Abstract
A family of bioreducible poly(disulfide amine)s, which differ in the length of polymethylene spacer [–(CH2)n–] in the main chain and the side chain, has been synthesized. These bioreducible poly(disulfide amine)s exhibit local environment specific degradability and are associated with lower cytotoxicity than branched poly(ethylenimine) (bPEI, 25kDa). These cationic polymers also show higher buffering capacity and protonation degree than bPEI, facilitating the endosomal escape of carried genetic materials. The transfection efficiency of these agents is polymethylene length dependent. Poly(cystaminebisacrylamide-spermine) [poly(CBA-SP);], poly(cystaminebisacrylamide-bis(3-aminopropyl)-1,3-propanediamine) [poly(CBA-APPD);], and poly(cyxtaminebisacrylamide-bis(3-aminopropyl)-ethylenediamine) [ploy(CBA-APED);] with longer propylene [–(CH2)3–] side spacer, demonstrate higher transfection efficacy than the counterpart poly(cystaminebisacrylamide-bis(2-aminoethyl)-1,3-propanediamine) [poly(CBA-AEPD);] and poly(cystaminebisacrylamide-triethylenetetramine) [poly(CBA-TETA);], which have shorter ethylene [–(CH2)2–] side spacer. The poly(CBA-SP), poly(CBA-APPD), poly(CBA-APED) with the main chain spacer of –(CH2)4–, –(CH2)3–, –(CH2)2– demonstrate similar transfection efficiency, indicating the length of polymer main chain spacer has less influence on transfection efficiency. However, with the same short ethylene [–(CH2)2–] side spacer, poly(CBA-AEPD), with the longer main chain oligomethylene units [–(CH2)3–], showed relatively higher transfection efficiency than poly(CBA-TETA), having shorter main chain oligomethylene units [–(CH2)2–]. Of these polymeric carriers, poly(CBA-SP) demonstrated the highest transfection in the C2C12 cell line, while poly(CBA-APED) showed the highest transfection in the Hela cell line. All of these agents showed greater transfection activity than commercialized bPEI 25kDa. The poly(disulfide amine)s are promising safe and efficient non-viral vectors for gene delivery.
Keywords: poly(disulfide amine)s, biodegradable, gene delivery, gene transfection
1. Introduction
The development of safe and efficient vectors is a major challenge in the development of gene therapy to treat human diseases[1, 2]. Gene delivery vectors are classified into viral and non-viral vectors, whose advantages and disadvantages have been well documented[3, 4]. Cationic polymers are one of the main categories of non-viral vectors, having received greater attention recently because of their inherent advantages, including non-immunogenicity, stability, capacity to carry large nucleic acid loads, and ease of manufacturing[5, 6]. By condensing nucleic acids into nanoparitcles through electrostatic interactions, polymeric carriers can protect nucleic acids from nuclease degradation, facilitate cellular uptake of polymer/DNA complexes (polyplexes) via endocytosis, and induce efficient gene expression[5, 7]. Over the last two decades, many cationic polymers have been synthesized for gene delivery, such as poly(ethylenimine) (PEI), poly(L-lysine) (PLL) and polyamidoamine dendrimers[8, 9]. The main drawback for these cationic polymers, however, is their high level of cytotoxicity, mostly due to their non-degradability and accumulation[10]. Therefore, the design of new vectors to overcome the problem of cytotoxicity and increase the efficiency of gene transfection has become mandatory to advance the development of clinically efficacious gene therapy.
To successfully deliver DNA into nucleus, polymeric vectors must have multiple functional groups to overcome a series of extra- and intracellular barriers. The first barrier is that polymers need to condense DNA and form stable and positively-charged polyplexes[1, 9, 10]. Previously, poly(amido amine)s (PAAs) with protonated amines bind DNA more strongly than their counterpart polymers lacking protonated amine groups[11]. The modification of poly(β-amino ester)s end groups from alcohol to primary amine groups also increases the polymers' DNA binding affinity[12, 13]. Protonatable primary amine groups in the pendant side chains allow for strong DNA binding ability and charge density and therefore the ability to condense DNA into nanosized particles extracellularly.
The second barrier to gene delivery is the ability of the polymer to escape from the endosomal-lysosomal pathway[3, 10]. Behr and others introduced the hypothesis of the “proton sponge effect” and defined polymers having good buffering capacity (as the percentage of amine groups becoming protonated between pH 7.4 and 5.1) as facilitating endosomal rupture via osmolysis and thereby inducing efficient gene expression[9, 14]. The combination of titratable amine groups with basic pKa values enables good buffering capacity of a polymer and enhances DNA binding capability. As a result, gene transfection efficiency increases by incorporating different amine groups into polymer structures. Our previously synthesized poly(amido ethylenimine)s containing different amine groups have demonstrated significant higher transfection efficiencies than bPEI 25kDa in a variety of cell lines[15, 16]. Poly(amido amine)s (PAAs) containing different amine groups showed high buffering capacities and similar or higher transfection efficiency than bPEI 25kDa in COS-7 cells[17].
To efficiently induce gene expression with low cytotoxicity, polymeric vectors should not only have superior DNA binding capability and buffering capacity, but also be able to be degraded into small and non-toxic molecules[4]. To achieve this goal of degradability, the use of disulfide bonds has received more attention in the design of “bioreducible” polymers[18, 19]. Polymers with disulfide bonds can form stable complexes with DNA in the extracellular oxidative environment, while readily release DNA intracellulary via the cleavage of the disulfide bonds by glutathione and other small redox molecules with free thiol groups (5 – 10 mM) through the efficient disulfide-thiol exchange reaction[20, 21]. As a result, gene expression is enhanced and cytotoxicity is significantly reduced. Previously, improved gene transfection efficiency and decreased cytotoxicity have been observed when using low molecular weight PEI with disulfide bonds[22, 23]. Recently, PAAs containing disulfide bonds have been demonstrated to have higher transfection efficiency and lower cytotoxicity than the counterpart PAAs without disulfide linkages[24].
To explore the studies of (1) side chain length and (2) main chain spacer on polymeric DNA binding capability, buffering capacity, protonation degree and basicity, biodegradability and gene transfection, we synthesized a family of biodegradable poly(disulfide amine)s as polymeric gene vectors. These poly(disulfide amine)s are synthesized with defined structures: two pendant primary amine groups at the side chains, two tertiary amine groups and one disulfide bond in the backbond in each repeating unit and different lengths of oligomethylene spacers [–(CH2)n–, n = 2-4] in the main and side chains. The characterization of the polymers includes the measurement of buffering capacity and particle size, DNA condensation and releasing ability, in vitro transfection efficiency, cytotoxicity and fluorescence-labeled cellular uptake. We hypothesized that chemical structures in polymer design may play important roles in gene transfection efficiency and cytotoxicity.
2. Materials and methods
2.1. Materials
All chemicals: spermine (SP, Sigma, St. Louis, MO), N,N′-bis(3-aminopropyl)-1,3-propanediamine (APPD, Sigma-Aldrich, St. Louis, MO), N,N′-bis(3-aminopropyl)-ethylenediamine (APED, Acros Organics, Fair Lawn, NJ), N,N′-bis(2-aminoethyl)-1,3-propanediamine (AEPD, Sigma-Aldrich, St. Louis, MO), triethylenetetramine (TETA, Sigma-Fluka, St. Louis, MO), N,N′-cystaminebisacrylamide (CBA, PolySciences, Warrington, PA), branced polyethylenimine (bPEI, Mw=25kDa, Sigma, St. Louis, MO), ethylenediamine (EDA, Sigma-Aldrich, St. Louis, MO), 2-acetyldimedone (Dde-OH, EMD Chemicals, Inc. Gibbstown, NJ), hydroxylamine hydrochloride (NH2OH·HCl, Sigma-Aldrich, St. Louis, MO), imidazole (Sigma-Aldrich, St. Louis, MO), N-methyl-2-pyrrolidinone (NMP, Sigma-Aldrich, St. Louis, MO), N,N-dimethylformamide (DMF, Sigma-Aldrich, St. Louis, MO), 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT, Sigma, St. Louis, MO), dimethyl sulfoxide (DMSO, Sigma-Aldrich, St. Louis, MO), dithiothreitol (DTT, Sigma-Aldrich, St. Louis, MO), and SYBR® Safe DNA gel stain (10,000×concentrate in DMSO, Invitrogn, Carlsbad, CA) were purchased in the highest purity and used without further purification. The plasmid pCMV-Luc, containing a firefly luciferase reporter gene, was amplified in E.coli DH5α and purified by standard Maxiprep kit (Invitrogen, Carlsbad, CA). Dulbecco's Modified Eagle's medium (DMEM), penicillin-streptomycin (P/S), fetal bovine serum (FBS), trypsin-like enzyme (TrypLE Express) and Dulbecco's phosphate buffered saline (PBS) were all purchased from Invitrogen-Gibco (Carlsbad, CA). Luciferase assay system with reporter lysis buffer was purchased from Promega (Madison, WI). The BCATM protein assay system was purchased from Thermo Scientific (Rockford, IL). YOYO-1 iodide (1 mM solution in DMSO) was purchased from Molecular Probes (Eugene, OR). Hela cells (human cervical cancer cell line) and C2C12 (mouse myoblast cell line) were purchased from the American Type Culture Collection (ATCC) and cultured according to recommended protocols.
2.2. Polymer synthesis
The synthesis of five novel poly(disulfide amine)s was shown in Scheme 1. The synthetic route of poly(cystaminebisacrylamide-spermine) [poly(CBA-SP);] is described here as the representative procedure. Briefly, SP (0.202g, 1 mmol) and Dde-OH (0.419 g, 2.3 mmol) were dissolved in 1 mL MeOH, stirring at room temperature for 24 h to let Dde group specifically protect primary amine groups in spermine. The next day, CBA (0.260g, 1 mmol) was added into above system in 1.5 mL of MeOH/diH2O (9/1 v/v). Polymerization was conducted at 60°C in the dark under nitrogen atmosphere for 3-4 days. Then, 10% mol excess EDA was added to consume any unreacted acrylamide groups, stirring at 60°C for at least additional 2 h. The product was precipitated in 40 mL anhydrous diethyl ether to get the intermediate polymer poly(CBA-SP-Dde). The Dde groups were removed in the deprotection mixture of NH2OH·HCl/Imidazole/NMP/DMF, stirring at room temperature for 4 h. The deprotection mixture was prepared as follows: 1.25 g (1.80 mmol) of NH2OH·HCl and 0.918 g (1.35 mmol) of imidazole were suspended in 5 mL of NMP. The mixture was sonicated until complete dissolution. Just before reaction, 5 volume of this solution was diluted with 1 volume of DMF to make the deprotection mixture[25]. After deprotection, the crude product was purified by dialysis (MWCO = 1000) against MilliQ water for 24 h, followed by lypholization to obtain poly(CBA-SP) as solid gel. poly(cystaminebisacrylamide-bis(3-aminopropyl)-1,3-propanediamine) [poly(CBA-APPD);], poly(cyxtaminebisacrylamide-bis(3-aminopropyl)-ethylenediamine) [ploy(CBA-APED);], poly(cystaminebisacrylamide-bis(2-aminoethyl)-1,3-propanediamine) [poly(CBA-AEPD);] and poly(cystaminebisacrylamide-triethylenetetramine) [poly(CBA-TETA);] were synthesized in the same way. These poly(disulfide amine)s were then analyzed by 1H NMR (400 MHz, D2O, δ, ppm) and the data were listed as below:
Scheme 1.
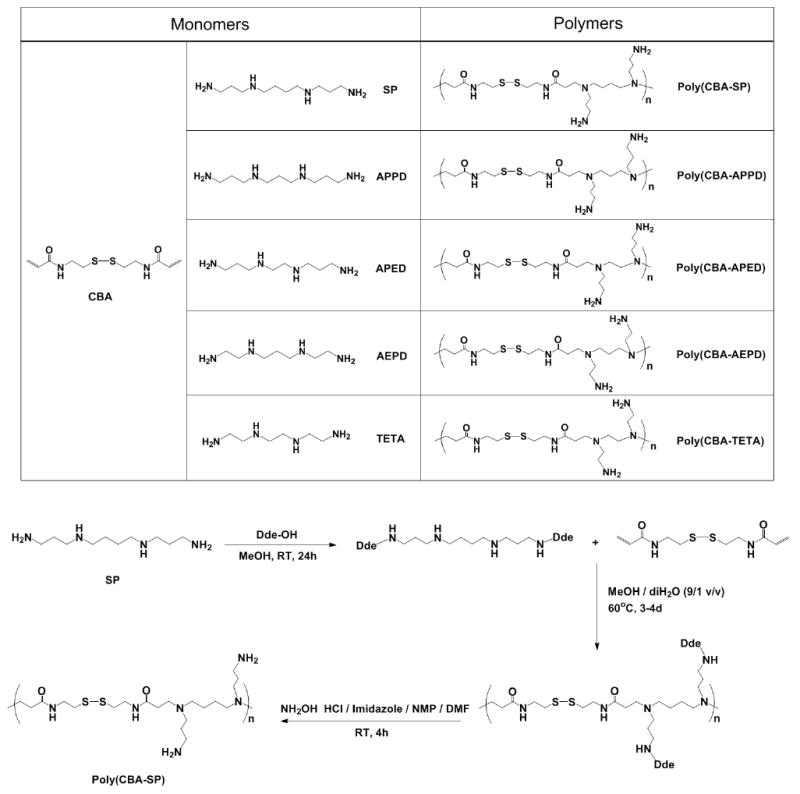
Synthesis of poly(disulfide amine)s using oligoamines and bisacryamide monomers ((poly(CBA-SP), poly(CBA-APPD), poly(CBA-APED), poly(CBA-AEPD) and poly(CBA-TETA)). The synthetic route of poly(CBA-SP) is shown as a representative procedure.
Poly(CBA-SP): δ = 3.37 (CONHCH2CH2SS, 4H), 2.87 (CONHCH2CH2SS, 4H), 2.77 (NCH2CH2CH2NH2, 4H), 2.70 (NHCOCH2CH2N, 4H), 2.53 (NCH2CH2CH2NH2, 4H), 2.47 (NCH2CH2CH2CH2N, 4H), 2.34 (NHCOCH2CH2N, 4H), 1.75 (NCH2CH2CH2NH2, 4H), 1.35 (NCH2CH2CH2CH2N, 4H).
Poly(CBA-APPD): δ = 3.36 (CONHCH2CH2SS, 4H), 2.86 (CONHCH2CH2SS, 4H), 2.71 (NHCOCH2CH2N, 4H; NCH2CH2CH2NH2, 4H), 2.46 (NCH2CH2CH2N, 4H), 2.36 (NCH2CH2CH2NH2, 4H), 2.30 (NHCOCH2CH2N, 4H), 1.70 (NCH2CH2CH2NH2, 4H), 1.53 (NCH2CH2CH2N, 2H).
Poly(CBA-APED): δ = 3.38 (CONHCH2CH2SS, 4H), 2.86 (CONHCH2CH2SS, 4H), 2.70 (NHCOCH2CH2N, 4H; NCH2CH2CH2NH2, 4H), 2.49 (NCH2CH2CH2NH2, 4H; NCH2CH2N, 4H), 2.30 (NHCOCH2CH2N, 2H), 1.50 (NCH2CH2CH2NH2, 4H).
Poly(CBA-AEPD): δ = 3.37 (CONHCH2CH2SS, 4H), 2.94 (CONHCH2CH2SS, 4H), 2.82 (NCH2CH2NH2, 4H), 2.70 (NHCOCH2CH2N, 4H), 2.60 (NCH2CH2NH2, 4H), 2.44 (NCH2CH2CH2N, 4H), 2.30 (NHCOCH2CH2N, 4H), 1.48 (NCH2CH2CH2N, 2H).
Poly(CBA-TETA): δ = 3.38 (CONHCH2CH2SS, 4H), 2.93 (CONHCH2CH2SS, 4H), 2.83 (NCH2CH2NH2, 4H), 2.69 (NHCOCH2CH2N, 4H), 2.62 (NCH2CH2NH2, 4H), 2.49 (NCH2CH2N, 4H), 2.30 (NHCOCH2CH2N, 4H). The 1H NMR spectra of all poly(disulfide amine)s are given in the Supporting Information.
2.3. Polymer characterization
The molecular weights of five poly(disulfide amine)s were determined by size exclusion chromatography (SEC) on an AKTA FPLC system (Amersham Biosciences, Piscataway, NJ) equipped with a superose 12 column, and UV and refractive index detectors, eluted with Tris buffer (20 mM, pH 7.4) at a rate of 0.5 mL/min. The molecular weights and polydispersity index (PDI =Mw/Mn) were calibrated with standard poly[N-(2-hydroxyproyl)methacrylamide;] (pHPMA). The FPLC spectra data of polymers are given in the Supporting Information.
2.4. Acid-base titration
The buffering capacity of poly(disulfide amine)s was determined by acid-base titration. An amount equal to 5 mmol of amine groups of poly(disulfide amine)s was dissolved in 10 mL of 0.1 M NaCl solution. The pH of polymer solutions was initially set to pH 11.0 by 0.1 M NaOH and titrated to pH 3.0 by using 0.01 M HCl. bPEI 25kDa and 0.1 M NaCl solutions were titrated in the same way as controls. The pH of the solutions was measured after each addition by a pH meter (Corning 340, Mettler-Toledo, Inc, Columbus, OH). Buffering capacity is defined as the percentage of amine groups that become protonated from pH 7.4 to 5.1 and calculated using the following equation[11]:
Here ΔVHCl is the volume of HCl solution (0.01 M) which brought the pH value of polymer solutions from 7.4 to 5.1, and N mol (5 mmol) is the total moles of protonable amine groups in poly(disulfide amine)s and bPEI. The relationship between pH and protonation degree of polymers (α) is determined from the obtained titration curve. Further, the apparent pKa of polymers was plotted against 1-α, while pKa = pH + log [α/(1-α);][26-28].
2.5. Dynamic light scattering (DLS)
The size of polymer/DNA polyplexes was measured using a dynamic light scattering detector (BI-200SM system, Brookhaven Instruments, Holtsville, NY) equipped with a 5 mW helium neon incident laser beam at 633 nm output wavelength. Polyplexes were prepared at weight ratios (w/w) of 1, 5, 10, 20 and 30 by mixing poly(disulfide amine)s and bPEI 25 kDa with 1 μg plasmid DNA in diH2O, followed by vortexing for 5 s and incubating at room temperature for 30 min. The polyplexes were then diluted in 2 mL of dust-free diH2O. Measurements were made at 25°C at a scattering angle of 90°. Each measurement was repeated triplicate. The particle sizes were expressed as effective diameters and mean values ± standard deviations.
2.6. Gel electrophoresis assay
Agarose gel (1%, w/v) containing 0.5 μg/mL SYBR® Safe DNA gel stain was prepared in TAE (Tris-Acetate-EDTA) buffer. Poly(disulfide amine)s/DNA complexes (0.5 μg DNA) were prepared in 20 μl of 20 mM HEPES buffer (pH 7.4) at w/w ratios of 0.1, 0.2, 0.5, 1, 2, 5, 10, 20 and 30, followed by vortexing for 5 s and incubating at room temperature for 30 min. bPEI 25kDa/DNA complexes (w/w 1:1) was prepared for comparison. The samples were mixed with 4 μl of 6 × loading dye and the mixtures were loaded onto an agarose gel. The gel was run at 100 V for 30 min and the location of DNA bands was visualized by a UV illuminator using a Gel Documentation Systems (Bio-Rad, Hercules, CA). The DNA release from polyplexes was evaluated by incubating polyplexes with 10 mM DTT at 37°C for 1 h. The samples were analyzed by gel electrophoresis as same manners.
2.7. In vitro transfection
Poly(disulfide amine)s mediated gene transfection was evaluated on Hela and C2C12 cell lines by using luciferase reporter gene (pCMV-Luc, 1.0 μg/mL) in the absence of serum. Cells were maintained in DMEM containing 10% FBS, streptomycin (100 μg/mL) and penicillin (100 units/mL) at 37°C with 5% CO2 humidified atmosphere. 24 h prior to transfection, cells were seeded in 24-well plates at an initial density of 4.5 × 104 cells/well. Poly(disulfide amine)s/DNA complexes were prepared in 20 mM HEPES buffer (pH 7.4) 30 min before transfection at w/w ratio of 1, 5, 10, 20 and 30. bPEI (25 kDa)/DNA complexes (w/w 0.6:1) transfected cells and non-treated cells were used as positive and negative controls. At the time of transfection, the medium in each well was replaced with fresh serum-free medium. Polyplexes were added into each well and incubated with the cells at 37°C for 4 h. Then the serum-free medium was replaced with 500 μL of fresh complete medium. With additional 44 h incubation, cells were washed with pre-warmed PBS, treated with 100 μL cell lysis buffer and subjected to a freezing-thawing cycle. Cellular debris was removed by centrifugation at 16,000 rpm for 2 min. The luciferase activity in cell lysate (25 μL) was measured using a luciferase assay kit (100 μL luciferase assay buffer) on a luminometer (Dynex Technologies Inc., Chantilly, VA). The relative luminescent unit (RLU) of luciferase expression was normalized against protein concentration in the cell extracts, measured by a BCA protein assay kit. All transfection assays were carried out in triplicate.
2.8. Cytotoxicity assay
In vitro cytotoxicity of poly(disulfide amine)s was evaluated using MTT assay in C2C12 cells. Polyplexes were prepared at w/w ratio of 1, 5, 10, 20 and 30 as described above 30 min before use with bPEI 25kDa as comparison. Cells were incubated with polyplexes in serum-free medium for 4 h followed by 20 h incubation in complete medium with 10% FBS. MTT solution in PBS (50 μL, 2 mg/mL) was then added and the cells were further incubated for 2 h. The medium was removed and 300 μL DMSO was added to each well to dissolve the formazan crystal formed by viable cells. The optical densities of each well were measured at 570 nm using a microplate reader (Model 680, Bio-Rad Lab, Hercules, CA) and expressed as a percentage relative to control cells (untreated cells). All cytotoxicity experiments were performed in triplicate. The relative cell viability was calculated according to the equation: ([Abs]sample-[Abs]blank)/([Abs]control-[Abs]blank)×100%.
2.9. Cellular uptake assay
The cellular uptake of polyplexes was examined by flow cytometry. Approximately 2 × 105 C2C12 cells were seeded in 12-well plates 24 h prior to study. YOYO-1 iodide tagged pCMV-Luc (1 molecule of YOYO-1 dye per 20 base pairs of nucleotide) was prepared 30 min before use. Then polyplexes were prepared by mixing poly(disulfide amine)s with YOYO-1 labeled plasmid DNA at w/w ratio of 1, 5, 10, 20 and 30 as described above. At the time of transfection, fluorescence labeled polyplexes (1.0 μg/mL DNA) was incubated with cells at 37°C for 4 h in serum-free medium. Then medium was removed by aspiration and the cells were washed with PBS, harvested by trypsin-like enzyme (TrypLE Express) and neutralized with serum containing medium. After centrifugation at 1500 rpm for 2 min, cells were fixed with 2% paraformaldehyde in PBS-0.5% BSA solution for 20 min at room temperature, and washed and resuspended in 0.3 mL ice-cold PBS. Samples were kept in ice and analyzed by a FACScan analyzer (BD Biosciences, San Jose, CA) at a minimum of 1 × 104 cells using FL1-height channel for YOYO-1 dye. Non-treated cells are used as control for calibration. Experiments were performed in duplicate. Data were analyzed by using Windows Multiple Document Interface software, version 2.9 (WinMDI, Microsoft, Redmond, Washington).
3. Results
3.1. Synthesis of poly(disulfide amine)s
Five biodegradable poly(disulfide amine)s with different length of oligomethylene spacer [–(CH2)n–] where n = 2-4 in the main chain and the side chain were synthesized in a two-step process shown in Scheme 1. First, 2-acetyldimedone (Dde-OH) was used to protect primary amines of five oligoamine monomers (SP, APPD, APED, AEPD, and TETA). Second, polymerizations were carried out between Dde-protected monomers and disulfide containing bisacrylamide based on Michael addition. The secondary amines in oligoamine monomers were converted into tertiary amines in the polymer main chains during polymerization. Pendant primary amines in polymer side chains were revealed by removing Dde protecting groups in NH2OH·HCl/Imidazole/NMP/DMF solution. The final polymer structures were confirmed by 1H NMR to contain one disulfide bond and two tertiary amine groups in the main chain and two pendant primary amine groups in the side chains in each repeating unit. Poly(CBA-SP), poly(CBA-APPD) and poly(CBA-APED) have propylene [–(CH2)3–] side chain spacer and butylene [–(CH2)4–], propylene [–(CH2)3–], and ethylene [–(CH2)2–] main chain spacers, respectively. Poly(CBA-AEPD) and poly(CBA-TETA) have shorter ethylene [–(CH2)2–] side chain spacer and shorter propylene [–(CH2)3–] and ethylene [–(CH2)2–] main chain spacers, respectively. The disappearance of signal peaks between δ 5 to 7 ppm indicated that the polymerization was complete and that acrylamide end groups did no longer exist in the final polymer products. The range of weight average molecular weight (Mw) of poly(disulfide amine)s was from 3.8 to 6.1 kDa with narrow polydispersities (PDI = 1.15 ∼ 1.33) (Table 1).
Table 1.
Characterization of bioreducible poly(disulfide amine)s: apparent number average molecular weight (Mn), apparent weight average molecular weight (Mw), polydispersity index (PDI), buffering capacity in pH range 7.4 – 5.1, protonation degree (α) and apparent pKa.
| Polymers | Mn (kDa)a | Mw (kDa)a | PDIa | Buffering Capacity (pH 5.1-7.4) (%)b | Protonation Degree (α, %) | Apparent pKa | ||
|---|---|---|---|---|---|---|---|---|
| pH 5.1 | pH 7.4 | pKa1 (α=25%) | pKa2 (α=75%) | |||||
| Poly(CBA-SP) | 4.16 | 4.81 | 1.16 | 38.0 | 76.5 | 52.5 | 9.53 | 5.85 |
| Poly(CBA-APPD) | 3.36 | 4.23 | 1.26 | 36.0 | 76 | 54.5 | 9.57 | 5.76 |
| Poly(CBA-APED) | 4.62 | 6.12 | 1.33 | 26.0 | 73 | 55 | 9.63 | 4.75 |
| Poly(CBA-AEPD) | 3.64 | 4.19 | 1.15 | 28.0 | 71.5 | 53.5 | 9.51 | 4.89 |
| Poly(CBA-TETA) | 3.17 | 3.80 | 1.20 | 28.0 | 72 | 55 | 9.55 | 5.05 |
| bPEI (25 kDa) | – | – | – | 22.0 | 69 | 52 | – | – |
FPLC Conditions: poly(disulfide amine)s are dissolved in 0.5 mL Tris buffer (pH 7.4) at a concentration of 25 mg/mL. Superose 12 column is used to measure the apparent molecular weights, which are used to calculate polydispersity. Flow rate is 0.5 mL/min. Standard calibration is calculated by using poly[N-(2-hydroxypropyl)methacrylamide;] (pHPMA).
Acid-base titration conditions: poly(disulfide amine)s and bPEI (5 mmol amino nitrogen atoms) in 10 mL of 0.1 M NaCl was titrated by 0.01 M HCl solution from pH 11.0 to 3.0. The absolute weight average molecular weight of bPEI is 25kDa.
3.2. Characterization of poly(disulfide amine)s and polyplexes
Buffering capacity is an important feature for cationic polymers, as set out in the “proton sponge” hypothesis, by facilitating the endosomal escape of polyplexes to mediate efficient gene expression[3, 9]. Poly(disulfide amine)s exhibited high buffering capacity, as 26-38% of amine groups become protonated in the endosomal-lysosomal pH range (7.4 to 5.1). This is higher than the buffering capacity of bPEI 25kDa (22%) and manifests as relatively flatter slopes in the acid-base titration curve (Figure 1A and Table 1). Poly(CBA-SP) and poly(CBA-APPD) have higher buffering capacity (38%, 36% respectively) than that of poly(CBA-APED), poly(CBA-AEPD) and poly(CBA-TETA) (26%, 28%, 28% respectively). The high buffering capacity of poly(disulfide amine)s may be attributed to the combined effect of the primary and tertiary amine groups in these polymers.
Figure 1.
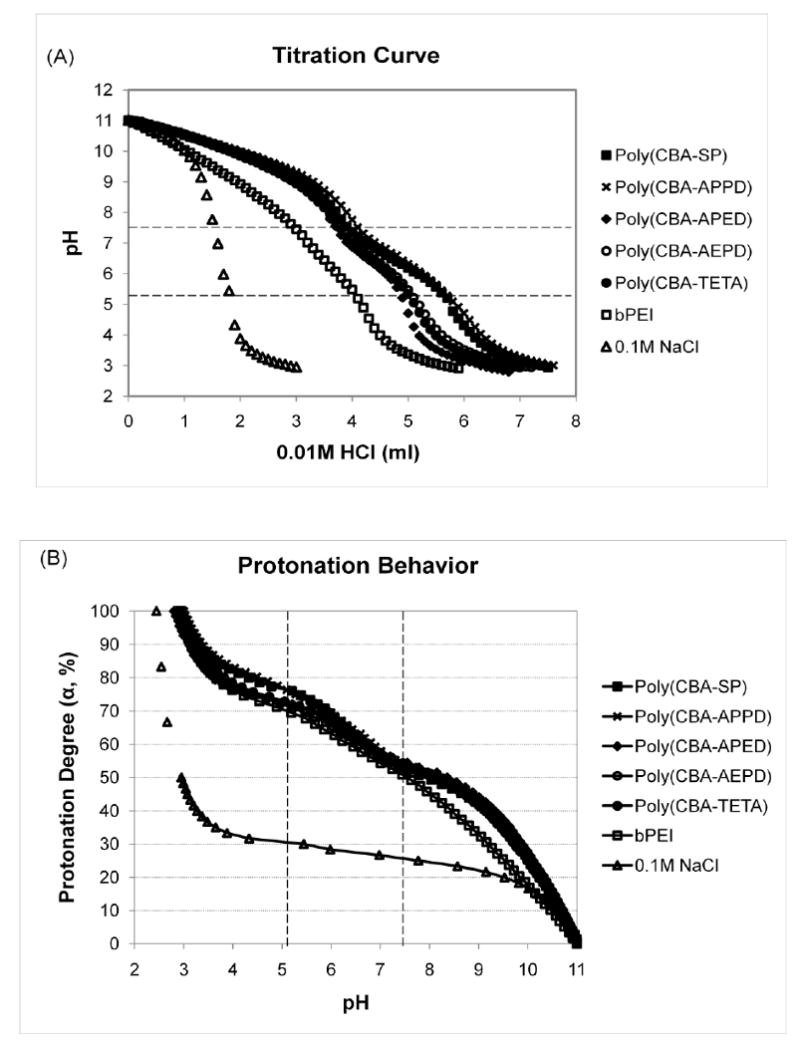
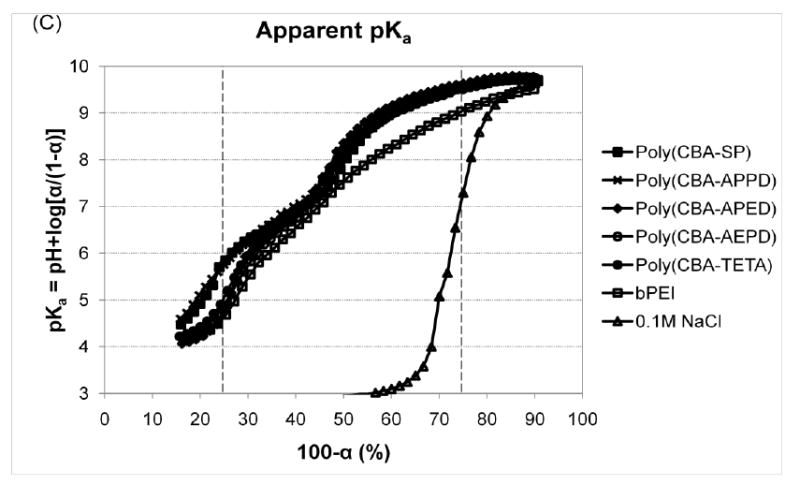
Buffering capacity and protonation behavior of poly(disulfide amine)s. (A) Titration curves of poly(disulfide amine)s. Polymers with 5 mmol amino nitrogen atoms were dissolved in 10 mL of 1.0 M NaCl. Solutions were initially set to pH 11.0 by 0.1 M NaOH and then titrated by 0.01 M HCl to pH 3.0. As a reference, the titration curves of bPEI (25 kDa) and 0.1 M NaCl were also presented. (B) Protonation degree (α) – pH curves of poly(disulfide amine)s in the pH range of 11 to 3. (C) Apparent pKa – (1-α) curves of poly(disulfide amine)s.
The protonation degree (α) of cationic polymers at neutral and endosomal acidic pH (from 7.4 to 5.1) is another important factor to determine polymer basicity (protonation ability) and for successful gene transfection[26]. The α of polymers can be estimated from titration curve in the pH range of 11 to 3 as shown in α-pH curve (Figure 1B and Table 1). At pH 7.4, the protonation degrees of five poly(disulfide amine)s and bPEI 25kDa are similar ranging from 52% to 55% due to the ready protonation of primary amine groups in polymer structures under neutral conditions. At pH 5.1, the protonation degree of poly(disulfide amine)s (71.5% - 76.5%) are relatively higher than that of bPEI (69%), showing higher degree of tertiary amine groups have been protonated than bPEI under the endosomal acidic conditions. These results indicated that poly(disulfide amine)s have relatively higher basicity than bPEI. In addition, poly(CBA-SP), poly(CBA-APPD), and poly(CBA-APED), have relatively higher protonation degree (76.5%, 76%, 73%, respectively) than that of poly(CBA-AEPD) and poly(CBA-TETA) (71.5% and 72%) at pH 5.1.
Due to the primary and tertiary amine groups in structures, poly(disulfide amine)s demonstrated two steps of protonation from pH 11 to 3. Apparent pKa values in the first and second protonations were defined as pKa1 (α = 25%) and pKa2 (α = 75%), respectively[26-28]. From the pKa/1-α curve (Figure 1C and Table 1), pKa1 for poly(disulfide amine)s is similar from 9.51 to 9.63, which is for the protonation of primary amine groups. In the second step of protonation, tertiary amine groups will be protonated (pKa2). It is noting that pKa2 of poly(CBA-SP) and poly(CBA-APPD) (5.85 and 5.76) are relatively higher than that of poly(CBA-APED), poly(CBA-AEDP) and poly(CBA-TETA) (4.75, 4.89, and 5.05, respectively). The poly(disulfide amine)s with longer propylene side chain spacer [–(CH2)3–] have higher basicity than the polymers with shorter enthylene side chain spacer [–(CH2)2–]. Therefore, one more methylene unit between two amine groups in the side chain and main chain of polymers may effectively enhance higher buffering capacity and basicity to facilitate the protonation during endosome-lysosome pathway.
To determine the size distribution of polyplexes with variant w/w ratios, dynamic light scattering (DLS) was used. As Fig. 2 shows, all poly(disulfide amine)s condensed DNA into particles as effective diameter below 200 nm at w/w ratio of 5:1. Significant size reduction to ∼150 nm was observed for polyplexes at w/w ratio of 10:1. The particle sizes reduced to ∼100 nm when w/w ratio was 20:1 for polyplexes. The results proved that cationic poly(disulfide amine)s can electrostatically condense negatively charged plasmid DNA into stable nanoparticles which are necessary for efficient cellular uptake.
Figure 2.
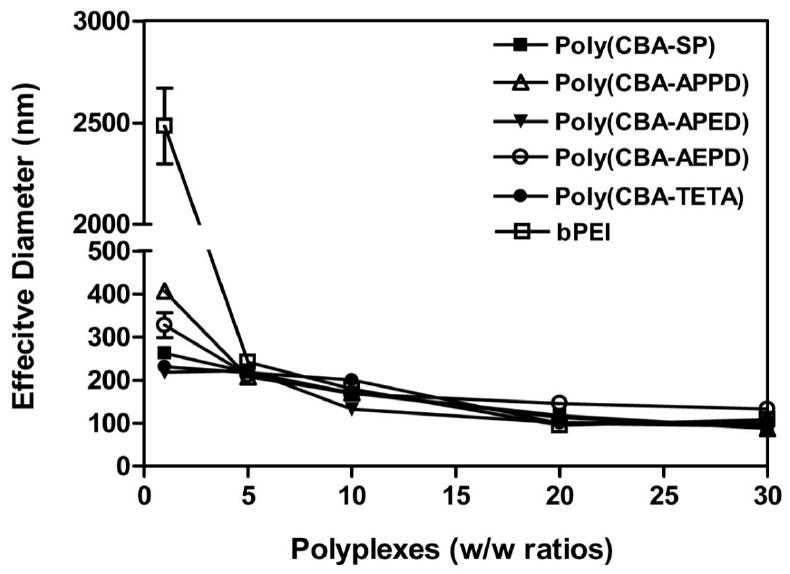
Average particle sizes of poly(disulfide amine)s/DNA polyplexes at w/w ratios of 1:1, 5:1, 10:1, 20:1 and 30:1. bPEI (25 kDa)/DNA polyplexes were measured at the same w/w ratios for comparison. The experiments were repeated three times and the data were represented as mean value ± standard deviations.
The formation of poly(disulfide amine)s/DNA polyplexes was also studied by gel retardation assay. As Fig.3A shows, poly(CBA-SP) and poly(CBA-APPD) completely retard DNA migration at a w/w ratio of 1:1. Poly(CBA-APED) partially retains DNA migration at a w/w ratio of 1:1 and completely retains DNA migration at a w/w ratio of 2:1. DNA was completely retained by poly(CBA-AEPD) and poly(CBA-TETA) at a w/w ratio of 2:1. In the presence of disulfide reducing agent DTT incubation at 37°C for 1 h, free DNA was released from all of the poly(disulfide amine)s/DNA polyplexes (Figure 3B). For the comparable non-degradable polymer bPEI 25kDa, there was no free DNA release from the bPEI polyplexes with DTT incubation. Agarose gel electrophoresis confirmed that all poly(disulfide amine)s have good DNA condensing capability at low w/w ratios. A longer side and main chain spacer allows a higher degree of protonation and thus a higher positive charge density of polymers. Consequently, the poly(disulfide amine)s with longer side and main chain spacer have a stronger ability to form stable complexes with DNA extracellularly than the polymers with shorter side and main chain spacer. In addition, these poly(disulfide amine)s are ready to release DNA intracellulaly via disulfide bonds cleavage by small redox molecules with free thiol groups through the efficient disulfide-thiol exchange reactions, thereby mediating efficient gene transfection.
Figure 3.
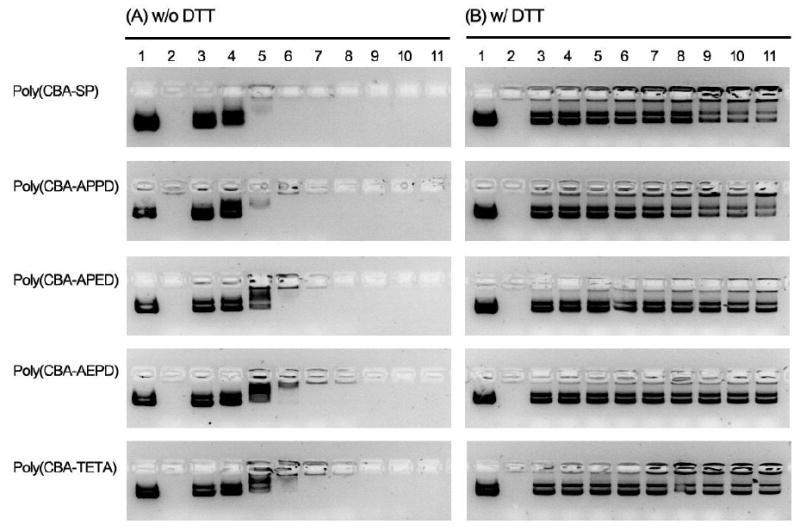
Gel retardation assay of poly(disulfide amine)s/DNA polyplexes at varying w/w ratios at the conditions of (A) without and (B) with DTT incubation (10.0 mM, 37°C, 1 h). Lane assignments correspond to polymer/DNA w/w ratios and represented as: lane 1 (0:1, plasmid DNA); lane 2 (bPEI (25kDa)/DNA 1:1); lane 3 (0.1:1); lane 4 (0.2:1); lane 5 (0.5:1); lane 6 (1:1); lane 7 (2:1); lane 8 (5:1); lane 9 (10:1); lane 10 (20:1); lane 11 (30:1).
3.3. In vitro gene transfection
Fig. 4 shows the luciferase expression levels mediated by poly(disulfide amine)s as a function of polymer/DNA w/w ratios. The goal of this experiment was to evaluate gene transfection efficiency of the poly(disulfide amine)s in different cell lines. Two model cell lines were chosen to approximate potential human diseases as targets for polymeric gene therapy. A Hela cancer cell line was used to evaluate gene transfection efficiency in cells with characteristics of human cervical carcinoma. A C2C12 myoblast cell line was to evaluate gene transfection efficiency in cells with characteristics of diseases of human muscle such as muscular dystrophy or ischemic heart disease. Luciferase expression as mediated by bPEI 25kDa was used as the control, demonstrating the optimal transfection without cytotoxicity at w/w ratio of 0.6:1 (N/P ∼ 5:1). Luciferase expression mediated by the poly(disulfide amine)s was much higher than that mediated by bPEI 25kDa from a w/w ratio of 5:1 to 30:1. In Hela cells, poly(CBA-APED) showed the highest transfection efficiency, up to 351-fold higher than that of bPEI 25kDa. In C2C12 cells, the highest luciferase expression was mediated by poly(CBA-SP), which was 146-fold higher than that of bPEI. It seems that transfection efficiency is oligomethylene length dependent for poly(disulfide amine)s. The poly(CBA-SP), poly(CBA-APPD), and ploy(CBA-APED), with the longer propylene [–(CH2)3–] side chain spacer, demonstrated higher transfection efficacy than their counterparts: poly(CBA-AEPD) and poly(CBA-TETA), which have shorter ethylene [–(CH2)2–] side chain spacer. Although poly(CBA-SP), poly(CBA-APPD), poly(CBA-APED) with different main chain spacer of –(CH2)4–, –(CH2)3–, –(CH2)2– demonstrated similar transfection efficiency in the two cell lines, the oligomethylene length of main chain spacer also had influence on transfection efficiency. With the same short ethylene [–(CH2)2–] side spacer, poly(CBA-AEPD) with longer propylene main chain spacer [–(CH2)3–] demonstrated higher luciferase expression than poly(CBA-TETA) with shorter ethylene main chain spacer[–(CH2)2–], which may due to higher level of buffering capacity, protonation degree of tertiary amine groups and basicity of poly(CBA-AEPD) caused by longer main chain spacer. The order of gene transfection efficiency among the poly(disulfide amine)s is poly(CBA-SP) ≈ poly(CBA-APPD) ≈ poly(CBA-APED) > poly(CBA-AEPD) > poly(CBA-TETA) > bPEI 25kDa. The results indicate that the chemical structures of the polymers influence their biophysical properties. The combination of primary and tertiary amines gives these polymers good DNA binding capability and buffering capacity, while the disulfide bonds in the structures of the polymers enable the release of free DNA efficiently from the polyplexes to induce efficient gene expression. Additionally, the longer side and main chain spacer gives these polymers higher degree of protonation, basicity and charge density so that the enhanced ability of transfecting cells and higher levels of gene expression are observed in poly(disulfide amine)s.
Figure 4.
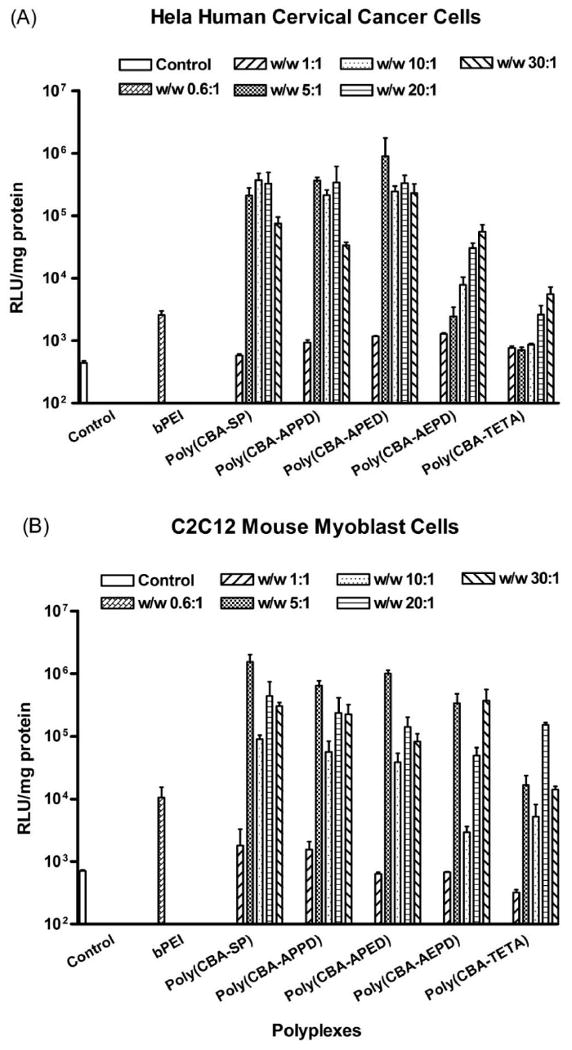
Transfection efficiency of poly(disulfide amine)s/pCMV-Luc polyplexes at varying w/w ratios in two different cell lines. (A) HeLa cells: human cervical cancer cell line; (B) C2C12 cells: mouse myoblast cell line. Control: non-treated cells. The transfection efficiency of bPEI (25kDa)/DNA at optimal w/w ratio of 0.6:1 was measured for comparison. The w/w ratios of poly(disulfide amine)s/DNA were 1:1, 5:1, 10:1, 20:1, and 30:1. Transfection experiments were repeated three times and the results were expressed as the relative luminescent unit (RLU) of luciferase reporter gene expression normalized by the total cell protein content in each well as mean values ± standard deviations.
3.4. Cytotoxicity
The in vitro cytotoxicity of the poly(disulfide amine)s was evaluated by MTT assay in C2C12 cells as a function of polymer/DNA w/w ratios (Figure 5). Results showed that all the poly(disulfide amine)s have overall lower cytotoxicity than bPEI 25kDa. Cells incubated with poly(CBA-SP), poly(CBA-APPD) and poly(CBA-APED) remained ∼ 80% and ∼ 50% viable at w/w ratio of 10:1 and 30:1. Poly(CBA-AEPD) and poly(CBA-TETA) exhibited no toxicity in the cells, which remained ∼ 100% viabile at a w/w ratio of 30:1. In contrast, only 12% of the cells incubated with bPEI 25kDa remained viable at a w/w ratio of 30:1. The results were similar with the Hela cells (data not shown). MTT assay demonstrated that poly(disulfide amine)s have a broad range of polymer/DNA w/w ratios where high level of transfection efficiency and low level of cytotoxicity are manifest.
Figure 5.
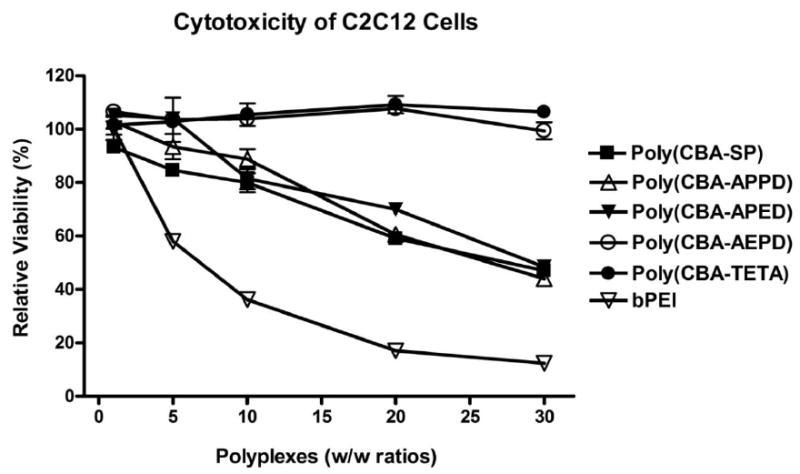
Relative cell viabilities of poly(disulfide amine)s/DNA polyplexes were measured as a function of w/w ratios in C2C12 cells. The polymer/DNA w/w ratios were set as 1:1, 5:1, 10:1, 20:1 and 30:1. The MTT assay of bPEI (25kDa)/DNA at the same w/w ratio was measured for comparison. Cytotoxicity was repeated three times and the data were represented as mean values ± standard deviations.
3.5. Transfection mechanism study of poly(disulfide amine)s
Using YOYO-1 iodide interclated pCMV-Luc, the cellular uptake of the polyplexes was estimated by flow cytometry in C2C12 cells as a function of the poly(disulfide amine)s/DNA w/w ratios (Figure 6). The results showed that the uptake of polyplexes increased with the increasing of w/w ratios from 1:1 to 30:1, with the fluorescent signals shifting to the stronger histogram area (M1 region). The proportion of cells taking up polyplexes (% cell count in R2 region) by poly(CBA-SP), poly(CBA-APPD) and poly(CBA-APED) was ∼ 99% at all w/w ratios (data not shown). At w/w ratio of 1:1, poly(CBA-AEPD) and poly(CBA-TETA) induced 77% and 71% cellular uptake. At a w/w ratio of 5 and above, poly(CBA-AEPD) and poly(CBA-TETA) increased cellular uptake to ∼ 99%. These results agreed with previous results that poly(disulfide amine)s can mediate high level of gene transfection in cells. Also, poly(CBA-SP), poly(CBA-APPD) and poly(CBA-APED), which contain the propylene side spacers [–(CH2)3–], can induce higher levels of cellular uptake and transfection efficiency than poly(CBA-AEPD) and poly(CBA-TETA), which contain the ethylene side spacers [–(CH2)2–]. The longer propylene main chain spacer [–(CH2)3–] also enable poly(CBA-AEPD) having relatively higher cellular uptake than poly(CBA-TETA) with shorter ethylene main chain spacer [–(CH2)2–].
Figure 6.
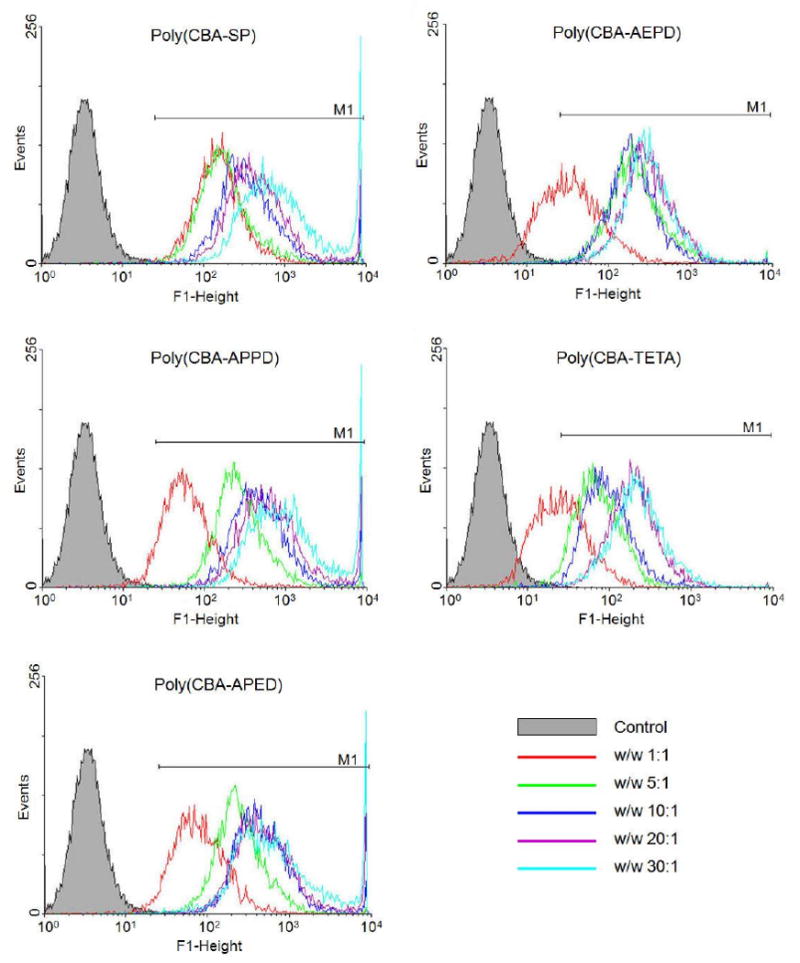
The cellular uptake of poly(disulfide amine)s/DNA polyplexes in C2C12 cells. Fluorescence histogram intensity correspond to polymer/DNA w/w ratios and represented as: closed grey peak (control, untreated cells); red line (1:1); green line (5:1); blue line (10:1); purple line (20:1); light blue line (30:1); M1 region (M1 gated fluorescent intensity).
4. Discussion
Gene therapy is defined as the delivery of genetic information to specific cells to direct the synthesis of specific proteins to treat a disease[29]. A key component of any gene therapy is the delivery vehicle. Through September 2008, 1472 clinical trials of gene therapy have been conducted worldwide (www.wiley.co.uk/genetherapy/clinical). Among these clinical trials, approximately 70% used viral vectors while less than 30% used non-viral methods to delivery DNA. While viral vectors allow for the potential for high gene transfection efficiency to cells, they suffer from a number of significant limitations including: 1) the potential for induced mutagenesis; 2) limited nucleic acid loading capacity; and 3) immunogenicity with repeat administration[2]. All of these factors limit the use of viral vectors in clinical applications. For these reasons, cationic polymers for gene delivery have been developed to overcome the inherent limitations of viral vectors. In general, synthetic polymeric vectors have previously had lower gene transfection efficiency than viruses but greater safety, stability, and ease of production and modification than viral vectors[10, 29].
In this study, a series of biodegradable poly(disulfide amine)s were synthesized in order to evaluate the effect of the structure-function relationship of this class of polymers on gene delivery. To develop polymers with superior DNA binding ability, basicity, protonation ability and buffering capacity, we introduced primary and tertiary amine groups into the polymer structures. To give the polymers the property of biodegradability, disulfide bonds were incorporated into the polymer backbones. To determine whether different length of oligomethylene spacers affect the transfection capacity of polymeric carriers, butylene [–(CH2)4–], propylene [–(CH2)3–], and ethylene [–(CH2)2–] spacers were introduced into the polymer main chain and side chains. The results confirmed our hypothesis that the functional groups within the polymer structures govern the DNA binding ability, protonation ability, basicity, buffering capacity and environmental biodegradability of the polymers, thereby improving gene transfection efficiency and lowering cytotoxicity.
DNA binding capability is an important feature of cationic polymers in determining capacity for gene delivery[30, 31]. A high positive charge density will condense DNA into nanosized particles and allow efficient cellular uptake through endocytosis. Since cytoplasm contains nucleases, cationic polymers with strong DNA binding ability will prevent nucleic acid degradation and inactivation[32]. Once internalized, the polyplexes traffic through the endosomal-lysosomal sorting pathway. Polymers that have a buffering capacity between pH 7.4 and 5.1 are able to mediate endosomal-lysosomal escape based on the “proton sponge effect”[9, 14]. This “proton spong effect” hypothesis was recently proved by Sonawane et al, showing that polymers with titratable amine groups such as PEI can increase the influx of H+ and the counterion Cl− into the endosome, resulting in swelling and rupture of the endosomal membrane by increasing osmotic pressure and mechanical perturbation[33]. Escape of the polyplexes is another major barrier and a requirement for delivery of DNA into the nucleus for efficient gene transfection. Previously, Wong et al. synthesized a series of pH sensitive polymers as gene carriers[34]. Their results showed that the polymers containing primary amine pendant groups have superior DNA binding and size-condensing ability and therefore superior gene transfection efficiency compared to the polymers containing only secondary and tertiary amine pendant groups. Zugates et al. used different amine groups to modify poly(β-amine ester)[12, 13]. They found that the modified poly(β-amine ester) with primary amine end groups has stronger DNA binding affinity and forms smaller nanoparticles with DNA and induces higher gene transfection efficiency than the counterpart poly(β-amine ester) with regular alcohol end groups. Lin et al. synthesized a series of poly(amido amine)s (PAAs) and found out that the polymers with primary, secondary and tertiary amine groups showed higher buffering capacity and gene transfection efficiencies than bPEI 25kDa[17]. In our study, the results of gel electrophoresis showed that poly(disulfide amine)s have strong DNA binding capability such that they can completely condense DNA into stable complexes at very low w/w ratios. The formed nanoparticles have an effective diameter below 200 nm as measured by dynamic light scattering. Based on the results of acid-base titration, all poly(disulfide amine)s have superior buffering capacity and protonation degree as compared to bPEI 25kDa. As a result, our poly(disulfide amine)s induced efficient cellular uptake of polyplexes and much higher luciferase expression than bPEI 25kDa in two model cell lines.
Disulfide bonds are another functional group used to solve the problem of the cytotoxicity of cationic polymers[24, 35]. The disulfide bonds can be cleaved in the cytoplasm by lots of small redox molecules with free thiol groups, including reduced glutathione, cysteine, cysteinylglycine, and homocysteine[21]. Since the concentration of these reducing agents is 2-20 μM in plasma, which is much lower than that in the cytoplasma (∼10 mM), the disulfide bonds are relatively stable in the extracellular environment, but readily be degradable in the intracellular environment through the efficient disulfide-thiol exchange reaction[20, 21]. This feature gives the disulfide bond-containing polymers the ability to form stable complexes with DNA extracellulary and release free genes intracellularly to induce efficient gene expression, while at the same time, to reduce cytotoxicity through polymer degradation. Oupicky et al. showed the disulfide-containing PLL have a much higher gene transfection efficiency than the counterpart non-degradable PLL[36]. Lin synthesized a series of bioreducible PAAs copolymers and found out that the polymers having higher numbers of disulfide bonds induced much higher transfection efficiency and lower cytotoxicity than the analogous polymers without disulfide bonds[24, 37]. Our previously synthesized poly(amido ethylenimine)s also mediated much higher transfection efficiency and lower cytotoxicity than bPEI 25kDa in different cell lines[15]. In this study, in the presence of DTT incubation, free DNA was released from poly(disulfide amine)s/DNA complexes in the gel electrophoresis. The poly(disulfide amine)s showed higher transfection efficiency and much lower cytotoxicity than bPEI 25kDa based on luciferase assay and MTT assay results.
The poly(disulfide amine)s with propylene [–(CH2)3–] side chains, such as poly(CBA-SP), poly(CBA-APPD) and poly(CBA-APED), showed higher DNA binding capability, buffering capacity and gene transfection efficiency than the counterpart polymers with ethylene [–(CH2)2–] side chains, e.g. poly(CBA-AEPD) and poly(CBA-TETA). The finding is that the relatively longer and more hydrophobic and flexible side groups will give these polymers higher transfection efficiency. Lin et al. synthesized a series of PAAs with different pendant functional groups, in which the polymers with longer alkyl chain have better gene transfection than the ones with shorter alkyl chains: pAPOL > pABOL > pMOPA[11]. Wong et al. found out that the polymers with longer carbonyl pendant groups had greater stability, DNA binding ability and gene transfection efficiency[34]. It is hypothesized that the longer hydrophobic residues promote interactions between the polyplexes and the target cell membrane, which results in efficient cellular uptake and higher gene transfection efficiency. In addition, the hydrophobic residues may increase the stability of the polyplexes, suggesting that alkylation may drive the polyplexes to a more thermodynamically favorable conformational arrangement so that DNA is prevented from being dissociated readily, even in the presence of competing polyanion[31, 34, 38]. As a result, the polymers with longer and hydrophobic side chains have the capacity for enhanced gene transfection. The longer oligomethylene spacer in main chain also benefits polymer gene transfection due to the enhanced polymer backbone flexibility, which may help polymer to form a more stable confirmation with DNA. Jones et al. observed that the polymer with the more flexible backbone of two tertiary amine and ethylene groups (pMBA-DMEDA) has better DNA binding ability and gene transfection than the polymers with only one tertiary amine group and rigid piperazine rings (pMBA-MMA and pMBA-2MP)[39].
The oligomethylene length in main chain and side chain may also influence the protonation degree, basicity, and buffering capacity of polymers. Primary amines of the poly(disulfide amine)s have similar pKa1 (9.51 - 9.63) and are almost completely protonated at physiological pH. It seems the oligomethylene spacer has little influence on basicity of primary amine groups. However, the protonation degree of nitrogens in tertiary amines in polymer main chain will be dependent on both the side chain and main chain oligomethylene length. Poly(CBA-SP) and poly(CBA-APPD) have the longest oligomethylene units in side chain and main chain and the highest buffering capacity, protonation degree and basicity of tertiary amines. Poly(CBA-APED) has propylene unit [–(CH2)3–] in side chain but ethylene unit [–(CH2)2–] in main chain, it has lower buffering capacity and protonation degree than the above two polymers. And poly(CBA-AEPD) and poly(CBA-TETA) have the shortest oligomethylene units in their side and main chain and the lowest buffering capacity and protonation degree. When two amine groups are separated by ethylene spacer [–(CH2)2–] in main chain and side chain, the strong electrostatic repulsion between two charged amino nitrogen atoms may decrease the protonation degree of tertiary amines in polymer main chain. The longer side chain and main chain spacer will reduce the repulsion between the charged amine groups and therefore increase the protonation degree and basicity of tertiary amine groups[26]. Taken together, these results show that the structure properties of the polymers influence their nucleic acid binding capability, charge-density, buffering capacity and gene transfection efficiency. Further investigation will be needed to fully understand the underlying mechanisms regarding how oligomethylene spacers in polymer side chain and main chain influence polymeric gene transfection efficiency.
Finally, another important feature of poly(disulfide amine)s is their cytotoxicity profile. Although poly(CBA-AEPD) and poly(CBA-TETA) have lower gene transfection efficiency, they have less cytotoxicity than poly(CBA-SP), poly(CBA-APPD) and poly(CBA-APED). As discussed above, the reason may be that the longer and more hydrophobic residues have more influence on the polyplexes-cell membrane interaction[34, 38, 40]. As a result, these hydrophobic residues may cause more cell membrane disruption and cytotoxicity. The important point, however, is that all the poly(disulfide amine)s are significant less cytotoxic than bPEI 25kDa. This low toxicity profile suggests that the biodegradable poly(disulfide amine)s will be a feasible, safe and promising gene vectors for future studies in gene delivery.
5. Conclusion
We have synthesized and evaluated a family of bioreducible poly(disulfide amine)s as polymeric gene vectors. The incorporation of multiple functional groups, such as amine groups and disulfide bonds, give these cationic polymers good DNA condensation ability, protonation ability, charge density, buffering capacity and intracellular biodegradability. The longer, more hydrophobic oligomethylene side chains and more flexible backbones of these polymers with [–(CH2)n–, where n = 2-4] increase gene transfection efficiency which may due to the enhanced buffering capacity, protonation degree of tertiary amine groups, and basicity and charge density of polymers. The results of these analyses: (1) identify a series of polymeric vectors with higher transfection efficiency and lower cytotoxicity than bPEI 25kDa; (2) reveal the structure-function relationships between different lengths of polymer backbones and side chains and the gene transfection efficiency of the polymers; and (3) describe a polymer family that is a safe and efficient vector system for gene delivery.
Supplementary Material
Acknowledgments
This work is supported by National Institutes of Health (NIH) grant HL065477 (SWK) and HL071541 (DAB). We thank Dr. Zheng-Rong Lu (University of Utah, Salt Lake City, UT) for the generous use of equipments and facilities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Green JJ, Langer R, Anderson DG. A Combinatorial Polymer Library Approach Yields Insight into Nonviral Gene Delivery. Acc Chem Res. 2008;41(6):749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma IM, Somia N. Gene therapy -- promises, problems and prospects. Nature. 1997;389(6648):239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 3.Lin C, Engbersen JF. Effect of chemical functionalities in poly(amido amine)s for non-viral gene transfection. J Control Release. 2008;132:267–272. doi: 10.1016/j.jconrel.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 4.Wang XL, Jensen R, Lu ZR. A novel environment-sensitive biodegradable polydisulfide with protonatable pendants for nucleic acid delivery. J Control Release. 2007;120(3):250–258. doi: 10.1016/j.jconrel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Huang L. Nonviral gene therapy: promises and challenges. Gene Ther. 2000;7(1):31–34. doi: 10.1038/sj.gt.3301110. [DOI] [PubMed] [Google Scholar]
- 6.Ou M, Wang XL, Xu R, Chang CW, Bull DA, Kim SW. Novel biodegradable poly(disulfide amine)s for gene delivery with high efficiency and low cytotoxicity. Bioconjug Chem. 2008;19(3):626–633. doi: 10.1021/bc700397x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho YW, Kim JD, Park K. Polycation gene delivery systems: escape from endosomes to cytosol. J Pharm Pharmacol. 2003;55(6):721–734. doi: 10.1211/002235703765951311. [DOI] [PubMed] [Google Scholar]
- 8.Park TG, Jeong JH, Kim SW. Current status of polymeric gene delivery systems. Adv Drug Deliv Rev. 2006;58(4):467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 10.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109(2):259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 11.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, et al. Novel bioreducible poly(amido amine)s for highly efficient gene delivery. Bioconjug Chem. 2007;18(1):138–145. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 12.Zugates GT, Tedford NC, Zumbuehl A, Jhunjhunwala S, Kang CS, Griffith LG, et al. Gene delivery properties of end-modified poly(beta-amino ester)s. Bioconjug Chem. 2007;18(6):1887–1896. doi: 10.1021/bc7002082. [DOI] [PubMed] [Google Scholar]
- 13.Zugates GT, Peng W, Zumbuehl A, Jhunjhunwala S, Huang YH, Langer R, et al. Rapid optimization of gene delivery by parallel end-modification of poly(beta-amino ester)s. Mol Ther. 2007;15(7):1306–1312. doi: 10.1038/mt.sj.6300132. [DOI] [PubMed] [Google Scholar]
- 14.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen LV, Chang CW, Kim WJ, Kim SW, Zhong Z, Lin C, et al. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery. Bioconjug Chem. 2006;17(5):1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 16.Christensen LV, Chang CW, Yockman JW, Conners R, Jackson H, Zhong Z, et al. Reducible poly(amido ethylenediamine) for hypoxia-inducible VEGF delivery. J Control Release. 2007;118(2):254–261. doi: 10.1016/j.jconrel.2006.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin C, Blaauboer CJ, Timoneda MM, Lok MC, van Steenbergen M, Hennink WE, et al. Bioreducible poly(amido amine)s with oligoamine side chains: synthesis, characterization, and structural effects on gene delivery. J Control Release. 2008;126(2):166–174. doi: 10.1016/j.jconrel.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Bulmus V, Woodward M, Lin L, Murthy N, Stayton P, Hoffman A. A new pH-responsive and glutathione-reactive, endosomal membrane-disruptive polymeric carrier for intracellular delivery of biomolecular drugs. J Control Release. 2003;93(2):105–120. doi: 10.1016/j.jconrel.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Pichon C, LeCam E, Guerin B, Coulaud D, Delain E, Midoux P. Poly[Lys-(AEDTP);]: a cationic polymer that allows dissociation of pDNA/cationic polymer complexes in a reductive medium and enhances polyfection. Bioconjug Chem. 2002;13(1):76–82. doi: 10.1021/bc015503o. [DOI] [PubMed] [Google Scholar]
- 20.Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134(3):489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 21.Lu ZR, Mohs AM, Zong Y, Feng Y. Polydisulfide Gd(III) chelates as biodegradable macromolecular magnetic resonance imaging contrast agents. Int J Nanomedicine. 2006;1(1):31–40. doi: 10.2147/nano.2006.1.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gosselin MA, Guo W, Lee RJ. Efficient gene transfer using reversibly cross-linked low molecular weight polyethylenimine. Bioconjug Chem. 2001;12(6):989–994. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 23.Lee Y, Mo H, Koo H, Park JY, Cho MY, Jin GW, et al. Visualization of the degradation of a disulfide polymer, linear poly(ethylenimine sulfide), for gene delivery. Bioconjug Chem. 2007;18(1):13–18. doi: 10.1021/bc060113t. [DOI] [PubMed] [Google Scholar]
- 24.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, et al. Linear poly(amido amine)s with secondary and tertiary amino groups and variable amounts of disulfide linkages: synthesis and in vitro gene transfer properties. J Control Release. 2006;116(2):130–137. doi: 10.1016/j.jconrel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Diaz-Mochon JJ, Bialy L, Bradley M. Full orthogonality between Dde and Fmoc: the direct synthesis of PNA--peptide conjugates. Org Lett. 2004;6(7):1127–1129. doi: 10.1021/ol049905y. [DOI] [PubMed] [Google Scholar]
- 26.Miyata K, Oba M, Nakanishi M, Fukushima S, Yamasaki Y, Koyama H, et al. Polyplexes from poly(aspartamide) bearing 1,2-diaminoethane side chains induce pH-selective, endosomal membrane destabilization with amplified transfection and negligible cytotoxicity. J Am Chem Soc. 2008;130(48):16287–16294. doi: 10.1021/ja804561g. [DOI] [PubMed] [Google Scholar]
- 27.Kanayama N, Fukushima S, Nishiyama N, Itaka K, Jang WD, Miyata K, et al. A PEG-based biocompatible block catiomer with high buffering capacity for the construction of polyplex micelles showing efficient gene transfer toward primary cells. ChemMedChem. 2006;1(4):439–444. doi: 10.1002/cmdc.200600008. [DOI] [PubMed] [Google Scholar]
- 28.Han M, Bae Y, Nishiyama N, Miyata K, Oba M, Kataoka K. Transfection study using multicellular tumor spheroids for screening non-viral polymeric gene vectors with low cytotoxicity and high transfection efficiencies. J Control Release. 2007;121(12):38–48. doi: 10.1016/j.jconrel.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Putnam D. Polymers for gene delivery across length scales. Nat Mater. 2006;5(6):439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 30.Chen DJ, Majors BS, Zelikin A, Putnam D. Structure-function relationships of gene delivery vectors in a limited polycation library. J Control Release. 2005;103(1):273–283. doi: 10.1016/j.jconrel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 31.Doody AM, Korley JN, Dang KP, Zawaneh PN, Putnam D. Characterizing the structure/function parameter space of hydrocarbon-conjugated branched polyethylenimine for DNA delivery in vitro. J Control Release. 2006;116(2):227–237. doi: 10.1016/j.jconrel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 32.Pouton CW, Seymour LW. Key issues in non-viral gene delivery. Adv Drug Deliv Rev. 2001;46(13):187–203. doi: 10.1016/s0169-409x(00)00133-2. [DOI] [PubMed] [Google Scholar]
- 33.Sonawane ND, Szoka FC, Jr, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278(45):44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 34.Wong SY, Sood N, Putnam D. Combinatorial Evaluation of Cations, pH-sensitive and Hydrophobic Moieties for Polymeric Vector Design. Mol Ther. 2009;13:1–11. doi: 10.1038/mt.2008.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, et al. Random and block copolymers of bioreducible poly(amido amine)s with high- and low-basicity amino groups: study of DNA condensation and buffer capacity on gene transfection. J Control Release. 2007;123(1):67–75. doi: 10.1016/j.jconrel.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Oupicky D, Parker AL, Seymour LW. Laterally stabilized complexes of DNA with linear reducible polycations: strategy for triggered intracellular activation of DNA delivery vectors. J Am Chem Soc. 2002;124(1):8–9. doi: 10.1021/ja016440n. [DOI] [PubMed] [Google Scholar]
- 37.Zhong Z, Song Y, Engbersen JF, Lok MC, Hennink WE, Feijen J. A versatile family of degradable non-viral gene carriers based on hyperbranched poly(ester amine)s. J Control Release. 2005;109(13):317–329. doi: 10.1016/j.jconrel.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 38.Thomas M, Klibanov AM. Enhancing polyethylenimine's delivery of plasmid DNA into mammalian cells. Proc Natl Acad Sci U S A. 2002;99(23):14640–14645. doi: 10.1073/pnas.192581499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones NA, Hill IR, Stolnik S, Bignotti F, Davis SS, Garnett MC. Polymer chemical structure is a key determinant of physicochemical and colloidal properties of polymer-DNA complexes for gene delivery. Biochim Biophys Acta. 2000;1517(1):1–18. doi: 10.1016/s0167-4781(00)00220-7. [DOI] [PubMed] [Google Scholar]
- 40.Wong SY, Putnam D. Overcoming limiting side reactions associated with an NHS-activated precursor of polymethacrylamide-based polymers. Bioconjug Chem. 2007;18(3):970–982. doi: 10.1021/bc0603790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


