublines. Oncology Res. 1995;7:103–111. [PubMed] [Google Scholar]
- 29.Isaacs RJ, Davies SL, Sandri MI, Redwood C, Wells NJ, Hickson ID. Physiological regulation of eukaryotic topoisomerase II. Biochim. Biophys. Acta. 1998;1400:121–137. doi: 10.1016/s0167-4781(98)00131-6. [DOI] [PubMed] [Google Scholar]
- 30.Dereuddre S, Delaporte C, Jacquemin-Sablon A. Role of topoisomerase II beta in the resistance of 9-OH-ellipticine-resistant Chinese hamster fibroblasts to topoisomerase II inhibitors. Cancer Res. 1997;57:4301–4308. [PubMed] [Google Scholar]
- 31.Sakaguchi A, Kikuchi A. Functional compatibility between isoform alpha and beta of type II DNA topoisomerase. J. Cell. Sci. 2004;117:1047–1054. doi: 10.1242/jcs.00977. [DOI] [PubMed] [Google Scholar]
- 32.Osheroff N. Effect of antineoplastic agents on the DNA cleavage/religation reaction of eukaryotic topoisomerase II: inhibition of DNA religation by etoposide. Biochemistry. 1989;28:6157–6160. doi: 10.1021/bi00441a005. [DOI] [PubMed] [Google Scholar]
- 33.Robinson MJ, Osheroff N. Effects of antineoplastic drugs on the post-strand-passage DNA cleavage/religation equilibrium of topoisomerase II. Biochemistry. 1991;30:1807–1813. doi: 10.1021/bi00221a012. [DOI] [PubMed] [Google Scholar]
- 34.Kaufmann SH. Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochim. Biophys. Acta. 1998;1400:195–211. doi: 10.1016/s0167-4781(98)00136-5. [DOI] [PubMed] [Google Scholar]
- 35.Kaufmann SH, Gore SD, Miller CB, Jones RJ, Zwelling LA, Schneider E, Burke PJ, Karp JE. Topoisomerase II and the response to antileukemic therapy. Leukemia Lymph. 1998;29:217–237. doi: 10.3109/10428199809068560. [DOI] [PubMed] [Google Scholar]
- 36.Rowley JD. The critical role of chromosome translocations in human leukemias. Ann. Rev. Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 37.Felix CA. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim. Biophys. Acta. 1998;1400:233–255. doi: 10.1016/s0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 38.Sordet O, Khan QA, Kohn KW, Pommier Y. Apoptosis induced by topoisomerase inhibitors. Curr. Med. Chem. Anti-Canc. Agents. 2003;3:271–290. doi: 10.2174/1568011033482378. [DOI] [PubMed] [Google Scholar]
- 39.Felix CA, Kolaris CP, Osheroff N. Topoisomerase II and the etiology of chromosomal translocations. DNA Repair (Amst) 2006;5:1093–1108. doi: 10.1016/j.dnarep.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 40.Keith WN, Tan KB, Brown R. Amplification of the topoisomerase II alpha gene in a non-small cell lung cancer cell line and characterisation of polymorphisms at the human topoisomerase II alpha and beta loci in normal tissue. Genes Chromo. Cancer. 1992;4:169–175. doi: 10.1002/gcc.2870040211. [DOI] [PubMed] [Google Scholar]
- 41.Keith WN, Douglas F, Wishart GC, McCallum HM, George WD, Kaye SB, Brown R. Co-amplification of erbB2, topoisomerase II alpha and retinoic acid receptor alpha genes in breast cancer and allelic loss at topoisomerase I on chromosome 20. Eur. J. Cancer. 1993;10:1469–1475. doi: 10.1016/0959-8049(93)90022-8. [DOI] [PubMed] [Google Scholar]
- 42.Skotheim RI, Kallioniemi A, Bjerkhagen B, Mertens F, Brekke HR, Monni O, Mousses S, Mandahl N, Soeter G, Nesland JM, Smeland S, Kallioniemi OP, Lothe RA. Topoisomerase-II alpha is upregulated in malignant peripheral nerve sheath tumors and associated with clinical outcome. J. Clin. Oncol. 2003;21:4586–4591. doi: 10.1200/JCO.2003.07.067. [DOI] [PubMed] [Google Scholar]
- 43.Chow KC, Macdonald TL, Ross WE. DNA binding by epipodophyllotoxins and N-acyl anthracyclines: implications for mechanism of topoisomerase II inhibition. Mol. Pharmacol. 1988;34:467–473. [PubMed] [Google Scholar]
- 44.Sullivan DM, Latham MD, Rowe TC, Ross WE. Purification and characterization of an altered topoisomerase II from a drug-resistant Chinese hamster ovary cell line. Biochemistry. 1989;28:5680–5687. doi: 10.1021/bi00439a051. [DOI] [PubMed] [Google Scholar]
- 45.Beck WT, Danks MK, Wolverton JS, Kim R, Chen M. Drug resistance associated with altered DNA topoisomerase II. Adv. Enzyme Regul. 1993;33:113–127. doi: 10.1016/0065-2571(93)90012-3. [Review] [DOI] [PubMed] [Google Scholar]
- 46.Nitiss JL. Using yeast to study resistance to topoisomerase II-targeting drugs. Cancer Chemother. Pharmacol. 1994;34:S6–13. doi: 10.1007/BF00684857. [DOI] [PubMed] [Google Scholar]
- 47.Vassetzky YS, Alghisi GC, Gasser SM. DNA Topoisomerase II Mutations and Resistance to Antitumor Drugs. BioEssays. 1995;17:767–774. doi: 10.1002/bies.950170906. [DOI] [PubMed] [Google Scholar]
- 48.Elsea SH, Hsiung Y, Nitiss JL, Osheroff N. A yeast type II topoisomerase selected for resistance to quinolones. Mutation of histidine 1012 to tyrosine confers resistance to nonintercalative drugs but hypersensitivity to ellipticine. J. Biol. Chem. 1995;270:1913–1920. doi: 10.1074/jbc.270.4.1913. [DOI] [PubMed] [Google Scholar]
- 49.Burden DA, Kingma PS, Froelich-Ammon SJ, Bjornsti M-A, Patchan MW, Thompson RB, Osheroff N. Topoisomerase II-etoposide interactions direct the formation of drug-induced enzyme-DNA cleavage complexes. J. Biol. Chem. 1996;46:29238–29244. doi: 10.1074/jbc.271.46.29238. [DOI] [PubMed] [Google Scholar]
- 50.Burden DA, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 51.Larsen AK, Skladanwski A. Cellular resistance to topoisomerase-targeted drugs: from drug uptake to cell death. Biochim. Biophys. Acta. 1998;1400:257–274. doi: 10.1016/s0167-4781(98)00140-7. [DOI] [PubMed] [Google Scholar]
- 52.Kingma PS, Burden DA, Osheroff N. Binding of etoposide to topoisomerase II in the absence of DNA: decreased affinity as a mechanism of drug resistance. Biochemistry. 1999;38:3457–3461. doi: 10.1021/bi982855i. [DOI] [PubMed] [Google Scholar]
- 53.Leroy D, Kajava AV, Frei C, Gasser SM. Analysis of etoposide binding to subdomains of human DNA topoisomerase II alpha in the absence of DNA. Biochemistry. 2001;40:1624–1634. doi: 10.1021/bi0019141. [DOI] [PubMed] [Google Scholar]
- 54.Wilstermann AM, Bender RP, Godfrey M, Choi S, Anklin C, Berkowitz DB, Osheroff N, Graves DO. Topoisomerase II-Drug Interaction Domains: Identification of Substituents on Etoposide That Interact with the Enzyme. Biochemistry. 2007;46:8217–8225. doi: 10.1021/bi700272u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang ZQ, Hu H, Chen HX, Cheng YC, Lee KH. Antitumor agents. 124. New 4 beta-substituted aniline derivatives of 6,7-O,O-demethylene-4'-O-demethylpodophyllotoxin and related compounds as potent inhibitors of human DNA topoisomerase II. Journal of Medicinal Chemistry. 1992;35:871–877. doi: 10.1021/jm00083a010. [DOI] [PubMed] [Google Scholar]
- 56.Daley L, Guminski Y, Demerseman P, Kruczynski A, Etievant C, Imbert T, Hill BT, Monneret C. Synthesis and antitumor activity of new glycosides of epipodophyllotoxin, analogues of etoposide, and NK 611. J Med Chem. 1998;41:4475–4485. doi: 10.1021/jm9800752. [DOI] [PubMed] [Google Scholar]
- 57.Kamal A, Laxman N, Ramesh G. Facile and efficient one-pot synthesis of 4beta-arylaminopodophyllotoxins: synthesis of DNA topoisomerase II inhibitors (NPF and W-68). Bioorg Med Chem Lett. 2000;10:2059–2062. doi: 10.1016/s0960-894x(00)00407-8. [DOI] [PubMed] [Google Scholar]
- 58.Worland ST, Wang JC. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1989;264:4412–4416. [PubMed] [Google Scholar]
- 59.Kingma PS, Greider CA, Osheroff N. Spontaneous DNA lesions poison human topoisomerase IIα and stimulate cleavage proximal to leukemic 11q23 chromosomal breakpoints. Biochemistry. 1997;36:5934–5939. doi: 10.1021/bi970507v. [DOI] [PubMed] [Google Scholar]
- 60.Fortune JM, Osheroff N. Merbarone inhibits the catalytic activity of human topoisomerase IIα by blocking DNA cleavage. J. Biol. Chem. 1998;273:17643–17650. doi: 10.1074/jbc.273.28.17643. [DOI] [PubMed] [Google Scholar]
- 61.Subramanian D, Kraut E, Staubus A, Young DC, Muller MT. Analysis of topoisomerase I/DNA complexes in patients administered topotecan. Cancer Res. 1995;55:2097–2103. [PubMed] [Google Scholar]
- 62.Byl JA, Cline SD, Utsugi T, Kobunai T, Yamada Y, Osheroff N. DNA topoisomerase II as the target for the anticancer drug TOP-53: mechanistic basis for drug action. Biochemistry. 2001;40:712–718. doi: 10.1021/bi0021838. [DOI] [PubMed] [Google Scholar]
- 63.Byl JA, Fortune JM, Burden DA, Nitiss JL, Utsugi T, Yamada Y, Osheroff N. DNA topoisomerases as targets for the anticancer drug TAS-103: primary cellular target and DNA cleavage enhancement. Biochemistry. 1999;38:15573–15579. doi: 10.1021/bi991791o. [DOI] [PubMed] [Google Scholar]
- 64.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 65.Bender RP, Lindsey RH, Jr., Burden DA, Osheroff N. N-acetyl-pbenzoquinone imine, the toxic metabolite of acetaminophen, is a topoisomerase II poison. Biochemistry. 2004;43:3731–3739. doi: 10.1021/bi036107r. [DOI] [PubMed] [Google Scholar]
- 66.O'Reilly EK, Kreuzer KN. A unique type II topoisomerase mutant that is hypersensitive to a broad range of cleavage-inducing antitumor agents. Biochemistry. 2002;41:7989–7997. doi: 10.1021/bi025897m. [DOI] [PubMed] [Google Scholar]
- 67.Loike JD. VP16−213 and podophyllotoxin. A study on the relationship between chemical structure and biological activity. Cancer Chemother. Pharmacol. 1982;7:103–111. doi: 10.1007/BF00254530. [DOI] [PubMed] [Google Scholar]
- 68.Long BH, Musial ST, Brattain MG. Comparison of cytotoxicity and DNA breakage activity of congeners of podophyllotoxin including VP16−213 and VM26: a quantitative structure-activity relationship. Biochemistry. 1984;23:1183–1188. doi: 10.1021/bi00301a024. [DOI] [PubMed] [Google Scholar]
- 69.Long BH. Structure-activity relationships of podophyllin congeners that inhibit topoisomerase II. NCI Monogr. 1987;4:123–127. [PubMed] [Google Scholar]
- 70.van Maanen JM, Retel J, de Vries J, Pinedo HM. Mechanism of action of antitumor drug etoposide: a review. J. Natl. Cancer Inst. 1988;80:1526–1533. doi: 10.1093/jnci/80.19.1526. [Review] [DOI] [PubMed] [Google Scholar]
- 71.Saulnier MG, Vyas DM, Langley DR, Doyle TW, Rose WC, Crosswell AR, Long BH. E-ring desoxy analogues of etoposide. J. Med. Chem. 1989;32:1418–1420. doi: 10.1021/jm00127a002. [DOI] [PubMed] [Google Scholar]
- 72.Sinha BK, Politi PM, Eliot HM, Kerrigan D, Pommier Y. Structure-activity relations, cytotoxicity and topoisomerase II dependent cleavage induced by pendulum ring analogues of etoposide. Eur. J. Cancer. 1990;26:590–593. doi: 10.1016/0277-5379(90)90084-7. [DOI] [PubMed] [Google Scholar]
- 73.Long BH. Mechanisms of action of teniposide (VM-26) and comparison with etoposide (VP-16). Semin. Oncol. 1992;19:3–19. [Review] [PubMed] [Google Scholar]
- 74.Long BH, Casazza AM. Structure-activity relationships of VP-16 analogues. Cancer Chemother. Pharmacol. 1994;34:S26–31. doi: 10.1007/BF00684860. [DOI] [PubMed] [Google Scholar]
- 75.Damayanthi Y, Lown JW. Podophyllotoxins: current status and recent developments. Curr. Med. Chem. 1998;5:205–252. [PubMed] [Google Scholar]
- 76.Lee KH. Anticancer drug design based on plant-derived natural products. J. Biomed. Sci. 1999;6:236–250. doi: 10.1007/BF02253565. [DOI] [PubMed] [Google Scholar]
- 77.Chen A, Shapiro MJ. Affinity NMR. Anal. Chem. 1999;71:669A–675A. doi: 10.1021/ac9907179. [DOI] [PubMed] [Google Scholar]
- 78.Roberts GC. NMR spectroscopy in structure-based drug design. Curr. Opin. Biotechnol. 1999;10:42–47. doi: 10.1016/s0958-1669(99)80008-1. [DOI] [PubMed] [Google Scholar]
- 79.Mayer M, Meyer B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angew. Chem. Int. Ed. Engl. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 80.Pellecchia M, Sem DS, Wuthrich K. NMR in drug discovery. Nat. Rev. Drug Discov. 2002;1:211–219. doi: 10.1038/nrd748. [DOI] [PubMed] [Google Scholar]
- 81.Jardine I, Strife RJ, Kozlowski J. Synthesis, 470-MHz 1H NMR spectra, and activity of delactonized derivatives of the anticancer drug etoposide. J. Med. Chem. 1982;25:1077–1081. doi: 10.1021/jm00351a014. [DOI] [PubMed] [Google Scholar]
- 82.Capranico G, Binaschi M. DNA sequence selectivity of topoisomerases and topoisomerase poisons. Biochim. Biophys. Acta. 1998;1400:185–194. doi: 10.1016/s0167-4781(98)00135-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.