Abstract
Receptor activator for nuclear factor-κB ligand (RANKL), a critical osteoclastogenic factor expressed in marrow stromal/preosteoblast cells is up-regulated in Paget’s disease of bone (PDB). We previously demonstrated that heat-shock factor-2 (HSF-2) is a downstream target of fibroblast growth factor-2 (FGF-2) signaling to induce RANKL expression in bone marrow stromal/preosteoblast cells. In this study, we identified a 2.5-fold increase in serum FGF-2 levels in patients (n = 8) with PDB compared with normal subjects (n = 10). We showed that HSF-2 co-immunoprecipitates with heat-shock protein-27 (HSP-27) and that FGF-2 stimulation significantly increased phospho-HSP-27 levels in marrow stromal cells. Confocal microscopy revealed HSF-2 colocalization with HSP-27 in unstimulated cells and HSF-2 nuclear translocation upon FGF-2 stimulation. We further show that FGF-2 stimulation significantly increased the levels of phosphorylated signal transducers and activators of the transcription (p-STAT-1) in these cells. Western blot analysis confirmed that small interfering RNA suppression of STAT-1 significantly decreased (3.2-fold) RANKL expression and promoter activity in FGF-2-stimulated cells. Chromatin immunoprecipitation assay revealed STAT-1 binding to a putative motif located far upstream (−8 kb) in the hRANKL gene promoter region. These results suggest STAT-1 is a downstream effector of FGF-2 signaling and that elevated levels of FGF-2 stimulates RANKL expression in PDB.
STAT-1 is a downstream effector of fibroblast growth factor-2 (FGF-2) signaling and elevated levels of FGF-2 stimulates RANK ligand expression in Paget’s disease of bone.
Paget’s disease of bone (PDB) is a chronic focal skeletal disorder that affects 2–3% of the population over the age of 60 yr. PDB is inherited as an autosomal dominant trait with genetic heterogeneity. Several mutations in the ubiquitin-associated domain of sequestosome 1 (SQSTM1/p62) have been identified, with the P392L amino acid substitution being the most common in patients with PDB (1); however, p62 proved neither necessary nor sufficient to cause PDB (2). Environmental factors such as paramyxoviruses are implicated in PDB (3,4), but viral etiology remains controversial because others have failed to detect expression of paramyxoviral transcripts (5,6). The disease is characterized by highly localized areas of bone turnover with increased osteoclast (OCL) activity followed by an exaggerated osteoblast response. The OCL in PDB are characterized by the presence of paramyxoviral nuclear inclusions and nucleocapsid transcripts. We have previously detected expression of measles virus nucleocapsid (MVNP) transcripts in OCL from patients with PDB. Receptor activator for nuclear factor-κB ligand ligand (RANKL), a critical OCL differentiation factor expressed by marrow stromal/osteoblast cells, is increased in PDB (7). Furthermore, enhanced levels of IL-6, macrophage colony-stimulating factor (M-CSF), and endothelin-1 have been associated with PDB (8,9). We recently identified increased serum levels (2- to 5-fold) of the inflammatory cytokine kininogen in patients with PDB compared with normal subjects; however, kininogen has no significant effect on RANKL gene expression (10).
Several osteotropic factors such as 1,25-dihydroxyvitamin D3, PTH, IL-1β, IL-11, and prostaglandin E2 induce OCL differentiation through enhanced expression of RANKL in marrow stromal/osteoblast cells (11,12), but the molecular mechanisms that regulate RANKL gene expression are unclear. IL-1β and TNF-α stimulate RANKL expression in human bone marrow stromal cells through activation of the p38 MAPK pathway (13). In addition, fibroblast growth factor-2 (FGF-2) has been shown to induce RANKL production through cyclooxygenase-2-mediated prostaglandin synthesis and by suppressing osteoclastogenesis inhibitory factor in osteoblastic cells (14). Similarly, lipopolysaccharide treatment increased RANKL expression through activation of Toll-like receptors in primary murine osteoblast cells (15). Furthermore, TGF-β has been shown to increase RANKL expression in activated T cells by increasing anti-CD3 (16). It has also been reported that PTH stimulates RANKL expression through the cAMP/protein kinase A/cAMP response element-binding protein cascade (17).
We previously demonstrated that heat-shock factor-2 (HSF-2) is a downstream target of FGF-2 signaling to induce RANKL expression in bone marrow stromal/preosteoblast cells (18). Heat-shock proteins (HSP) are molecular chaperones expressed in cells in response to a variety of stimuli such as temperature and stimulation of membrane-bound receptors by hormones/cytokines and other chemical factors. Heat-shock transcription factors (HSF), which bind to the heat-shock-responsive element (HSE), modulate expression of HSP and several other genes including the TNF-α family (19,20). More recently, we further demonstrated that DACH1, the human homolog of Drosophila dachshund gene, which interacts with the nuclear corepressor (NCoR), negatively regulates RANKL gene expression and suppresses FGF-2-enhanced RANKL gene expression. We found that HSF-2 co-immunoprecipitated with DACH1 and that FGF-2 stimulation significantly increased HSF-2 binding to DACH1 (21). Therefore, RANKL expression is regulated by complex regulatory mechanisms operative in stromal/preosteoblast cells.
FGF and their receptors are important in both normal bone remodeling and pathological disorders of bone (22). FGF signaling induced the MAPK cascade and signal transducers and activators of the transcription (STAT) signaling pathway (23). Although multiple osteotropic factors including FGF-2 are known to modulate RANKL gene expression in the bone microenvironment, the transcriptional regulatory mechanisms operative in marrow stromal/preosteoblast cells are not well established in pathological conditions such as PDB. The present study aims to delineate the molecular mechanisms of RANKL expression in stromal/preosteoblast cells in response to elevated levels of FGF-2 associated with PDB. Our results suggest that STAT-1 is a downstream effector of FGF-2 signaling and that elevated levels of FGF-2 stimulate RANKL expression in PDB.
Results
FGF-2 levels are elevated in patients with PDB
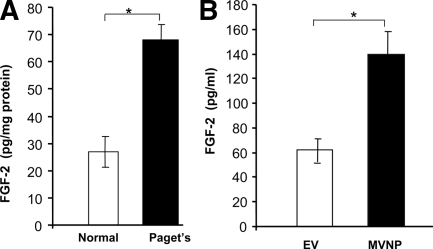
Enhanced levels of IL-6, M-CSF, RANKL, and endothelin-1 have been associated with PDB (8,9). RANKL is a critical osteoclastogenic factor expressed in stromal/preosteoblastic cells. We and others have reported FGF-2 stimulation of RANKL expression in bone marrow stromal/preosteoblast cells (18,24). Therefore, we further quantified the level of FGF-2 in serum from patients with PDB in comparison with normal subjects. ELISA analysis revealed a significant increase in the level of FGF-2 (2.5-fold) in serum samples obtained from patients with PDB (n = 8) when compared with normal subjects (n = 10) (Fig. 1A).
Figure 1.
A, FGF-2 levels in serum samples from normal subjects (n = 10) and patients with PDB (n = 8) serum. B, Human bone marrow cells were transduced with control EV or MVNP retrovirus expression vectors and cultured for OCL formation. Serum-free CM was collected for 24 h. FGF-2 was measured by ELISA. Values are expressed as mean ± sd. *, P < 0.05.
We have previously identified expression of measles virus nucleocapsid (MVNP) transcript in bone marrow cells and peripheral blood-derived monocytes from patients with PDB (9). Additional studies indicated that retroviral expression of MVNP in OCL progenitor cells resulted in osteoclasts with the Pagetic phenotype (25). We therefore further tested whether conditioned medium (CM) obtained from MVNP-transduced OCL stimulates FGF-2 production. Normal human bone marrow mononuclear cells were transduced with empty vector (EV) and the MVNP gene and cultured to form OCL as described in Materials and Methods. After 24 h, the CM collected was analyzed for FGF-2 levels. As shown in Fig. 1B, FGF-2 production significantly increased (2.4-fold) in CM collected from the MVNP-transduced OCL culture compared with mock (EV)-transduced cells. These data suggest that MVNP expression in OCL in PDB results in elevated levels of FGF-2, which implicates FGF-2 stimulation of RANKL expression in stromal/preosteoblast cells.
MVNP CM enhances nuclear translocation of HSF-2 and RANKL expression
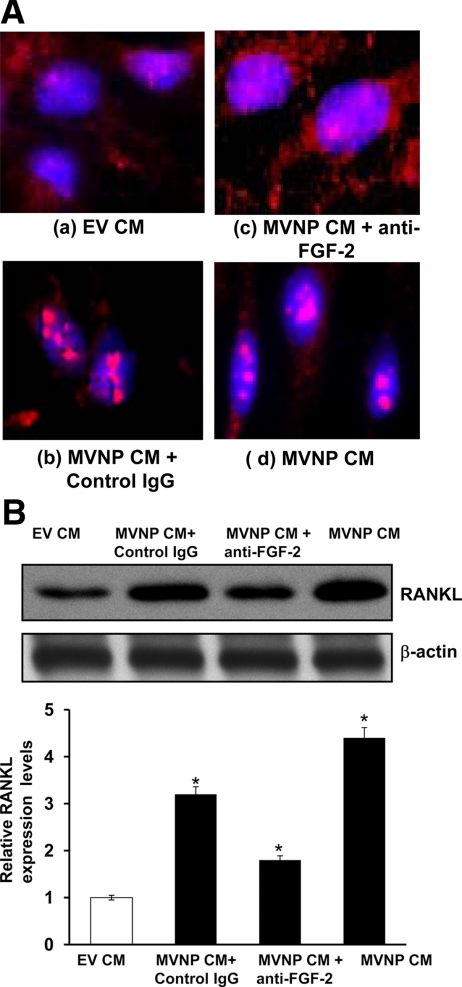
Previously, we identified that HSF-2 is a downstream target molecule for FGF-2 signaling to enhance RANKL expression in marrow stromal/preosteoblast cells. FGF-2 induces nuclear translocation of HSF-2, which binds to HSE present in the RANKL gene promoter to modulate RANKL gene expression in these cells (18). Therefore, we further tested MVNP CM to induce HSF-2 translocation to the nucleus in human bone marrow stromal cells. Cells were stimulated with 20% of MVNP CM for 24 h and immunostained with anti-HSF-2 antibody. Confocal microscopy revealed that MVNP CM induced nuclear translocation of HSF-2 in bone marrow stromal/preosteoblast cells. HSF-2 nuclear translocation was inhibited in these cells in the presence of anti-FGF-2 neutralizing antibody (Fig. 2A). In contrast, cells treated with control EV CM showed cytosolic localization of HSF-2 in the presence and absence of anti-FGF-2 antibody. Furthermore, we examined whether MVNP CM to stimulates RANKL expression in human bone marrow stromal cells. Cells were stimulated with 20% of MVNP CM for 48 h, and total cell lysates obtained were analyzed by Western blot for RANKL expression. As shown in Fig. 2B, MVNP CM significantly increased (4.7-fold) RANKL expression compared with control EV CM. RANKL expression was significantly decreased in cells treated with anti-FGF-2 neutralizing antibody. Taken together, these results suggest that MVNP expression elevates FGF-2, which enhances nuclear translocation of HSF-2 and induces RANKL expression in bone marrow stromal/preosteoblast cells.
Figure 2.
MVNP CM induces HSF-2 nuclear translocation and enhances RANKL expression in bone marrow stromal cells. A, Bone marrow stromal cells were treated with 20% of MVNP CM for 24 h. Immunostaining for HSF-2 is shown as detected by Alexa 568-conjugated antigoat and nuclear staining by 4′,6-diamidino-2-phenylindole. a, Control cells show HSF-2 expression in the cytosol; b MVNP CM treated with control IgG; c, anti-FGF-2 antibody blocks the nuclear translocation of HSF-2 in MVNP CM-treated cells; d, MVNP CM induces nuclear translocation of HSF-2. B, Bone marrow stromal cells were treated with EV and MVNP CM (20%) for 48 h. Total cell lysates obtained were analyzed by Western blot for RANKL expression. The band intensity was quantified by NIH ImageJ program. The values are expressed as mean ± sd. *, P < 0.05.
HSF-2 interacts with HSP-27 in human bone marrow stromal cells
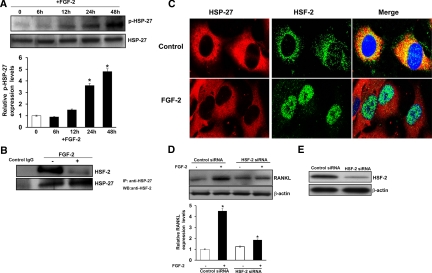
HSP are molecular chaperones that modulate HSF activation (26). Activated HSF translocate to the nucleus and modulate gene expression (19,20). HSP-27 has been shown to be regulated in bone marrow stromal/preosteoblast cells (27). Thus, we measured phospho- (p-)HSP-27 in human bone marrow stromal/preosteoblast cells in response to FGF-2 treatment in a time-dependent manner. Immunoblot analysis of total cell lysates obtained from bone marrow-derived stromal cells stimulated with FGF-2 (4 ng/ml) over time (0–48 h) revealed a significant increase in p-HSP-27 (4.8-fold) at 48 h, compared with untreated control cells (Fig. 3A). Coimmunoprecipitation assay further demonstrated that HSP-27 binds with HSF-2 in unstimulated control cells; however, it significantly decreased in response to FGF-2 treatment of these cells (Fig. 3B). Consistent with these results, confocal microscopy analysis revealed cytosolic colocalization of HSF-2 and HSP-27 in unstimulated cells and that HSF-2 alone is translocated to the nucleus in response to FGF-2 stimulation (Fig. 3C). These results indicate FGF-2-specific regulation of HSP-27 and HSF-2 activation in marrow stromal/preosteoblast cells. Furthermore, small interfering RNA (siRNA) suppression of HSF-2 significantly decreased RANKL expression compared with control, nonspecific siRNA-transfected cells (Fig. 3D). siRNA suppression of HSF-2 expression was confirmed by Western blot analysis (Fig. 3E). These results are consistent with our previous findings that HSF-2 is a downstream target of FGF-2 signaling to induce RANKL expression in bone marrow stromal cells (18).
Figure 3.
FGF-2 increases phosphorylation of HSP-27 in stromal/preosteoblast cells. A, Human bone marrow-derived stromal cells were stimulated with FGF-2 (4 ng/ml) for the indicated time point, and total cell lysates obtained were analyzed by Western blot for p-HSP-27. B, Coimmunoprecipitation of HSP-27 with HSF-2. Normal human bone marrow-derived stromal/preosteoblast cells were stimulated with and without FGF-2 (4 ng/ml) for 24 h. Total cell lysates were immunoprecipitated using anti-HSP-27 antibody. Immunoprecipitates were analyzed by Western blot using anti-HSF-2 antibody. Protein content was normalized with respect to total HSF-2 expression in these cells. C, Colocalization of HSF-2 with HSP-27 in human bone marrow-derived stromal cells. Cells were stimulated with FGF-2 (4 ng/ml) for 24 h and analyzed with confocal microscopy. Immunostaining for HSP-27 and HSF-2 is detected by Alexa 568-conjugated antigoat IgG and Alexa 488-conjugated antirabbit IgG, respectively, and nuclear staining by DRAQ5. The merged image demonstrates colocalization of HSF-2 and HSP-27 in untreated control cells and nuclear translocation of HSF-2 in FGF-2-stimulated cells. D, siRNA suppression of HSF-2 inhibits FGF-2-stimulated RANKL expression. Bone morrow stromal cells were transiently transfected with control nonspecific siRNA and HSF-2 siRNA. Cells were stimulated with FGF-2 (4 ng/ml) for 48 h. Total cell lysates obtained were analyzed by Western blot. The band intensity was quantified by NIH ImageJ program. The values are expressed as mean ± sd. *, P < 0.05. E, siRNA suppression of HSF-2 was confirmed by Western blot analysis.
FGF-2 activation of STAT-1 regulates RANKL expression in stromal/preosteoblast cells
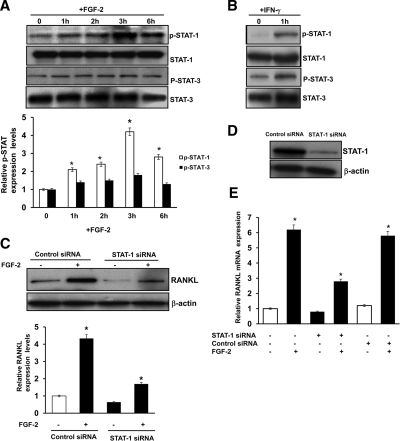
Because STAT molecules are implicated in FGF-2 signaling in osteoblast cells and bone development (28), we further examined the participation of STAT in FGF signaling to enhance RANKL expression. Bone marrow stromal/preosteoblast cells were stimulated (0–6 h) with FGF-2 (4 ng/ml), and total cell lysates obtained were analyzed by Western blot for p-STAT-1/3 and STAT-1/3 expression. As shown in Fig. 4A, FGF-2 stimulation significantly increased the levels of p-STAT-1 (4.0-fold) at 3 h; however, no significant change in p-STAT-3 occurred in these cells. Furthermore, there was no significant change in the levels of STAT-1/3 expression with and without FGF-2 treatment of these cells. These data suggest FGF-2 activation of STAT-1 in bone marrow stromal/preosteoblast cells. Interferon-γ-activated Jurkat T cells were used as positive controls for STAT-1/3 activation (Fig. 4B). To further determine the role of STAT-1 in FGF-2-induced RANKL expression, we transiently transfected siRNA against STAT-1 into marrow stromal cells and stimulated them with FGF-2 (4 ng/ml) for 48 h. siRNA suppression of STAT-1 decreased (3.2-fold) RANKL expression in FGF-2-stimulated cells (Fig. 4C). In contrast, cells transfected with a control, nonspecific scrambled siRNA exhibited no significant change in the levels of RANKL expression in these cells. siRNA suppression of STAT-1 expression was confirmed by Western blot analysis (Fig. 4D). We next examined the role of STAT-1 on FGF-2-stimulated RANKL mRNA expression. Bone marrow stromal cells were transfected with control, nonspecific siRNA and STAT-1 siRNA, and cells were stimulated with FGF-2 (4 ng/ml) for 48 h. Total RNA isolated from these cells was analyzed with real-time PCR for RANKL mRNA expression (Fig. 4E). RANKL mRNA was significantly decreased in FGF-2-stimulated cells transfected with STAT-1 siRNA. No significant changes were observed in control siRNA-transfected cells.
Figure 4.
STAT-1 participation in FGF-2 stimulation of RANKL expression. A, FGF-2 increases phosphorylation of STAT-1 in stromal/preosteoblast cells. Bone marrow stromal cells were stimulated with FGF-2 (4 ng/ml) for the indicated time. Total cell lysates were analyzed by Western blot for p-STAT-1/3 and STAT-1/3 expression. B, Jurkat T cells were stimulated with interferon-γ (50 ng/ml) for 1 h. Total cell lysates were subjected to Western blot analysis for p-STAT-1/3 expression as a positive control (C). siRNA suppression of STAT-1 inhibits FGF-2-stimulated RANKL expression in bone marrow stromal cells. Bone marrow-derived stromal cells were transiently transfected with control and STAT-1 siRNA. Cells were stimulated with FGF-2 (4 ng/ml), and total cell lysates obtained after 48 h were analyzed by Western blot. The band intensity was quantified by NIH ImageJ program. The values are expressed as mean ± sd. *, P < 0.05. D, siRNA suppression of STAT-1 expression was confirmed by Western blot analysis. E, STAT-1 modulates FGF-2-stimulated RANKL mRNA expression in human bone marrow cells. Cells were transfected with control siRNA and STAT-1 siRNA. Cells were stimulated with and without FGF-2 (4 ng/ml) for 48 h, and total RNA was isolated was analyzed by real-time PCR for RANKL mRNA expression (*, P < 0.05).
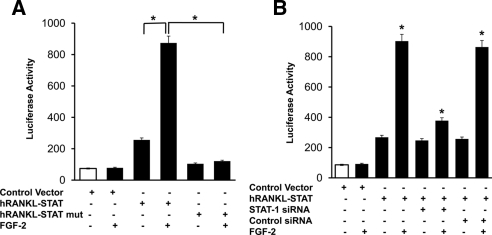
We then examined the role of STAT binding element in transcriptional regulation of RANKL gene expression in a human bone marrow-derived stromal cell line (SAKA-T). We used the SAKA-T cell line rather than primary bone marrow stromal cells to obtain high DNA transfection efficiency (>80%). We identified STAT binding element (ATTTGGGGAA) at −8 kb upstream in the hRANKL promoter region by web-based TFSEARCH analysis for transcription factor binding sites. Because long-range enhancer elements have been shown to play an important role in modulating RANKL gene expression (29), we further examined the potential role of STAT binding element to modulate RANKL gene promoter activity in response to FGF-2 stimulation. The STAT binding region in the hRANKL gene upstream sequence was subcloned into the pGL2 promoter-luciferase reporter vector. The resulting plasmid, hRANKL-pSTAT, was used as a template to generate STAT mutant reporter plasmid (hRANKL-pSTAT mut) by site-directed mutagenesis as described in Materials and Methods. hRANKL-pSTAT, hRANKL-pSTAT mut reporter plasmids, and a control EV were transiently transfected into SAKA-T stromal cells and cultured in the presence and absence of FGF-2 (4 ng/ml) for 48 h. Total cell lysates obtained from these cells were analyzed for luciferase activity. As shown in Fig. 5A, FGF-2 stimulation increased (4-fold) hRANKL-pSTAT promoter activity compared with unstimulated control cells. In contrast, hRANKL-pSTAT mut transfected cells demonstrated no significant change in the promoter activity, suggesting that mutation in the STAT binding site abrogated FGF-2 stimulation of hRANKL promoter activity in these cells. FGF-2 stimulation did not affect luciferase activity levels in control EV-transfected cells. To further confirm that STAT-1 regulate RANKL gene transcription, hRANKL-pSTAT reporter plasmid was transiently cotransfected with siRNA against STAT-1. The cells were cultured in the presence and absence of FGF-2 (4 ng/ml) for 48 h. Interestingly, hRANKL-pSTAT promoter activity was significantly decreased (2.4-fold) in FGF-2-stimulated cells transfected with STAT-1 siRNA (Fig. 5B). Transfection efficiency was normalized by coexpression of pRSV β-galactosidase plasmid, and β-galactosidase activity was measured in these cells. Results suggest that STAT-1 mediates FGF-2 stimulation of RANKL gene expression in bone marrow stromal/preosteoblast cells.
Figure 5.
STAT-1 modulation of hRANKL-pSTAT promoter activity. A, hRANKL-pSTAT, hRANKL-pSTAT mut reporter plasmids, and a control EV were transiently transfected into SAKA-T human bone-derived stromal cell line and stimulated with and without FGF-2 (4 ng/ml) for 48 h. B, SAKA-T stromal cells were cotransfected with hRANKL-pSTAT reporter construct with siRNA against STAT-1 and control scrambled siRNA. Cells were stimulated with and without FGF-2 (4 ng/ml) for 48 h. Total cell lysates obtained were assayed for luciferase activity. The transfection efficiency was normalized by β-galactosidase activity coexpressed in these cells. *, P < 0.05.
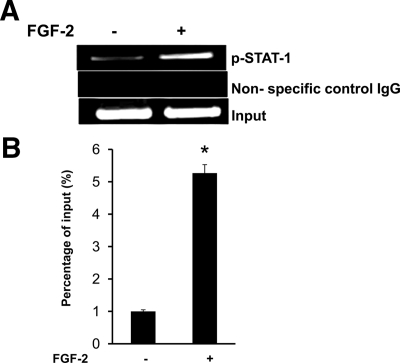
We next confirmed STAT-1 binding to the hRANKL gene promoter element by ChIP assay. SAKA-T bone marrow stromal cells stimulated with FGF-2 were analyzed by chromatin immunoprecipitation (ChIP) with anti-pSTAT-1 antibody as described in Materials and Methods. PCR analysis of chromatin immune complexes was then performed with hRANKL gene-specific primers for the STAT binding region. As shown in Fig. 6, A and B, FGF-2 significantly stimulated STAT-1 binding to the RANKL promoter region. A low level of STAT-1 binding at basal level could be due to transformed phenotype of SAKA-T cells. We have detected no amplification using primer sets specific for an additional segment in the hRANKL promoter region that lack STAT binding element in the absence or presence of FGF-2 stimuli (data not shown). Thus, our results suggest STAT-1 is a downstream effector of FGF-2 signaling to stimulate RANKL expression in PDB.
Figure 6.
ChIP assay for STAT-1 binding to the hRANKL gene promoter region. A, Normal human bone marrow stromal cells were stimulated with and without FGF-2 (4 ng/ml) for 48 h, and ChIP assay was performed using anti-p-STAT-1 (Tyr701) antibody as described in Materials and Methods. B, ChIP assay was quantified by real-time PCR. Data represent triplicate studies and are shown as mean ± sd. *, P < 0.05.
Discussion
High levels of RANKL expression in the bone microenvironment plays an important role in osteoclastogenesis and high bone turnover associated with PDB. Immature osteoblasts are the major responders to RANKL-inducing cytokines, suggesting that the relative proportions of immature and mature osteoblasts in the local microenvironment may control the degree of bone resorption at specific sites (30). Abnormal OCL in PDB are characterized by expression of MVNP transcripts and are shown to produce high levels of IL-6 (31). However, IL-6 increases RANKL expression in murine systems but does not induce RANKL expression in human marrow stromal cells or osteoblasts (32). These results suggest that the high levels of IL-6 produced in the Pagetic lesions do not directly induce RANKL expression in marrow stromal cells and that other cytokines may be responsible. Increased FGF-2 expression in OCL was observed in PDB compared with normal bone (33). Our results are consistent with the finding that serum FGF-2 is elevated in patients with PDB compared with normal subjects. Also, our findings that MVNP stimulated FGF-2 production in OCL cultures suggest a potential pathophysiological role for MVNP expression in OCL in PDB. We previously reported that HSF-2 is a downstream target for FGF-2 to enhance RANKL expression in marrow stromal/preosteoblast cells (18). However, FGF-2 has been shown to affect multiple stages during osteoblast differentiation. FGF-2 promotes human bone marrow stromal cell proliferation and maintenance of osteogenic precursors (34).
We have shown that FGF-2 treatment did not induce RANKL expression in HSF2−/− stromal/preosteoblast cells. Furthermore, HSF-2 deficiency resulted in a rapid induction of alkaline phosphatase and osteocalcin expression in stromal/preosteoblast cells (35). Therefore, it is possible that HSP-27 and HSF-2 activation may have pleiotropic effects on gene expression associated with osteoblastic proliferation and differentiation. FGF-2 has been shown to stimulate OCL formation in mouse bone marrow cultures by mechanisms that require prostaglandin synthesis (36). However, our results do not delineate whether a prostaglandin pathway is involved in HSP-27 and HSF-2 activation in response to FGF-2 treatment to stromal/preosteoblastic cells. Reactive oxygen species have been shown to promote HSF-2 binding to an HSE in the hRANKL promoter, thereby stimulating RANKL gene expression (37). In addition, Sp1 and Sp3 have been reported to regulate basal RANKL gene transcription in stromal/osteoblast cells (38). The mouse RANKL gene 5′-flanking region contains a CCAAT box, Runx2/Cbfa1, and vitamin D-responsive element (VDRE) motifs (39,40). Mouse RANKL expression has also been shown to be up-regulated by calcemic hormones such as 1,25-dihydroxyvitamin D3 and PTH; however, these transcriptional regulatory regions are located long range (−76 kb in mouse and −96 kb in human) relative to the transcription site in a region of high evolutionary conservation that is designated as the distal control region (17,41). O’Brien et al. (39) found that cotransfection of cells with Runx2/Cbfa1 and a RANKL reporter construct did not increase RANKL expression. However, Runx2/Cbfa1 potentiates PTH stimulation of RANKL through a site in the distal control region. Our findings that the STAT binding motif presents −8 kb upstream to the start codon and that STAT-1 binds to this region in response to FGF-2 stimulation further support previous reports that long-range enhancer elements may play an important role in modulating RANKL gene expression in bone marrow stromal/preosteoblast cells. We have previously characterized proximal hRANKL gene promoter region (<2.0 kb) with respect to FGF-2 stimulation and HSF-2 transcriptional control; however, the proximal promoter lacks a putative STAT binding element (18). Srivastava et al. (42) have shown that STAT5 regulates the proximal mouse RANKL gene promoter. Recently, microarray studies identified elevated Dkk-1 in PDB (43). It has been reported that Dkk1 attenuates Wnt3a-dependent inhibition of RANKL in mesenchymal stem cells treated with 1,25-dihydroxyvitamin D3 (44). In addition to genetic control of the mouse RANKL promoter, epigenetic control has been demonstrated. Kitazawa and Kitazawa (45) found that methylation of the CpG loci occurred around the transcription start site in late-passage cells and that methylation of the sites in the RANKL promoter silenced promoter activity. Therefore, RANKL expression could be modulated by complex cytokine-mediated transcriptional regulatory mechanisms in Pagetic lesions. In summary, our results suggest that STAT-1 is a downstream effector of FGF-2 signaling and that elevated FGF-2 stimulates RANKL expression in PDB.
Materials and Methods
Reagents and antibodies
Cell culture and DNA transfection reagents were purchased from Invitrogen (Carlsbad, CA). FGF-2 and anti-human RANKL antibody were obtained from R&D Systems Inc. (Minneapolis, MN). Phosho-STAT-1 (Tyr701; catalog no. 9171L) and p-STAT-3 (Tyr705; catalog no. 9131S) was purchased from Cell Signaling (Danvers, MA). Antigoat-HSF-2, siRNA, and peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Super signal enhanced chemiluminescence (ECL) reagent was obtained from Amersham Bioscience (Piscataway, NJ), and nitrocellulose membranes were purchased from Millipore (Bedford, MA). A luciferase reporter assay system was obtained from Promega (Madison, WI). Protease inhibitor cocktail was purchased from Sigma Chemical Co. (St. Louis, MO).
Retroviral expression of MVNP in bone marrow cells
We have previously developed a retroviral plasmid construct, pILXAN#1 that transcribes MVNP mRNA expression under the control of 5′-LTR viral promoter elements. The recombinant plasmid construct was transfected into the PT67 amphotropic packaging cell line using the Lipofectamine reagent (Invitrogen). Stable clonal cell lines producing MVNP recombinant retrovirus at high titer (1 × 106 virus particles/ml) were established by selecting for resistance to neomycin (600 μg/ml). Similarly, a control retrovirus producer cell line was established by transfecting the cells with the pLXSN EV. Producer cell lines were maintained in DMEM containing 10% fetal calf serum (GIBCO, Gaithersburg, MD), 100 U/ml each of streptomycin and penicillin, 4 mm l-glutamine, and high glucose (4.5 g/liter). Normal human bone marrow-derived mononuclear cells were transduced with EV or MVNP retroviral supernatants (20%) from the producer cell lines in 4 μg/ml polybrene for 24 h at 37 C in a 5% CO2 incubator as described earlier (25). The cells (4 × 105/ml) were cultured in methylcellulose (Stem Cell Technologies, Vancouver, Canada) and incubated for 7 d to form CFU-GM as described (46), and purified colony-forming unit-granulocyte macrophage were cultured to form OCL in the presence of 10 ng/ml M-CSF and 100 ng/ml RANKL (R&D Systems) for 1 wk. At the end of the culture period, the OCL culture media were removed and serum-free CM collected after 24 h from MVNP- and EV-transduced OCL cultures was used for additional experiments.
ELISA
FGF-2 in serum samples from age-matched normal subjects, patients with PDB (mean age 70 ± 10 yr), and cell culture CM were assayed by ELISA (R&D Systems). All the patients had either mono- or polyostotic PDB with elevated levels of alkaline phosphatase activity. All human samples were obtained following the Institutional Review Board-approved protocol at the Medical University of South Carolina. In brief, CM were collected from MVNP-transduced human OCL cultures as described and concentrated with an Amicon filter. Total protein content was determined by bicinchoninic acid protein assay kit (Pierce, Rockford, IL). FGF-2 levels measured by ELISA were normalized according to the total protein content in all samples analyzed.
Coimmunoprecipitation assay
Human bone marrow-derived stromal/preosteoblast cells were obtained as described (18). Cells were stimulated with and without FGF-2 (4 ng/ml) for 24 h and then lysed in a lysis buffer [50 mm HEPES (pH 7.5), 250 mm NaCl, 0.2 mm EDTA, 10 μm NaF, 0.5% Nonidet P-40]. Total cell lysates were immunoprecipitated using anti-HSP-27 antibody. Immunoprecipitates were subjected to SDS-PAGE and Western blot analysis using anti-HSF-2 antibody.
Confocal microscopy
Human bone marrow stromal cells were cultured (1 × 103/well) in a Lab-Tek four-well chamber slides (Nunc, Rochester, NY). The cells were stimulated with and without FGF-2 (4 ng/ml) or 20% of CM collected from MVNP-transduced human OCL cultures for 24 h and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. The cells were permeabilized with 0.1% Triton X-100 for 10 min at room temperature and blocked for 1 h with PBS containing 2% horse serum. Cells were immunostained with primary antibodies against HSF-2 or HSP-27 in PBS containing 2% horse serum and incubated for 3 h at room temperature. After extensive washing with PBS, cells were incubated with Alexa 488-conjugated antirabbit IgG and Alexa 568-conjugated antigoat IgG in PBS containing 2% horse serum for 1 h at room temperature. Nuclear staining was performed with DRAQ5, and localization of HSP-27 and HSF-2 was visualized by confocal microscopy (LSM 510; Carl Zeiss, Inc., Thornwood, NY).
Quantitative real-time RT-PCR
RANKL mRNA expression in bone marrow stromal/preosteoblast cells was measured by real-time RT-PCR as described previously (21). Briefly, total RNA was isolated from bone marrow stromal cells transfected with control siRNA and STAT-1 siRNA. Cells were stimulated with and without FGF-2 (4 ng/ml) for 48 h, using RNAzol reagent (Biotecx Labs, Houston, TX). A reverse transcription reaction was performed using poly-dT primer and Moloney murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA) in a 25-μl reaction volume containing total RNA (2 μg), 1× PCR buffer, and 2 mm MgCl2, at 42 C for 15 min followed by 95 C for 5 min. The quantitative real-time PCR was performed using IQ SYBR Green Supermix in an iCycler (iCycler iQ single-color Real Time PCR detection system; Bio-Rad, Hercules, CA). The primer sequences used to amplify glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were 5′-CCTACCCCCAATGTATCCGTTGTG-3′ (sense) and 5′-GGAGGAATGGGAGTTGCTGTTGAA-3′ (antisense) and for hRANKL mRNA were 5′-ACCAGCATCAAAATCCCAAG-3′ (sense) and 5′-TAAGGAGTTGGAGACCT-3′ (antisense). Thermal cycling parameters were 94 C for 3 min, followed by 40 cycles of amplifications at 94 C for 30 sec, 60 C for 1 min, 72 C for 1 min, and 72 C for 5 min as the final elongation step. Relative levels of RANKL mRNA expression were normalized in all the samples analyzed with respect to GAPDH amplification.
hRANKL gene promoter activity assay
Normal human bone marrow stromal SAKA-T cells (18) were cultured in α-MEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin/streptomycin. Cells were maintained in a humidified atmosphere with 5% CO2 at 37 C. DNA transfections were performed using Lipofectamine transfection reagent (Invitrogen) according to the manufacturer’s protocol. The STAT binding element (ATTTGGGGAA) in the hRANKL promoter region (−8015 to −8243 kb) was PCR amplified and subcloned into the pGL2 promoter vector (Promega) containing SV40 heterologous promoter at KpnI and BglII restriction enzyme sites. The resulting plasmid construct is termed hRANKL-pSTAT. The transfection efficiency was normalized by cotransfection with 0.2 μg pRSV β-gal plasmid and measuring β-galactosidase activity in the cells. LacZ cytochemical activity staining (Invitrogen Inc., San Diego, CA) indicated a DNA transfection efficiency (>80%) in SAKA-T cells. Cells were cultured in the presence or absence of FGF-2 (4 ng/ml) for 48 h. The cell monolayer was washed twice with PBS and incubated at room temperature for 15 min with 0.3 ml cell lysis reagent (Promega). The monolayer was scraped and spun briefly in a microfuge to pellet the debris. A 20-μl aliquot of each sample was then mixed with 100 μl of the luciferase assay reagent. Light emission was measured for 10 sec of integrated time using a Sirius Luminometer (Promega).
Site-directed mutagenesis
Site-directed mutagenesis of STAT binding element in the hRANKL-pSTAT plasmid was performed using the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The STAT primers (mutant bases underlined) used were 5′-TTAGTAGATTCAGGGAAGTCTGAA-3′ (sense) and 5′-TTCAGACTTCCCTGAATCTACTAA-3′ (antisense). The hRANKL-pSTAT mut plasmid construct developed was confirmed by DNA sequence analysis.
siRNA interference
Human bone marrow stromal/preosteoblast cells were seeded (5 × 105 cells per well) in six-well plates and supplemented with α-MEM containing 10% fetal calf serum. One day after seeding, cells were transfected with double-stranded control, nonspecific siRNA and STAT-1 siRNA (20 nm) by the oligofectamine (Invitrogen) method. Cells were cultured with or without FGF-2 (4 ng/ml) for 48 h. Cells were lysed in a buffer containing 20 mm Tris-HCl (pH 7.4), 1% Triton X-100, 1 mm EDTA, 1.5 mm MgCl2, 150 mm NaCl, 0.1 mm Na3VO4, and 1× protease inhibitor cocktail. Protein content of the samples was measured using the bicinchoninic acid protein assay reagent (Pierce). Samples (20 μg protein) were then subjected to SDS-PAGE using 12% Tris-HCl gels, and blot transferred onto a nitrocellulose membrane, and immunoblotted with anti-RANKL and anti-STAT-1 antibody. Bands were detected using an enhanced chemiluminescence detection system. Band intensity was quantified by densitometric analysis using the NIH ImageJ Program.
ChIP assay
ChIP was performed using the ChIP assay kit (Upstate, Temecula, CA). SAKA-T human bone marrow stromal cells were treated with or without FGF-2 (4 ng/ml) for 48 h. Cells were cross-linked with a 1% final concentration of formaldehyde at 37 C for 10 min. Crude nuclei were collected by centrifugation at 4000 × g for 3 min, washed once with ice-cold PBS, and resuspended in SDS lysis buffer. Soluble chromatin was prepared by sonication with a Branson-250 digital sonifier (Branson Ultrasonics, Danbury, CT) to an average DNA length of 200–1000 bp. Approximately 5 × 105 cell equivalents (one sixth) of the sheared soluble chromatin was precleared with blocked protein G agarose, and 10% of the precleared chromatin was set aside for input as positive control. Samples were immunoprecipitated overnight at 4 C using p-STAT-1 antibody or equivalent concentrations of normal rabbit IgG as a negative control. Immune complexes were pulled down using protein G agarose, washed, and eluted twice with 250 μl elution buffer (0.1 m NaHCO3, 1% SDS), and cross-linking was reversed in 200 mm NaCl at 65 C overnight with 20 μg ribonuclease A. To analyze the STAT binding region to the hRANKL promoter, immunoprecipitated chromatin DNA samples were amplified by PCR using primer pairs for the STAT binding region in the hRANKL promoter (−8096 to −7979 bp): 5′-TTTACAGCAATGAGCAGACCT-3′ (sense) and 5′-CAGGATGCATGGGATTACCT-3′ (antisense). Primers used to amplify the hRANKL promoter region (−7383 to −7213 bp) that lack the STAT binding element were 5′-ACCAGTCTGGGCAACATAGG-3′ (sense) and 5′-GCTAGAGTGCAGTGGTGCAA-3′ (antisense). Immune precipitated DNA samples or input DNA fractions were analyzed by real-time quantitative PCR analysis as described above. Thermal cycling parameters were 35 cycles of 94 C for 30 sec, 55 C for 30 sec, and 72 C for 30 sec with a final extension at 72 C for 5 min. The percentage of chromatin immunoprecipitated DNA relative to input was calculated and shown as mean ± sd from three independent experiments. PCR products were subjected to electrophoresis by using 2% agarose gels and visualized by ethidium bromide.
Statistical analysis
Results are presented as mean ± sd for three independent experiments and were compared by Student’s t test. Values were considered significantly different for P < 0.05.
Footnotes
This work was supported by National Institutes of Health Grant AR 049363 and a U.S. Department of Defense Medical Research Award (DAMD 17-03-1-0763).
Disclosure Summary: The authors declare no conflict of interest.
First Published Online June 25, 2009
Abbreviations: ChIP, Chromatin immunoprecipitation; CM, conditioned medium; EV, empty vector; FGF-2, fibroblast growth factor-2; HSE, heat-shock-responsive element; HSF, heat-shock transcription factor; HSP, heat-shock protein; M-CSF, macrophage colony-stimulating factor; OCL, osteoclast; PDB, Paget’s disease of bone; RANKL, receptor activator for nuclear factor-κB ligand; siRNA, small interfering RNA; STAT, signal transducers and activators of the transcription.
References
- Hocking LJ, Lucas GJ, Daroszewska A, Mangion J, Olavesen M, Cundy T, Nicholson GC, Ward L, Bennett ST, Wuyts W, Van Hul W, Ralston SH 2002 Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget’s disease. Hum Mol Genet 11:2735–2739 [DOI] [PubMed] [Google Scholar]
- Seton M 2008 Paget’s disease: epidemiology and pathophysiology. Curr Osteoporos Rep 6:125–129 [DOI] [PubMed] [Google Scholar]
- Roodman GD, Windle JJ 2005 Paget disease of bone. J Clin Invest 115:200–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant A, Smielewska M, Patel N, Akunowicz JD, Saria EA, Delaney JD, Leach RJ, Seton M, Hansen MF 2009 Somatic mutations in SQSTM1 detected in affected tissues from patients with sporadic Paget’s disease of bone. J Bone Miner Res 24:484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich MH, Hobson RP, Grabowski PS, Zurbriggen A, Cosby SL, Dickson GR, Fraser WD, Ooi CG, Selby PL, Crisp AJ, Wallace RG, Kahn S, Ralston SH 2000 A negative search for a paramyxoviral etiology of Paget’s disease of bone: molecular, immunological, and ultrastructural studies in UK patients. J Bone Miner Res 15:2315–2329 [DOI] [PubMed] [Google Scholar]
- Matthews BG, Afzal MA, Minor PD, Bava U, Callon KE, Pitto RP, Cundy T, Cornish J, Reid IR, Naot D 2008 Failure to detect measles virus ribonucleic acid in bone cells from patients with Paget’s disease. J Clin Endocrinol Metab 93:1398–1401 [DOI] [PubMed] [Google Scholar]
- Menaa C, Reddy SV, Kurihara N, Maeda H, Anderson D, Cundy T, Cornish J, Singer FR, Bruder JM, Roodman GD 2000 Enhanced RANK ligand expression and responsivity of bone marrow cells in Paget’s disease of bone. J Clin Invest 105:1833–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale SD, Schulze E, Smith R, Athanasou NA 2002 The influence of serum cytokines and growth factors on osteoclast formation in Paget’s disease. QJM 95:233–240 [DOI] [PubMed] [Google Scholar]
- Reddy SV 2004 Etiology of Paget’s disease and osteoclast abnormalities. J Cell Biochem 93:688–696 [DOI] [PubMed] [Google Scholar]
- Tsuruga E, Rao DS, Baatz JE, Reddy SV 2006 Elevated serum kininogen in patients with Paget’s disease of bone: a role in marrow stromal/preosteoblast cell proliferation. J Cell Biochem 98:1681–1688 [DOI] [PubMed] [Google Scholar]
- Nakashima T, Kobayashi Y, Yamasaki S, Kawakami A, Eguchi K, Sasaki H, Sakai H 2000 Protein expression and functional difference of membrane-bound and soluble receptor activator of NF-κB ligand: modulation of the expression by osteotropic factors and cytokines. Biochem Biophys Res Commun 275:768–775 [DOI] [PubMed] [Google Scholar]
- Lee SK, Kalinowski J, Jastrzebski S, Lorenzo JA 2002 1,25(OH)2 vitamin D3-stimulated osteoclast formation in spleen-osteoblast cocultures is mediated in part by enhanced IL-1α and receptor activator of NF-κB ligand production in osteoblasts. J Immunol 169:2374–2380 [DOI] [PubMed] [Google Scholar]
- Rossa C, Ehmann K, Liu M, Patil C, Kirkwood KL 2006 MKK3/6-p38 MAPK signaling is required for IL-1β and TNF-α-induced RANKL expression in bone marrow stromal cells. J Interferon Cytokine Res 26:719–729 [DOI] [PubMed] [Google Scholar]
- Nakagawa N, Yasuda H, Yano K, Mochizuki S, Kobayashi N, Fujimoto H, Shima N, Morinaga T, Chikazu D, Kawaguchi H, Higashio K 1999 Basic fibroblast growth factor induces osteoclast formation by reciprocally regulating the production of osteoclast differentiation factor and osteoclastogenesis inhibitory factor in mouse osteoblastic cells. Biochem Biophys Res Commun 265:158–163 [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Matsuguchi T, Tsuboi N, Mitani A, Tanaka S, Matsuoka M, Yamamoto G, Hishikawa T, Noguchi T, Yoshikai Y 2001 Gene expression of osteoclast differentiation factor is induced by lipopolysaccharide in mouse osteoblasts via Toll-like receptors. J Immunol 166:3574–3579 [DOI] [PubMed] [Google Scholar]
- Wang R, Zhang L, Zhang X, Moreno J, Celluzzi C, Tondravi M, Shi Y 2002 Regulation of activation-induced receptor activator of NF-κB ligand (RANKL) expression in T cells. Eur J Immunol 32:1090–1098 [DOI] [PubMed] [Google Scholar]
- Fu Q, Manolagas SC, O'Brien CA 2006 Parathyroid hormone controls receptor activator of NF-κB ligand gene expression via a distant transcriptional enhancer. Mol Cell Biol 26:6453–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccisana JL, Kawanabe N, Kajiya H, Koide M, Roodman GD, Reddy SV 2004 Functional role for heat shock factors in the transcriptional regulation of human RANK ligand gene expression in stromal/osteoblast cells. J Biol Chem 279:10500–10507 [DOI] [PubMed] [Google Scholar]
- Singh IS, Viscardi RM, Kalvakolanu I, Calderwood S, Hasday JD 2000 Inhibition of tumor necrosis factor-α transcription in macrophages exposed to febrile range temperature. A possible role for heat shock factor-1 as a negative transcriptional regulator. J Biol Chem 275:9841–9848 [DOI] [PubMed] [Google Scholar]
- Snoeckx LH, Cornelussen RN, Van Nieuwenhoven FA, Reneman RS, Van Der Vusse GJ 2001 Heat shock proteins and cardiovascular pathophysiology. Physiol Rev 81:1461–1497 [DOI] [PubMed] [Google Scholar]
- Sundaram K, Mani SK, Kitatani K, Wu K, Pestell RG, Reddy SV 2008 DACH1 negatively regulates the human RANK ligand gene expression in stromal/preosteoblast cells. J Cell Biochem 103:1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca F, Baron J 1999 Control of bone growth by fibroblast growth factors. Trends Endocrinol Metab 10:61–65 [DOI] [PubMed] [Google Scholar]
- Sahni M, Ambrosetti DC, Mansukhani A, Gertner R, Levy D, Basilico C 1999 FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes Dev 13:1361–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Okada Y, Saito K, Tanaka Y 2004 Induction of RANKL expression and osteoclast maturation by the binding of fibroblast growth factor 2 to heparan sulfate proteoglycan on rheumatoid synovial fibroblasts. Arthritis Rheum 50:2450–2458 [DOI] [PubMed] [Google Scholar]
- Kurihara N, Reddy SV, Menaa C, Anderson D, Roodman GD 2000 Osteoclasts expressing the measles virus nucleocapsid gene display a pagetic phenotype. J Clin Invest 105:607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians ES, Zhou Q, Renard J, Benjamin IJ 2003 Heat shock proteins in mammalian development. Semin Cell Dev Biol 14:283–290 [DOI] [PubMed] [Google Scholar]
- Takai S, Tokuda H, Matsushima-Nishiwaki R, Hanai Y, Kato K, Kozawa O 2006 Phosphatidylinositol 3-kinase/Akt plays a role in sphingosine 1-phosphate-stimulated HSP27 induction in osteoblasts. J Cell Biochem 98:1249–1256 [DOI] [PubMed] [Google Scholar]
- Xiao L, Naganawa T, Obugunde E, Gronowicz G, Ornitz DM, Coffin JD, Hurley MM 2004 Stat1 controls postnatal bone formation by regulating fibroblast growth factor signaling in osteoblasts. J Biol Chem 279:27743–27752 [DOI] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Shevde NK, Pike JW 2007 Transcriptional control of receptor activator of nuclear factor-κB ligand by the protein kinase A activator forskolin and the transmembrane glycoprotein 130-activating cytokine, oncostatin M, is exerted through multiple distal enhancers. Mol Endocrinol 21:197–214 [DOI] [PubMed] [Google Scholar]
- Thomas GP, Baker SU, Eisman JA, Gardiner EM 2001 Changing RANKL/OPG mRNA expression in differentiating murine primary osteoblasts. J Endocrinol 170:451–460 [DOI] [PubMed] [Google Scholar]
- Kurihara N, Zhou H, Reddy SV, Garcia Palacios V, Subler MA, Dempster DW, Windle JJ, Roodman GD 2006 Expression of measles virus nucleocapsid protein in osteoclasts induces Paget’s disease-like bone lesions in mice. J Bone Miner Res 21:446–455 [DOI] [PubMed] [Google Scholar]
- Hofbauer LC, Khosla S, Dunstan CR, Lacey DL, Boyle WJ, Riggs BL 2000 The roles of osteoprotegerin and osteoprotegerin ligand in the paracrine regulation of bone resorption. J Bone Miner Res 15:2–12 [DOI] [PubMed] [Google Scholar]
- Mills BG, Frausto A 1997 Cytokines expressed in multinucleated cells: Paget’s disease and giant cell tumors versus normal bone. Calcif Tissue Int 61:16–21 [DOI] [PubMed] [Google Scholar]
- Martin I, Muraglia A, Campanile G, Cancedda R, Quarto R 1997 Fibroblast growth factor-2 supports ex vivo expansion and maintenance of osteogenic precursors from human bone marrow. Endocrinology 138: 4456–4462 [DOI] [PubMed] [Google Scholar]
- Kajiya H, Ito M, Ohshima H, Kenmotsu S, Ries WL, Benjamin IJ, Reddy SV 2006 RANK ligand expression in heat shock factor-2 deficient mouse bone marrow stromal/preosteoblast cells. J Cell Biochem 97:1362–1369 [DOI] [PubMed] [Google Scholar]
- Hurley MM, Lee SK, Raisz LG, Bernecker P, Lorenzo J 1998 Basic fibroblast growth factor induces osteoclast formation in murine bone marrow cultures. Bone 22:309–316 [DOI] [PubMed] [Google Scholar]
- Bai XC, Lu D, Liu AL, Zhang ZM, Li XM, Zou ZP, Zeng WS, Cheng BL, Luo SQ 2005 Reactive oxygen species stimulates receptor activator of NF-κB ligand expression in osteoblast. J Biol Chem 280:17497–17506 [DOI] [PubMed] [Google Scholar]
- Liu J, Yang H, Liu W, Cao X, Feng X 2005 Sp1 and Sp3 regulate the basal transcription of receptor activator of nuclear factor κB ligand gene in osteoblasts and bone marrow stromal cells. J Cell Biochem 96:716–727 [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Kern B, Gubrij I, Karsenty G, Manolagas SC 2002 Cbfa1 does not regulate RANKL gene activity in stromal/osteoblastic cells. Bone 30:453–462 [DOI] [PubMed] [Google Scholar]
- Kitazawa R, Kitazawa S, Maeda S 1999 Promoter structure of mouse RANKL/TRANCE/OPGL/ODF gene. Biochim Biophys Acta 1445:134–141 [DOI] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW 2006 Activation of receptor activator of NF-κB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol 26:6469–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Matsuda M, Hou Z, Bailey JP, Kitazawa R, Herbst MP, Horseman ND 2003 Receptor activator of NF-κB ligand induction via Jak2 and Stat5a in mammary epithelial cells. J Biol Chem 278:46171–46178 [DOI] [PubMed] [Google Scholar]
- Naot D, Bava U, Matthews B, Callon KE, Gamble GD, Black M, Song S, Pitto RP, Cundy T, Cornish J, Reid IR 2007 Differential gene expression in cultured osteoblasts and bone marrow stromal cells from patients with Paget’s disease of bone. J Bone Miner Res 22:298–309 [DOI] [PubMed] [Google Scholar]
- Fujita K, Janz S 2007 Attenuation of WNT signaling by DKK-1 and -2 regulates BMP2-induced osteoblast differentiation and expression of OPG, RANKL and M-CSF. Mol Cancer 6:71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa S, Kitazawa R 2002 Epigenetic control of mouse receptor activator of NF-κB ligand gene expression. Biochem Biophys Res Commun 293:126–131 [DOI] [PubMed] [Google Scholar]
- Koide M, Kurihara N, Maeda H, Reddy SV 2002 Identification of the functional domain of osteoclast inhibitory peptide-1/hSca. J Bone Miner Res 17:111–118 [DOI] [PubMed] [Google Scholar]