Abstract
GTP cyclohydrolase I (GCH-1) is the rate-limiting enzyme in the biosynthesis of tetrahydrobiopterin, an essential cofactor for nitric oxide synthase and aromatic amino acid hydroxylase. To explore the interactome of GCH-1, we established a HEK293 cell line stably expressing tetracycline-inducible FLAG-GCH-1. FLAG-GCH-1 and associated proteins were immunoprecipitated and analyzed by liquid chromatography/tandem mass spectrometry. Twenty-nine proteins, derived from different subcellular components such as cytosol, membranes, nucleus and mitochondria were identified to interact with GCH-1. Cell fractionation studies also showed that GCH-1 was present in the cytosol, membranes and nucleus. Gene ontology analysis revealed that GCH-1 interactome was involved in a variety of biological processes such as signal transduction, apoptosis, metabolism, transport and cell organization. To our knowledge, this study is the first to provide a comprehensive analysis of the GCH-1 interactome. Findings expand the number and diversity of proteins that are known to associate with GCH-1.
Keywords: GTP Cyclohydrolase I, proteomic, interacting proteins, tetrahydrobiopterin, nitric oxide synthase, aromatic amino acid hydroxylase
INTRODUCTION
GTP cyclohydrolase I (GCH-1) is the rate-limiting enzyme in the biosynthesis of tetrahydrobiopterin (BH4), an essential cofactor for the nitric oxide synthase (NOS) isozymes and aromatic amino acid hydroxylase [1]. In diabetic rats, GCH-1 gene transfer reverses BH4 deficiency and increases nitric oxide (•NO) synthesis in endothelial cells and isolated vessels [2]. GCH-1 deficient hlp-1 mice develop pulmonary hypertension and right ventricular hypertrophy because of loss of BH4 and reduced •NO production [3]. Mutation of GCH-1 has been identified as playing a causal role in neurological diseases such as dopa-reponsive dystonia (DRD) and atypical severe phenylketonuria (PKU) [4] because BH4 modulates aromatic acid hydroxylase activity, which is involved in the synthesis of catecholamines and serotonin. Another recent report showed that single nucleotide polymorphisms in GCH-1 were associated with increased susceptibility of patients to develop neuropathic and inflammatory pain [5].
GCH-1 is regulated at the level of transcription, translation and post-translation. GCH-1 mRNA and protein expression can be induced by cytokines such as LPS, interferon-γ, IL-1β and TGF-β. In addition, kinase activators and hormones have been observed to increase GCH-1 mRNA and GCH-1 activity [6, 7]. GCH-1 feedback regulating protein (GFRP) mediates BH4 feedback inhibition and phenylalanine feedforward stimulation of GCH-1 activity via direct interactions with the enzyme [8]. Phosphorylation and ubiquitinization have been recently reported to regulate GCH-1 activity [9, 10].
It is well known that interactions among proteins significantly impact specific biological functions. Activator of Heat Shock 90 kDa Protein and GFRP were found to interact with GCH-1 [8, 11]. Largely, however, the protein partners of GCH-1 remain unknown. The objective of our study was to define the GCH-1 interactome as a means of identifying potential new functions and mechanisms of regulation for GCH-1.
Materials and methods
GCH-1 expressing cell line
The GCH-1 expressing cell line was established using the Flp-In system and the Flp-In™ T-REx™-293 cells (Invitrogen). The Flp-In™ T-REx™-293 cell line stably expresses a Tet repressor that allows tetracycline to induce Tet operator regulated gene expression. GCH-1 cDNA from Sprague Dawley rats was cloned into the pcDNA5/FRT/TO/Topo/TA vector (Invitrogen) with FLAG-tag fused to the N-terminal of the GCH-1 open reading frame. The generated plasmid GCH-1/pcDNA5 was cotransfected into the HEK 293 cell line with the pOG44 plasmid using Lipofectamine™ 2000 (Invitrogen). Transfected cells were selected and individual colonies were established and confirmed for tetracycline regulated expression of GCH-1 by western blot analysis using anti-FLAG antibody (Sigma).
Immunoprecipitation and western blot analysis
The established GCH-1-HEK cells were treated with or without tetracycline for 24 hours and lysed in MOPS buffer and the cell lysates were immunoprecipitated with anti-FLAG antibody as we described previously [12]. The precipitated proteins were eluted with ImmunoPure IgG elution buffer (Pierce Biotechnology) for liquid chromatography/tandem mass spectrometry (LC/MS/MS) assay or separated by SDS-PAGE for western blot analysis.
Sample preparation for mass spectrometry analysis
The eluted samples from the immunoprecipitation were polymerized into a polyacrylamide gel with acrylamide/bis (30%T/2.67%C), ammonium persulfate and TEMED. Gel pieces were fixed in methanol/acetic acid in water for 1 hour, followed by two washes in each of water, 50% acetonitrile and in 50% acetonitrile in 50mM ammonium bicarbonate, pH 8.0. Gel pieces were then dried using a speedvac (Savant). 200 μl 100 mM ammonium bicarbonate, pH 8.0 containing 1 μg Trypsin (Promega) was then added to each gel piece and incubated overnight at 37°C. Afterwards, each gel piece was extracted twice with 70% acetonitrile in 0.1% formic acid. Extracted peptides were resuspended in 6M guanidine-HCl, 5mM potassium phosphate, pH 6.5, and purified and concentrated using a C18 ZipTip (Millipore,).
LC/MS/MS
Peptide mixtures purified above were suspended in 5% acetonitrile in 0.1% formic acid in water and analyzed using an LTQ nano-spray LC/MS with a linear ion trap (Thermo-Fisher, San Jose, CA). Peptide mixtures are separated using a C18 reverse phase column (0.75 Å internal diameter and a flow rate of 1 μl/min) in line with the mass spectrometer. The mobile phases consist of 0.1% formic acid containing 5% acetonitrile (A) and 0.1% formic acid in 95% acetonitrile (B), respectively. Peptides were resolved using a 180-min linear gradient, followed by 60 minutes of equilibration in solvent A. The ions eluted from the column were electrosprayed at a voltage of 1.75 kV. The MS/MS data were searched using the SEQUEST (Version 27; Thermo Fisher) against the UniProt database (www.uniprot.org/). Five independent experiments were performed.
GCH-1 activity assay
After washing with PBS, the cells were scraped into Tris-HCl (50 mM) buffer pH 7.4 containing 1 mM magnesium chloride, 0.1M potassium chloride, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF and protease inhibitors cocktail (Sigma. Cat. P8340). Cell lysates were incubated with 1 mM GTP, 50 μg/ml bovine serum albumin, 1 mM magnesium chloride, 0.1 M potassium chloride, 1 mM EDTA, 1 mM EDTA and 1 mM dithiothreitol in 50 mM Tris-HCl pH 7.4 in a total volume of 200 μl for 1 h at 37°C, 8 μl of 5 N HCl was added and the samples were oxidized with 2% KI/1% I2 for 1 h at R.T. Then samples were decolorized with 5 μl of 20% ascorbic acid and incubated with 0.8U alkaline phosphatase for 1 hour. Samples were centrifuged for 2 min at 10,000 g and neopterin quantified by fluorescence plate reader (Turner Biosystems) at 360/465 nm (Ex/Em) using authentic neopterin standards (Schircks laboratories).
BH4 assay
HEK293 cells were washed with cold DPBS twice, scraped and pelleted. Pellets were immediately lysed in 150μl 50mM phosphate buffer pH 2.6 containing 0.2mM DTPA (Sigma) and 1mM DTE (Sigma). Samples were centrifuged at 12,500 rpm for 10 min at 4°C, and supernatants were filtered through a 10kD cutoff column (Millipore) and analyzed by an HPLC with a electrochemical detector (ESA Biosciences CoulArray® system Model 542) using a Synergi Polar-RP column eluted with argon degassed 50mM phosphate buffer (pH 2.6). Multi-channel coulometric detection was set between 0 and 600 mV. One channel was set at −250 mV to verify the reversibility of BH4 oxidative peak detection. Calibration curves were made by summation of the peak areas collected at 0 and 150 mV for BH4. Intracellular concentrations were calculated using authentic BH4 (10nM–100nM) standards and expressed as a function of cell protein.
Cellular fraction protein preparation
Cytoplasmic and nuclear proteins were prepared by using NE-PER® Nuclear and Cytoplasmic Extraction Reagents (Pierce Biotechnology) according to the manufacturer’s instructions. Briefly, the cell pellet was sequentially added with 200μl CER I and 11μl CERII and vortexed on the highest setting for 5 min. The supernatant containing the cytoplasmic fraction was collected and stored. The pellet was then resuspended with 100μl NER and spun for 15 seconds every 10 minutes, for a total of 40 minutes. Afterward, nuclear extracts were isolated by centrifugation to spin off insoluble material. Membrane proteins were prepared by using Mem-PER Eukaryotic Membrane Protein Extraction Kit (Pierce Biotechnology, 89826) and purified by PAGEprep™ Advance Kit (Pierce Biotechnology). As per manufacturer’s instructions’, cells were lysed with reagent A and then solubilized with a second detergent (mixture of Reagent B and C). The mixture was incubated at 37°C to separate the hydrophobic from hydrophilic proteins by phase partitioning. The extracted membrane proteins were passed through PAGE-prep Resin in the presence of dimethylsulfoxide to remove detergent from the samples. The extracted cytoplasmic, nuclear and membrane proteins were used for GCH-1 activity assays as well as for western blot analysis.
Data analysis
The proteins identified by LC/MS/MS were analyzed using Visualize software (Medical College of Wisconsin). The proteins were screened and analyzed using the following criteria: 1) the unique proteins that only appeared in the tetracycline-induced cells, 2) at least 4 spectral counts unique to the protein of interest, 3) appeared in at least two independent experiments out of five, and 4) the proteins with false discovery rate (FDR) less than 5%. Apropos, UniProtKB/Swiss-Prot Release 55.2 and UniProtKB/TrEMBL Release 38.2 software together with PubMed were used to annotate the Uniprot IDs of the identified proteins with their corresponding gene ontology (GO) annotations. Data are expressed mean ± SD. Comparisons between 2 groups were by 2-way ANOVA, followed by pos-hoc test with Duncan’s Multiple Range test. A value of P < 0.05 was considered statistically significant.
Results and discussion
Generation and characterization of inducible stable cell lines expressing FLAG-tagged GCH-1
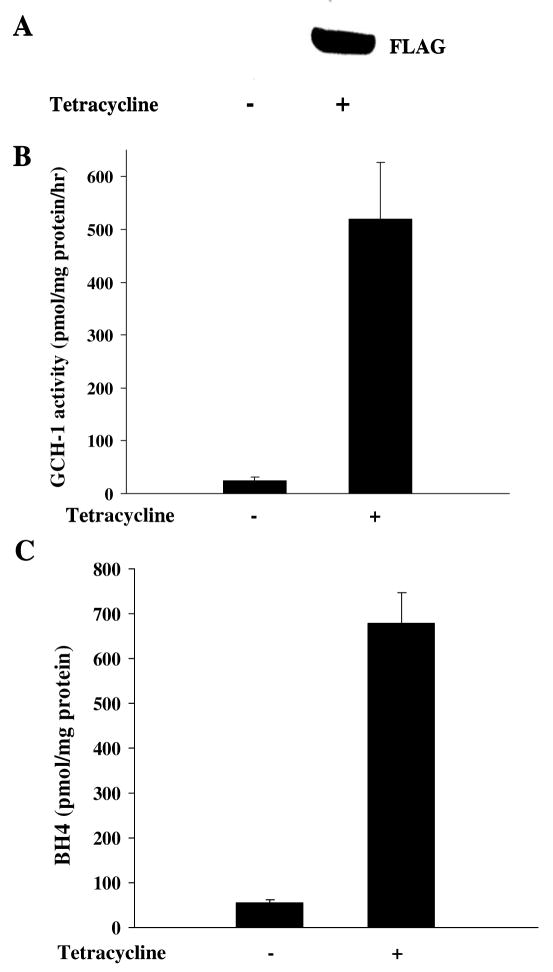
As shown in Fig 1A, GCH-1 expression was dramatically increased in the FLAG-GCH-1-HEK cells after tetracycline stimulation. To determine whether GCH-1 was functional, we measured enzyme activity and BH4 levels in these cells. As can be seen, endogenous GCH-1 activity was quite low (24.6±2.1 pmmol neopterin/mg protein/hr); while tetracycline stimulation substantially increased GCH-1 activity (519.6±37.4 pmmol neopterin/mg/hr) about 20 fold (Figure 1B). Similarly, tetracycline stimulation increased GCH-1 dependent BH4 production from 54.4±7.9 pmmol/mg protein to 679.3±67.6 pmmol/mg protein (Figure 1C). These data demonstrated that FLAG-GCH-1 expression and enzyme activity are dramatically increased by tetracycline in our stably GCH-1 transfected HEK 293 cells.
Figure 1.
Characterization of the cell line stably expressing FLAG-tagged rat GCH-1. FLAG-GCH-1 HEK 293 cells were stimulated with or without tetracycline (1μg/ml) for 24 hours. (A) Whole cell extracts were analyzed by Western blot analysis for the expression of GCH-1 using anti-FLAG antibody. (B) Cells were lysed for GCH-1 activity assay. (C) Cells were lysed and intracellular BH4 levels were measured by HPLC. Data are expressed as mean±SD (n=3). *P<0.05 vs. the cells without tetracycline treatment.
Identification of GCH-1-interacting proteins by LC/MS/MS
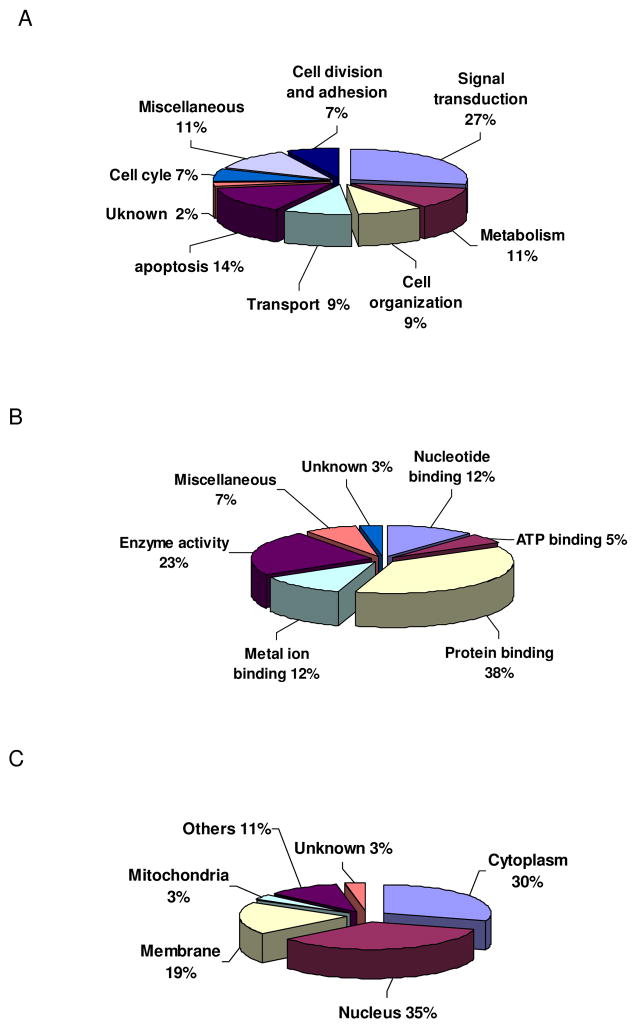
Twenty-nine proteins, including GCH-1, were identified (Table 1). Using GO classification, we discovered that the GCH-1 interacting proteins were involved not only in metabolism but also in many other molecular and cellular functions as shown in Fig 2A. Interestingly, a large amount of the interacted-proteins could bind proteins (37%) and had enzyme activity (23%) such as transferase, peptidase, hydrolase and kinase. Some of the proteins could also bind metal ion, or nucleotide or ATP. MS-based proteomic analysis of the epitope-tag affinity purified protein complexes has been shown to be an effective method for detailing protein interactions [13]. Using this approach in our study, we were able to identify, for the first time, twenty-nine proteins that interact with GCH-1 in a mammalian cell line. Moreover, besides metabolism, most of these identified proteins are also involved in a variety of cell functions, indicating that GCH-1 may have versatile functions.
Table 1.
GCH-1 interactome identified by LC/MS/MS.
| Protein Names | Accession | Scan count | Protein function |
|---|---|---|---|
| N-acetyltransferase-like protein | Q9H0A0 | 47 | Histone acetyltransferase |
| Protein FAM21C | Q9Y4E1 | 32 | Unknown |
| Thyrotropin-releasing hormone degrading ectoenzyme | Q9UKU6 | 29 | Inactivation of TRH |
| Molecule interacting with CasL protein 1 | Q8TDZ2 | 17 | Cytoskeletal regulator |
| Rho-GTPase-activating protein 7 | Q96QB1 | 16 | Cytoskeletal reorganization |
| NKG2-A/B-activating NK receptor | P26715 | 15 | Receptor fo rMHC recognition |
| Proprotein convertase subtilisin/kexin type 9 | Q8NBP7 | 15 | Dfferentiation of cortical neurons |
| Spectrin beta chain, brain 3 | Q9H254 | 13 | Cytoskeletal anchoring |
| Maspin precursor | P36952 | 11 | Tumor suppressor |
| Death-associated protein kinase 3 | O43293 | 10 | Regulator of apoptosis |
| Peroxisome assembly protein 12 | O00623 | 10 | Import into peroxisomes |
| Protocadherin 15 precursor | Q96QU1 | 9 | Cell-adhesion |
| USP6 N-terminal-like protein | Q92738 | 9 | Receptor trafficking |
| Apoptosis inhibitor 5 | Q9BZZ5 | 9 | Antiapoptotic factor |
| Centromere protein H | Q9H3R5 | 8 | Assembly of kinetochore |
| CENP-F kinetochore protein | P49454 | 7 | Chromosome segregation |
| Separin | Q14674 | 7 | Caspase-like protease |
| Latrophilin-2 precursor | O95490 | 6 | Regulation of exocytosis |
| Cyclic AMP-dependent transcription factor ATF-6 alpha | P18850 | 5 | Transcription factor |
| Huntingtin-interacting protein 1-related protein | O75146 | 5 | Component of clathrin-coated pits |
| Polyribonucleotide nucleotidyl-transferase | Q8TCS8 | 5 | mRNA degradation |
| Rho guanine nucleotide exchange factor 3 | Q9NR81 | 5 | GEF for Rho GTPase |
| Baculoviral IAP repeat-containing protein 3 | Q13489 | 4 | Apoptotic suppressor |
| Centromeric protein E | Q02224 | 4 | Microtubule motor |
| Splicing factor 3A subunit 3 | Q12874 | 4 | ‘A’ complex assembly |
| Afamin precursor | P43652 | 4 | Transport |
| Histone H2B | P62807 | 4 | Component of nucleosome |
| Interferon regulatory factor 1 | P10914 | 4 | Transcription factor |
| Proteoglycan-4 precursor | Q92954 | 4 | Boundary lubrication |
Figure 2.
Gene ontology of GCH-1-interacting proteins. (A) Biological process of GCH-1-interating proteins. (B) Molecular functions of GCH-1-interacting proteins. (C) Cellular location of GCH-1-interacting proteins.
As a key enzyme in de novo BH4 biosynthesis, GCH-1 is critical for maintaining coupled NOS activity and aromatic amino acid hydroxylation [1], pain sensitivity and chronicity [5] and immune responses. These diverse biological processes make GCH-1 an important regulator in neurological, cardiovascular and inflammatory diseases. Up to now, however the details concerning the cellular and molecular mechanisms regulating GCH-1 activity have been limited. In this study, we found that GCH-1 interacted with many signal transduction proteins such as receptors (NKG2-A/B-actvating NK receptor and latrophilin-2), kinase (Death-associated protein kinase 3), adaptors (Rho-GTPase-activating protein 7 and Rho-guanine nucleotide exchange factor 3) and transcriptional factors (ATF-6 alpha and interferon regulatory factor 1). Rho GTPase is well-known for its regulation of cytoskeleton reorganization which is critical for cell division, movement and axon guidance [14]. In Drosophila, the mutation of GCH-1 correlates nuclear division defects in embryo [15]; and in baby hamster kidney cells, GCH-1 mutation at 201 induces intracellular inclusion bodies [16]. Thus, it would be interesting to study whether GCH-1 is regulated by Rho GTpase to function in cellular dynamics. In addition, it is well established that GCH-1 gene expression is induced by interferon γ and increased neopterin levels are indicated as an inflammatory marker [1]. The interaction of GCH-1 with interferon regulatory factor 1 may indicate that besides being regulated by interferon γ, GCH-1 may also coordinate with interferon γ to regulate immune response. One possible explanation of GCH-1 interaction with signal transduction molecules is that these molecules may be responsible for regulating GCH-1. For example, if GCH-1, bound to a membrane receptor, is released upon receptor activation, this could link cell signaling to acute BH4 synthesis. An alternative explanation is that GCH-1 itself may serve as a signaling molecule, which may or may not be related directly to BH4 function. We also observed that GFRP, the known regulating protein of GCH-1, was not detected with GCH-1; one explanation might be that GFRP is very weakly expressed in HEK 293 cells (realtime-PCR data, not shown) make it difficult to be found using our proteomic approaches.
BH4 is a critical cofactor for aromatic hydroxylases. In this study, 23% of GCH-1 interacted proteins have enzyme activity, especially, hydrolase activity. For example, Molecule Interacting with CasL protein 1 (MICALs) is structurally similar to aromatic hydroxylases and amine oxidases that reduce molecular oxygen to H2O2 in the presence of NADPH [17]. As its enzyme activity is similar to NOS, MICALS may also be BH4 dependent by interaction with GCH-1. Support for this type of relationship comes from a report that GCH-1 co-localizes with aromatic hydroxylases in the brain [18]. Therefore, GCH-1 protein interactions likely afford an advantage by providing highly localized concentrations of BH4 for maintaining optimal enzyme activity for hydroxylases.
Although GCH-1 has also been reported to be involved in cell cycle regulation, cell proliferation and apoptosis, the molecular mechanisms by which it is involved remain unclear. In rat thymocytes, cell cycle progression is associated with a transient increase in GCH-1 activity, which peaks at the cell’s entry into the S phase [19]. Interestingly, we found that GCH-1 could interact with centromere protein H which increases at the S phase [20]. GCH-1 has also been reported to play a protective role in cell survival. Inhibition of GCH-1 by 2,4-diamino-6-hydroxypyrimidine decreases viability of PC12 cell numbers by increasing •NO toxicity [21], accelerates stress-induced endothelial cell death [22] and increases neuronal vulnerability to hypoxia-induced mitochondrial damage and necrosis [23]. With GCH-1 appearing to play so many key roles in cell cycle and programmed cell death, it is important to note that we also discovered that GCH-1 associates with anti-apoptotic proteins, apoptosis inhibitor 5 and 2. Such protein-protein interactions may contribute to the protective effect of GCH-1 in maintaining cell viability in a number of different conditions.
Subcellular localization of GCH-1 and its interacted proteins
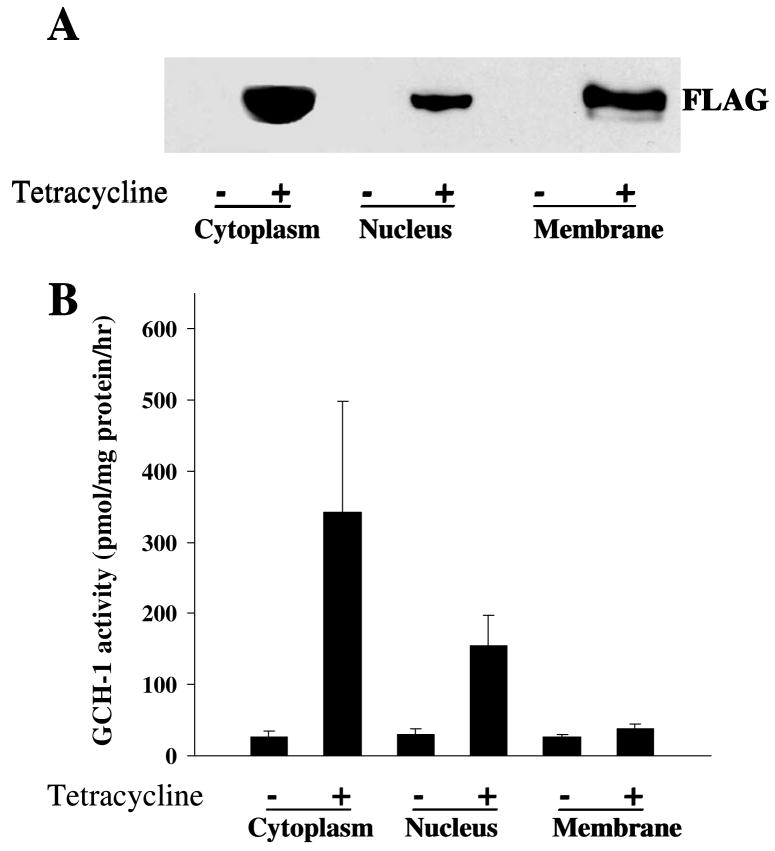
Interestingly, the identified proteins were not only in the cytoplasm (30%) but also in the nucleus (35%), membrane (19%) and mitochondria (3%) as shown in Figure 2C. If GCH-1 interacts with proteins from all of these different subcellular locations, it seems logical that GCH-1 should also be present in these different locations. Our cell fractionation studies together with western blot analysis confirmed that GCH-1 is expressed in the nucleus, cell membranes and the cytosol (Fig 3A). Furthermore, GCH-1 activity was higher in the cytoplasm and the nucleus than that in the membranes as shown in Fig 3B. Although GCH-1 activity in cell membranes was increased by tetracycline stimulation, no statistical difference was observed in cell membranes from cells with/without tetracycline stimulation. GCH-1 and its GCH-1-interacting proteins diversely distribute in the cytoplasm, membranes, nucleus, and mitochondria, suggesting that GCH-1 may locate or translocate among the cytoplasm, nucleus and cell membranes to interact with other regulating proteins. It is reported that GCH-1 was distributed in both the cytosol and the nucleus in epidermal keratinocytes and melanocytes [24]. Moreover, in COS-1 cells, over-expression of GCH-1 resulted in the detection of active enzyme in the nucleus [25]. Thus, the significance of our findings in the differences in enzyme activity and locations remains to be determined. Such diversity in association partners may have broad implications for the cell biology of GCH-1.
Figure 3.
Subcellular distribution of GCH-1 interacting proteins. Cell fractional proteins (cytoplasmic, nuclear and membrane proteins) from GCH-1 HEK cells were prepared using specific extraction reagents from Pierce Biotechnology. (A) 30μg proteins from each fraction were loaded on the SDS-PAGE and immunoblotted with anti-FLAG. (B) 50μl cell lysates from each fraction were analyzed for GCH-1 activity and normalized by their protein concentration. n=3/experiment
Conclusion
Using mass spectrometry-based proteomics, we identified twenty-nine proteins that interact with GCH-1. As the proteins come from multiple locations and are responsible for a broader scope of functions than previously recognized, our data suggest that GCH-1’s role in cell biology may be much greater than previously considered. Further characterization of these protein: protein interactions and their biological significance will be required before we can develop a comprehensive understanding of the functions of GCH-1 and the mechanisms of GCH-1 related diseases such as DRD and PKU.
Acknowledgments
This work was supported by National Institutes of Health Grants HL71214 (to K. A. P.) and HL080468 (to Y. S.). The administrative assistance of Meghann Sytsma and Anne Laulederkind are gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thony B, Auerbach G, Blau N. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem J. 2000;347(Pt 1):1–16. [PMC free article] [PubMed] [Google Scholar]
- 2.Meininger CJ, Cai S, Parker JL, Channon KM, Kelly KA, Becker EJ, Wood MK, Wade LA, Wu G. GTP cyclohydrolase I gene transfer reverses tetrahydrobiopterin deficiency and increases nitric oxide synthesis in endothelial cells and isolated vessels from diabetic rats. Faseb J. 2004;18:1900–1902. doi: 10.1096/fj.04-1702fje. [DOI] [PubMed] [Google Scholar]
- 3.Nandi M, Miller A, Stidwill R, Jacques TS, Lam AA, Haworth S, Heales S, Vallance P. Pulmonary hypertension in a GTP-cyclohydrolase 1-deficient mouse. Circulation. 2005;111:2086–2090. doi: 10.1161/01.CIR.0000163268.32638.F4. [DOI] [PubMed] [Google Scholar]
- 4.Ichinose H, Nagatsu T. Molecular genetics of hereditary dystonia--mutations in the GTP cyclohydrolase I gene. Brain Res Bull. 1997;43:35–38. doi: 10.1016/s0361-9230(96)00353-x. [DOI] [PubMed] [Google Scholar]
- 5.Tegeder I, Costigan M, Griffin RS, Abele A, Belfer I, Schmidt H, Ehnert C, Nejim J, Marian C, Scholz J, Wu T, Allchorne A, Diatchenko L, Binshtok AM, Goldman D, Adolph J, Sama S, Atlas SJ, Carlezon WA, Parsegian A, Lotsch J, Fillingim RB, Maixner W, Geisslinger G, Max MB, Woolf CJ. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med. 2006;12:1269–1277. doi: 10.1038/nm1490. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Huang FL, Feng Q, Liu J, Fan SX, McKenna TM. Overexpression of protein kinase C alpha enhances lipopolysaccharide-induced nitric oxide formation in vascular smooth muscle cells. J Cell Physiol. 1998;176:402–411. doi: 10.1002/(SICI)1097-4652(199808)176:2<402::AID-JCP19>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 7.Tabraue C, Diaz Penate R, Gallardo G, Hernandez I, Quintana J, Lopez Blanco F, Gonzalez Reyes J, Fanjul LF, Ruiz de Galarreta CM. Induction of guanosine triphosphate-cyclohydrolase by follicle-stimulating hormone enhances interleukin-1 beta-stimulated nitric oxide synthase activity in granulosa cells. Endocrinology. 1997;138:162–168. doi: 10.1210/endo.138.1.4854. [DOI] [PubMed] [Google Scholar]
- 8.Yoneyama T, Hatakeyama K. Decameric GTP cyclohydrolase I forms complexes with two pentameric GTP cyclohydrolase I feedback regulatory proteins in the presence of phenylalanine or of a combination of tetrahydrobiopterin and GTP. J Biol Chem. 1998;273:20102–20108. doi: 10.1074/jbc.273.32.20102. [DOI] [PubMed] [Google Scholar]
- 9.Widder JD, Chen W, Li L, Dikalov S, Thony B, Hatakeyama K, Harrison DG. Regulation of Tetrahydrobiopterin Biosynthesis by Shear Stress. Circ Res. 2007;101:830–838. doi: 10.1161/CIRCRESAHA.107.153809. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Wu Y, Song P, Zhang M, Wang S, Zou MH. Proteasome-dependent degradation of guanosine 5′-triphosphate cyclohydrolase I causes tetrahydrobiopterin deficiency in diabetes mellitus. Circulation. 2007;116:944–953. doi: 10.1161/CIRCULATIONAHA.106.684795. [DOI] [PubMed] [Google Scholar]
- 11.Swick L, Kapatos G. A yeast 2-hybrid analysis of human GTP cyclohydrolase I protein interactions. J Neurochem. 2006;97:1447–1455. doi: 10.1111/j.1471-4159.2006.03836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, Shi Y, Wang J, Jones D, Weilrauch D, Ying R, Wakim B, Pritchard KA., Jr A Heat Shock Protein 90 Binding Domain in Endothelial Nitric-oxide Synthase Influences Enzyme Function. J Biol Chem. 2007;282:37567–37574. doi: 10.1074/jbc.M706464200. [DOI] [PubMed] [Google Scholar]
- 13.Chang IF. Mass spectrometry-based proteomic analysis of the epitope-tag affinity purified protein complexes in eukaryotes. Proteomics. 2006;6:6158–6166. doi: 10.1002/pmic.200600225. [DOI] [PubMed] [Google Scholar]
- 14.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Reynolds ER, Ranganayakulu G, O’Donnell JM. A maternal product of the Punch locus of Drosophila melanogaster is required for precellular blastoderm nuclear divisions. J Cell Sci. 1994;107( Pt 12):3501–3513. doi: 10.1242/jcs.107.12.3501. [DOI] [PubMed] [Google Scholar]
- 16.Chiou YW, Hwu WL, Lee YM. Hsp27 decreases inclusion body formation from mutated GTP-cyclohydrolase I protein. Biochim Biophys Acta. 2008;1782:169–179. doi: 10.1016/j.bbadis.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Nadella M, Bianchet MA, Gabelli SB, Barrila J, Amzel LM. Structure and activity of the axon guidance protein MICAL. Proc Natl Acad Sci U S A. 2005;102:16830–16835. doi: 10.1073/pnas.0504838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dassesse D, Hemmens B, Cuvelier L, Resibois A. GTP-cyclohydrolase-I like immunoreactivity in rat brain. Brain Res. 1997;777:187–201. doi: 10.1016/s0006-8993(97)01111-6. [DOI] [PubMed] [Google Scholar]
- 19.Schott K, Brand K, Hatakeyama K, Kagamiyama H, Maier J, Werner T, Ziegler I. Control of cell-cycle-associated tetrahydrobiopterin synthesis in rat thymocytes. Exp Cell Res. 1992;200:105–109. doi: 10.1016/s0014-4827(05)80077-1. [DOI] [PubMed] [Google Scholar]
- 20.Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshimura K, Murakami Y, Tanaka J, Yamamoto M, Kato Y. Effect of tetrahydrobiopterin on nitric oxide synthase-containing cells in the rat hippocampus. Neurosci Res. 2004;50:161–167. doi: 10.1016/j.neures.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Ishii M, Shimizu S, Yamamoto T, Momose K, Kuroiwa Y. Acceleration of oxidative stress-induced endothelial cell death by nitric oxide synthase dysfunction accompanied with decrease in tetrahydrobiopterin content. Life Sci. 1997;61:739–747. doi: 10.1016/s0024-3205(97)00538-9. [DOI] [PubMed] [Google Scholar]
- 23.Delgado-Esteban M, Almeida A, Medina JM. Tetrahydrobiopterin deficiency increases neuronal vulnerability to hypoxia. J Neurochem. 2002;82:1148–1159. doi: 10.1046/j.1471-4159.2002.01055.x. [DOI] [PubMed] [Google Scholar]
- 24.Chavan B, Gillbro JM, Rokos H, Schallreuter KU. GTP cyclohydrolase feedback regulatory protein controls cofactor 6-tetrahydrobiopterin synthesis in the cytosol and in the nucleus of epidermal keratinocytes and melanocytes. J Invest Dermatol. 2006;126:2481–2489. doi: 10.1038/sj.jid.5700425. [DOI] [PubMed] [Google Scholar]
- 25.Elzaouk L, Laufs S, Heerklotz D, Leimbacher W, Blau N, Resibois A, Thony B. Nuclear localization of tetrahydrobiopterin biosynthetic enzymes. Biochim Biophys Acta. 2004;1670:56–68. doi: 10.1016/j.bbagen.2003.10.015. [DOI] [PubMed] [Google Scholar]