Abstract
Advanced prostate cancers preferentially metastasize to bone, suggesting that this tissue produces factors that provide a suitable microenvironment for prostate cancer cells. Recently, it has become clear that even in antiandrogen-resistant cancers, the androgen receptor (AR)-signaling axis is required for prostate cancer progression. Therefore, we hypothesized that AR may be involved in the regulation of pathways that are responsible for the homing of prostate cancer cells to select microenvironments. In support of this hypothesis, we have determined that chemokine (C-X-C motif) receptor 4 (CXCR4), the receptor for the chemokine CXCL12, is up-regulated in prostate cancer cells in response to androgens. Given that the levels of CXCL12 are elevated at sites of known prostate cancer metastases such as bone, these results suggest that androgens may influence prostate cancer metastasis. Specifically, we demonstrate that androgens increase the levels of both CXCR4 mRNA and functional protein in LNCaP prostate cancer cells. Importantly, androgens enhanced the migration of LNCaP cells toward a CXCL12 gradient, an effect that could be blocked by the specific CXCR4 antagonist AMD3100. Interestingly, CXCR4 is not directly regulated by androgens but rather is positively up-regulated by Krüppel-like factor 5 (KLF5), a transcription factor that we have shown to be an early, direct target of AR. Further, KLF5 is both required and sufficient for androgen-mediated CXCR4 expression and migration toward CXCL12. Taken together, these findings demonstrate that AR can utilize the CXCL12/CXCR4 axis through induction of KLF5 expression to promote prostate cancer progression and highlight the potential utility of CXCR4 antagonists as prostate cancer therapeutics.
Androgens increase the migration of prostate cancer cells towards CXCL12-rich environments by increasing CXCR4 levels through the direct AR-mediated expression of the transcription factor KLF5.
Prostate cancer is the most commonly diagnosed malignancy in American men and is second only to lung cancer in terms of cancer mortalities in men (1). Early/localized prostatic tumors are most often successfully treated by surgery alone (i.e. radical prostatectomy). As with many cancers, however, the treatment of advanced disease state requires a systemic intervention to inhibit the growth and spread of secondary metastasis. The majority of prostate cancers express the androgen receptor (AR) and rely on androgens for growth and survival (2). Hence, androgen ablation therapies are the standard of care for late-stage disease. While 80% of patients with local or metastatic prostate cancer initially respond favorably to androgen ablation therapy, most patients experience a relapse of the disease in 1–2 yr (2). Although hormone-refractory disease is unresponsive to androgen-deprivation therapy, AR-regulated signaling pathways remain active and have been shown to be necessary for cancer progression. Thus, it is now considered that AR itself and the processes downstream of the receptor remain viable targets for therapeutic intervention.
Androgens exhibit mitogenic activities in prostate cancer cells. Recently, however, it has become evident that AR also has a significant role in metastasis. Specifically, in a phase III clinical trial in patients undergoing radiotherapy for prostate carcinoma with or without long-term administration of adjuvant goserelin (an LHRH analog that blocks testicular androgen production), it was found that 27% of the patients receiving goserelin had distant metastases in comparison with 37% of the patients receiving radiotherapy alone (3). Likewise, the European Organization for Research and Treatment of Cancer reported that 27% of patients who received radiotherapy alone developed distant metastases in comparison with only 10.6% of patients that received long-term adjuvant goserelin treatment (4). This suggests that androgen ablation therapy not only inhibits the growth of the primary tumor but also reduces progression to metastatic disease.
During the later stages of metastasis, tumor cells leave the vasculature and eventually extravasate into surrounding tissues. Whereas many cancers simply metastasize to regions that physically favor cell retention due to high circulation or close proximity, some cancers preferentially extravasate to unique microenvironments. For example, there is usually a high degree of bone metastasis in patients with advanced prostate cancer (5). Therefore, we were interested in defining factors, enriched in the bone microenvironment, that could be responsible for the homing of AR-driven prostate metastases.
Chemokine (C-X-C motif) receptor 4 (CXCR4) is a cell surface receptor for the chemokine CXCL12 (formerly known as stromal-cell derived factor-1) and plays an important role in the homing of hematopoietic stem cells to the bone (6). This is due to the fact that CXCL12 levels are relatively high in the bone microenvironment. Recently, induction of the CXCL12/CXCR4 axis has been implicated in the progression and metastasis of prostate cancer (7,8,9,10,11,12,13). Additionally, the expression level of CXCR4 itself is elevated in a number of advanced cancers (14,15,16). Interestingly, in a series of microarray studies performed by our laboratory, we determined that the chemokine receptor CXCR4 was positively up-regulated by androgens in cellular models of AR-positive prostate cancer (17). We therefore hypothesized that AR drives prostate cancer progression, in part, through the up-regulation of CXCR4, leading to the migration of cancer cells toward microenvironments in which CXCL12 is expressed. In this study, we demonstrate that AR utilizes a novel mechanism that involves the transcription factor Krüppel-like factor 5 (KLF5) to up-regulate CXCR4 expression and that this facilitates the migration of LNCaP prostate cancer cells toward CXCL12.
Results
Androgens increase CXCR4 mRNA and protein levels in an AR-dependent manner
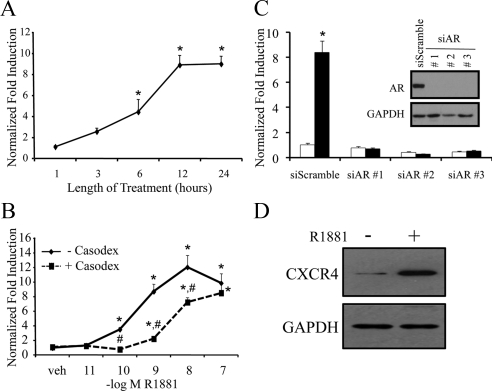
LNCaP human prostate carcinoma cells have previously been shown to be a reliable in vitro model with which to study androgen-regulated migration (18,19). Microarray analysis performed in our laboratory revealed that the CXCR4 mRNA is regulated by androgens in LNCaP cells (17). To validate our microarray studies, we performed quantitative RT-PCR (qPCR) to demonstrate that the synthetic androgen R1881 increased CXCR4 mRNA levels with a maximal induction after a 12-h treatment (Fig. 1A). The androgen-mediated induction of CXCR4 expression occurred in a dose-dependent manner with significant induction of mRNA observed after treatment with 100 pm R1881. Maximal induction occurred at 10 nm R1881, resulting in an approximately 12-fold increase in expression (Fig. 1B). Importantly, this dose-dependent increase in CXCR4 levels could be suppressed by pretreatment with the antiandrogen Casodex, indicating that, indeed, AR was necessary for androgen-mediated CXCR4 expression (Fig. 1B). Casodex suppression was overcome when cells were cotreated with a saturating concentration of R1881 (Fig. 1B), demonstrating the competitive ligand-specific effect. Furthermore, three separate small interfering RNAs (siRNAs) targeting AR completely blocked the androgen-mediated up-regulation of CXCR4 mRNA (Fig. 1C). Importantly, androgens also increased CXCR4 protein levels (Fig. 1D), an effect that was also dependent on AR (supplemental Fig. S1 published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). Collectively, these studies confirm that CXCR4 is an AR target in prostate cancer cells.
Figure 1.
Androgens up-regulate CXCR4 transcript and protein levels through the AR. LNCaP cells were treated with (A) 10 nm R1881 for the indicated time points or (B) pretreated with or without 10 μm Casodex for 30 min before addition of indicated doses of R1881 for an additional 24 h. C, LNCaP cells were transiently transfected with Stealth siRNAs targeting AR (siARs nos. 1–3) or a negative control (siScramble) at a final concentration of 100 nm. Cells were treated 48 h later with vehicle (white) or 10 nm R1881 (black) for 24 h. Inset, Western blot control confirming AR knockdown with siRNAs using GAPDH as a loading control. A–C, After treatment, cells were lysed, RNA isolated, and reverse transcribed. The expression of CXCR4 was assessed using qPCR and normalized to 36B4 levels. Results are expressed as fold induction over vehicle-treated cells + se (n = 3). *, P < 0.05 indicates significant changes from vehicle-treated cells. #, P < 0.05 indicates significant changes from cells treated without Casodex. D, LNCaP cells were treated with vehicle or 10 nm R1881 for 24 h and then subjected to Western blot analysis (n = 3). Blots were probed for CXCR4 (clone 12G5) or GAPDH (loading control). veh, Vehicle.
Androgens increase cell surface levels of CXCR4 protein
In order for CXCR4 to be functional, it must be expressed, translated, and translocated to the cell surface, where it acts as a receptor for its ligand CXCL12. To determine whether androgen treatment led to an increase in cell surface levels of CXCR4, we performed flow cytometry using a phycoerythrin (PE)-conjugated antibody that recognizes the extracellular surface of CXCR4. As shown in Fig. 2A, treatment with the synthetic androgen R1881 increased CXCR4 surface levels in a dose-dependent manner with levels starting to significantly increase at 1 nm R1881. Androgen-mediated CXCR4 cell surface levels were detectable at 18 h after treatment and increased even further up to 48 h (Fig. 2B). The overall percentage of cells that were stained with the CXCR4 antibody in this assay was relatively low because addition of the receptor antibody results in a very rapid internalization of the receptor (20,21,22,23,24). Nevertheless, although semiquantitative, the results of this assay confirm that androgens increase the expression of CXCR4 on the surface of LNCaP cells.
Figure 2.
Androgens increase cell surface levels of CXCR4. LNCaP cells were treated with ethanol (veh) or (A) increasing concentrations of R1881 for 24 h or (B) 10 nm R1881 for the indicated time points. After treatment, cells were dissociated and resuspended at a concentration of 1000 cells/ml in PBS and 1% BSA. Anti-CXCR4-PE or isotype control-PE (IgG) conjugates (100 μl) were incubated with the cells for 30–45 min on ice, followed by two washes and resuspension in PBS and 1% BSA. Cells expressing surface CXCR4 were then detected using flow cytometry. Results are expressed as percent gated cells + se (n = 3). *, P < 0.05 indicates significant changes from vehicle-treated cells.
Androgens increase migration toward a CXCL12 gradient in a CXCR4-dependent manner
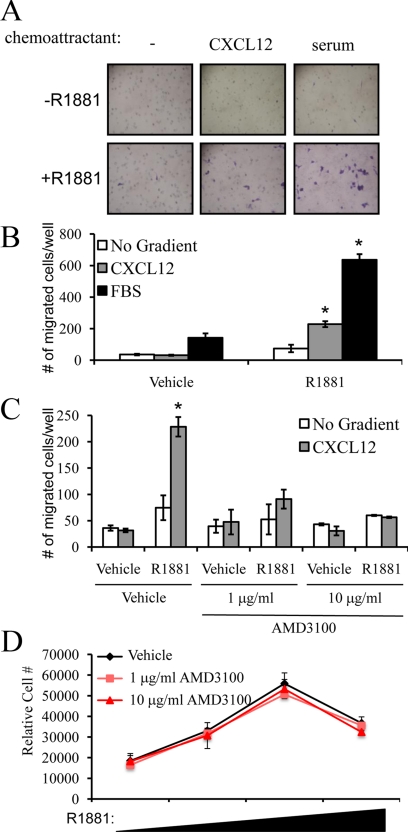
We next wanted to determine whether the androgen-mediated increase in CXCR4 resulted in enhanced migration toward the chemoattractant CXCL12. To test this, a Boyden dual-chamber migration assay was used. In the presence of a CXCL12 gradient, R1881 treatment led to significant LNCaP migration (Fig. 3, A and B). Not surprisingly, the absolute level of migration toward CXCL12 was less than toward full serum, the latter of which probably contains additional chemoattractants (Fig. 3, A and B). Whereas CXCL12 alone was able to promote androgen-mediated LNCaP migration in the Boyden chamber assay, it, unlike serum, did not induce significant LNCaP invasion (data not shown). Importantly, the selective CXCR4 antagonist, AMD3100 (25), blocked R1881-mediated LNCaP migration toward CXCL12, confirming the role of CXCR4 in this process (Fig. 3C). Although AMD3100 completely inhibited migration toward CXCL12, it did not completely block migration toward serum (data not shown), indicating that serum contains other important chemoattractants.
Figure 3.
Androgen increases the migration of LNCaP prostate cancer cells toward a CXCL12 gradient. A and B, Seeded cells were treated with or without 10 nm R1881 for 24 h followed by dissociation and reseeding of cells in the top chamber for a Boyden dual chamber migration assay. Fresh medium with or without 10 nm R1881 was added to the top and bottom chambers while either no chemoattractant, 5% FBS (serum), or 400 ng/ml CXCL12 was added to the bottom chamber. After 16 h, migrated cells were fixed, stained with crystal violet, and (A) imaged at ×100 magnification and (B) counted in three different microscopic fields and added together. The results are expressed as mean ± se (n = 3). *, P < 0.05 indicates significant changes from vehicle-treated cells. C, Migration assay was performed as described in panels A and B, except cells were pretreated for 1 h with vehicle, 1 or 10 μg/ml of the selective CXCR4 antagonist AMD3100 before treatment with or without 10 nm R1881. The following day, migration was assessed toward no gradient or 400 ng/ml CXCL12 as described in panel B. D, LNCaP cells were plated in 96-well plates and grown for 3 d. Cells were treated with vehicle (ethanol) or increasing concentrations of R1881 (0.01, 0.1, 1 nm) and the indicated doses of AMD3100 or vehicle (water) on d 3, d 5, and d 7. On d 10, cells were lysed and the relative number of cells was measured with the fluorescent DNA binding dye FluoReporter Blue. Each sample was performed in triplicate, and results from a representative experiment are shown. Results are expressed as relative cell number ± se (n = 3).
It has been shown by others that the CXCL12/CXCR4 axis is involved in cancer cell growth (15,26,27,28,29). However, in our hands, AMD3100, although having a significant effect on migration, had no effect on R1881-mediated LNCaP cell growth (Fig. 3, C and D). Taken together, these results confirm a specific role for the CXCR4-signaling axis in androgen-mediated LNCaP cell migration.
The CXCR4 gene is not a direct transcriptional target of AR
We next sought to define the mechanism by which AR regulates CXCR4 expression. To determine whether this was a direct AR effect, we cotreated LNCaP cells with or without the translational inhibitor cycloheximide in conjunction with vehicle or R1881. Using qPCR, we found that cycloheximide completely blocked R1881-induced CXCR4 mRNA expression while having no effect on serum and glucocorticoid-regulated kinase 1 (SGK1) (Fig. 4), an established primary target of AR (30). These results indicate that the ligand-dependent action of AR on the CXCR4 gene is indirect and requires intermediate protein synthesis.
Figure 4.
CXCR4 is not a direct target of AR. LNCaP cells were pretreated for 1 h with vehicle (dimethylsulfoxide) or 1 μg/ml cycloheximide followed by vehicle or 10 nm R1881 for 16 h. SGK1 or CXCR4 mRNA levels were quantitated using qPCR and normalized to 36B4. Results are expressed as fold induction over vehicle (no R1881)-treated cells ± se (n = 3). *, P < 0.05 indicates significant changes from vehicle-treated cells.
Krüppel-like factor 5 is both necessary and sufficient for androgen-mediated CXCR4 expression and migration toward a CXCL12 gradient
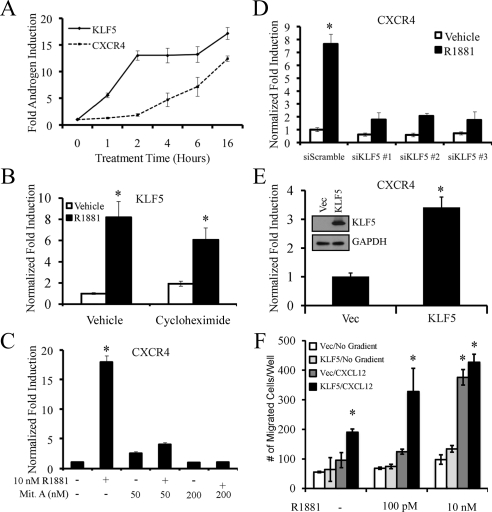
In search of a potential transcription factor that could mediate the androgen-induced CXCR4 expression, we reanalyzed the microarray data we had generated from prostate cancer cells treated for 6 h with an androgen (17). Specifically we used gene ontology term analysis to screen for transcription factors the expression of which preceded that of CXCR4. Among the factors identified, one in particular, Krüppel-like factor 5 (KLF5), was selected for further analysis because it has previously been implicated in cancer progression and cell migration (31,32,33,34,35). R1881 increased KLF5 mRNA levels rapidly within 1 h, which preceded the expression of CXCR4 (Fig. 5A). Furthermore, cotreatment of LNCaP cells with cycloheximide indicated that KLF5, unlike CXCR4 (Fig. 4), was a direct AR target (Fig. 5B). Using siRNAs targeting AR, we confirmed that, indeed, KLF5 was an AR target gene (supplemental Fig. S2). KLF5 is a member of the Sp superfamily of transcription factors that bind to GC-rich DNA elements (36). To determine whether GC-rich regions are important for androgen-dependent CXCR4 induction, cells were cotreated with androgens and mithramycin A, a compound that binds GC-rich regions and inhibits transcription factor binding at these sites. At concentrations previously shown to inhibit GC-rich-mediated gene expression (37), mithramycin A blocked R1881-mediated CXCR4 expression (Fig. 5C), but did not alter the R1881-mediated expression of the direct AR-target gene SGK1 (supplemental Fig. S3). To specifically implicate KLF5, we knocked down KLF5 expression using three independent siRNAs targeting KLF5 (supplemental Fig. S4A). Knockdown of KLF5 significantly inhibited R1881-mediated CXCR4 expression (Fig. 5D) but did not alter the expression of other well-characterized AR-regulated genes such as FKBP51 (supplemental Fig. S4B). To complement our siRNA studies, we overexpressed either a GAL4[DNA-binding domain (DBD)] negative control (Vec) or KLF5 by creating stable cell lines using retroviral techniques. Using this approach we demonstrated that overexpression of KLF5 significantly increased both CXCR4 expression (Fig. 5E) and migration toward a CXCL12 gradient (Fig. 5F). Interestingly, overexpression of KLF5 also appeared to sensitize LNCaP cells to lower levels of androgens, suggesting that patients with high KLF5 levels, and thus CXCR4, may be more prone to develop metastases from prostate cancers that have elevated AR activity, as is commonly seen in hormone-refractory diseases (38). Collectively, these data indicate that KLF5 is a direct androgen target that is important for AR-mediated CXCR4 expression and migration. Studies are ongoing to determine the exact mechanism by which KLF5 regulates CXCR4 expression.
Figure 5.
KLF5 is a direct AR target gene that is necessary for androgen-mediated CXCR4 expression and migration. A, LNCaP cells were treated for indicated time points with vehicle or 10 nm R1881. KLF5 or CXCR4 mRNA levels were quantitated using qPCR and normalized to 36B4. Results shown are fold R1881 induction ± se (n = 3). B, LNCaP cells were pretreated for 1 h with vehicle (dimethylsulfoxide) or 1 μg/ml cycloheximide followed by vehicle or 10 nm R1881 for 16 h. KLF5 mRNA levels were quantitated using qPCR and normalized to 36B4. Results are expressed as fold induction over vehicle (no R1881)-treated cells ± se (n = 3). *, P < 0.05 indicates significant changes from vehicle-treated cells. C, LNCaP cells were pretreated for 30 min with vehicle (dimethylsulfoxide) or indicated concentrations of mithramycin A (Mit. A) followed by vehicle or 10 nm R1881 for 16 h. CXCR4 mRNA levels were quantitated using qPCR and normalized to 36B4. Results are expressed as fold induction over vehicle-treated cells ± se (n = 3). *, P < 0.05 indicates significant changes from vehicle-treated cells. D, LNCaP cells were transfected for 48 h with siRNAs targeting a scramble sequence or KLF5 and then treated overnight with vehicle or 10 nm R1881. CXCR4 mRNA levels were quantitated using qPCR and normalized to 36B4. Results are expressed as fold induction over vehicle (siScramble)-treated cells ± se (n = 3). *, P < 0.05 indicates significant changes from vehicle (siScramble)-treated cells. E, LNCaP cells stably expressing either GAL4(DBD) (Vec) or KLF5 were harvested for RNA. CXCR4 mRNA levels were quantitated using qPCR and normalized to 36B4. Results are expressed as fold induction over negative control (Vec) cells ± se (n = 2). *, P < 0.05 indicates significant changes from control cells. Inset, Western blot control confirming KLF5 expression using GAPDH as a loading control. F, LNCaP-Vec and LNCaP-KLF5 stable cell lines treated with indicated concentrations of R1881 were subjected to a migration assay as described in Fig. 3 using either no gradient or 400 ng/ml CXCL12 as a chemoattractant. The results are expressed as mean ± se (n = 2). *, P < 0.05 indicates significant changes from vehicle-treated LNCaP-Vec cells.
Identification of a functional AR-binding site within intron 1 of KLF5 and demonstration of the role of KLF5-CXCR4 in a model of hormone-refractory prostate cancer
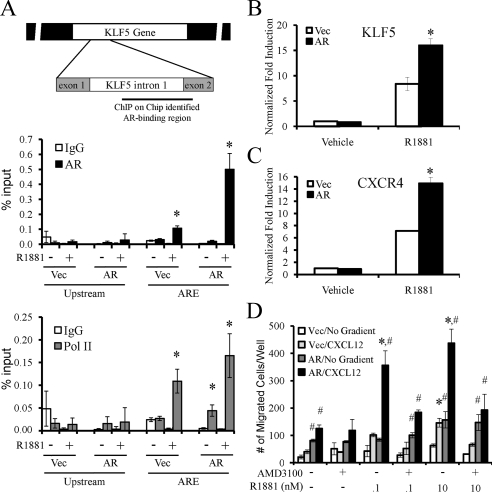
Given that KLF5 is a direct target of the AR, we next wanted to identify the androgen response element(s) [ARE(s)] within the KLF5 gene. Mining previous chromatin immunoprecipitation (ChIP) on Chip data (39) we found one potential AR-binding site within intron 1 of the KLF5 gene (Fig. 6A, top). No other AR binding was detected within the KLF5 gene or within 100 kb in either direction of the KLF5 gene. Consistent with CXCR4 not being a direct target of AR, we did not detect any potential AR-binding sites within the CXCR4 gene or 100 kb in either direction of the CXCR4 gene. Using chromatin immunoprecipitation assays, we demonstrate receptor binding to the identified region in the KLF5 intron 1 in a ligand-dependent manner (Fig. 6A, middle). Furthermore, we could also immunoprecipitate RNA polymerase II (Pol II) to this region in a ligand-dependent manner, demonstrating this is an active region of transcription (Fig. 6A, bottom). In support of these findings, LNCaP cells engineered to express higher levels of AR (supplemental Fig. S5) demonstrated increased AR binding to this site and subsequent increased Pol II recruitment (Fig. 6A). This corresponded with a simultaneous increase in both KLF5 (Fig. 6B) and CXCR4 mRNA levels (Fig. 6C) and CXCR4-mediated migration toward CXCL12 (Fig. 6D). The AR-overexpressing cells exhibited both a higher basal rate of migration and responded to lower concentrations of R1881 than their negative control counterparts (Fig. 6D). These results align with those seen in the KLF5-overexpressing cells (Fig. 5F) and therefore support our proposed AR-KLF5-CXCR4 mechanism.
Figure 6.
Identification of an AR-binding site within KLF5 intron 1 and evaluation of the impact of AR overexpression on KLF5-CXCR4-mediated migration. A, LNCaP-SRα (Vec) or LNCaP-SRαAR (AR) cells were treated with vehicle or 10 nm R1881 for 4 h. Cross-linked chromatin was immunoprecipitated with indicated antibodies. The precipitated DNA was amplified using primers spanning a region identified using ChIP on Chip data as a potential AR-binding site (indicated in top schematic) or a distal upstream region (negative control). The results are presented as percent input ± se (n =3). *, P < 0.05 indicates significant changes from IgG controls. B and C, LNCaP-SRα (Vec) or LNCaP-SRαAR (AR) cells were treated with vehicle or 10 nm R1881 for 16 h. The expression of KLF5 (B) or CXCR4 (C) was then assessed using qPCR and normalized to 36B4 levels. Results are expressed as fold induction over vehicle-treated cells ± se (n = 3). *, P < 0.05 indicates significant changes from LNCaP-SRα cells. D, LNCaP-SRα or LNCaP-SRαAR cells pretreated for 1 h with vehicle (water) or 10 μg/ml AMD3100 before treatment with indicated concentrations of R1881 were subjected to a migration assay as described in Fig. 3 using either no gradient or 400 ng/ml CXCL12 as a chemoattractant. The results are expressed as mean ± se (n = 2). *, P < 0.05 indicates significant changes from double vehicle-treated (water, ethanol) cells. #, P < 0.05 indicates significant changes from LNCaP-SRα cells receiving identical treatments.
To determine the functionality of the AR-binding site identified, we cloned overlapping regions of KLF5’s intron 1 and tested their ability to confer androgen responsiveness to an enhancerless luciferase reporter gene. Of three initial constructs spanning intron 1, only the 3′-most fragment, 2167 bp in length, was androgen responsive (Fig. 7A). This androgen response could be blocked by pretreatment with 10 μm Casodex (Fig. 7B). Deletion analysis further narrowed down the androgen-responsive region to a 328-bp stretch of DNA included in our 1367-bp KLF5 enhancer construct but not in our 1039-bp construct (Fig. 7C). Within the 328-bp fragment we found the sequence AGAACGttgTGTTTA that was most similar to the consensus androgen response element (ARE) sequence AGAACAnnnTGTTCT. Deletion of only these 15 bp completely eliminated androgen responsiveness, confirming this to be the exact ARE (Fig. 7D). Importantly, we confirmed this enhancer to be AR dependent in an AR-negative cell line (HepG2) transfected with an AR expression construct (Fig. 7E), indicating the AR cofactors needed for KLF5 expression exist in other cell/tissue types.
Figure 7.
Characterization of the functional ARE within KLF5 intron 1. A, Various KLF5 intron 1 luciferase reporter constructs (depicted in top model) were transfected into LNCaP cells and then treated overnight with 0, 0.1, 1, or 10 nm R1881. After treatment, cells were harvested and assayed for luciferase activity. Luciferase values were normalized to β-galactosidase control. Data are the mean relative light units (RLUs) ± sem for one representative experiment performed in triplicate (n = 3). *, P < 0.05 indicates significant changes from vehicle-treated cells. B, A KLF5 enhancer (2167 bp; fragment “C” from Fig. 7A) luciferase reporter construct was transfected into LNCaP cells and then pretreated for 30 min with vehicle or 10 μm Casodex followed by treatment overnight with vehicle or various concentrations of R1881 as indicated. After treatment, cells were harvested and assayed for luciferase activity. Luciferase values were normalized to β-galactosidase control. Data are the mean relative light units (RLUs) ± sem for one representative experiment performed in triplicate (n = 3). *, P < 0.05 indicates significant changes from vehicle-treated cells. C–E, KLF5 enhancer deletion constructs were transfected alone into LNCaP cells (C and D) or with an AR expression construct into HepG2 cells (E) and then treated overnight with vehicle or 10 nm R1881. After treatment, cells were harvested and assayed for luciferase activity. Luciferase values were normalized to β-galactosidase control. Data are the mean relative light units (RLUs) ± sem for one representative experiment performed in triplicate (n = 3). *, P < 0.05 indicates significant changes from vehicle-treated cells. Emp Vec, Empty vector.
Discussion
Prostate cancer preferentially metastasizes to bone. In fact, 90% of patients with advanced disease have osseous metastases (5). Previous studies aimed at investigating the link between AR and invasion/metastasis used androgen-independent cell lines that do not express endogenous AR, such as DU145 and PC3, in which AR cDNA was transfected and reexpressed. When AR is overexpressed in these cell lines, their invasive capacity is inhibited and the addition of androgens represses their growth and enhances apoptosis, contrary to what is commonly known for androgens in vivo (40,41). Therefore, reintroducing AR into these cells creates an artificial system that may not accurately reflect in vivo prostate cancer biology. It is likely that these androgen-independent and AR-negative prostate cancer cell lines have developed mutations that enable them to functionally compensate for the loss of AR. In support of this, an androgen-independent subline of the LNCaP cell line, LNCaP C4-2B, selected for its ability to form osteoblastic skeletal metastases, expresses higher basal levels of the AR-regulated prostate specific antigen, indicating that the AR-signaling axis is still active in these cells and has elevated levels of CXCR4 and CXCL12 (9,42). In this study, we demonstrated in prostate cancer cells expressing endogenous AR that the expression of CXCR4 is up-regulated by androgens and promotes migration toward a CXCL12 gradient. Furthermore, overexpression of AR (LNCaP-SRαAR cells), as is commonly found in prostate cancer, also increases the expression of CXCR4 and CXCR4-mediated migration. Conversely, because of CXCR4’s low expression in benign prostate, we think it is unlikely to play a physiological role in the normal prostate. Interestingly, although we were able to demonstrate androgen induction of KLF5 in AR-positive LNCaP prostate cancer cells and AR-negative HepG2 hepatoma cells transfected with an AR expression vector, we were unable to detect androgenic regulation of either KLF5 or CXCR4 in two other AR-positive prostate cancer cell lines, LAPC4 and VCaP (data not shown). This indicates the AR-CXCR4 signaling axis may be important for a subset of prostate cancers. However, data extracted from the Oncomine database demonstrate that CXCR4 expression levels are increased in advanced prostate cancers (16). Hence, it is possible that we have simply not yet found another suitable AR-positive prostate cancer cell model to study this process.
Recently, our laboratory has demonstrated that SGK3, a protein involved in the regulation of the turnover of CXCR4 (43), is up-regulated in androgen-treated cells (30). Specifically, SGK3-mediated phosphorylation of the ubiquitin ligase, AIP4, inhibits the ubiquitination of CXCR4, thus stabilizing the receptor (43,44). Therefore, in addition to increasing the transcript levels of CXCR4, androgens may also stabilize the CXCR4 protein. Additionally, the phosphatidylinositol 3-kinase (PI3K) pathway (which activates SGK3) is stimulated downstream of CXCR4 activation by CXCL12, suggesting the presence of a feed-forward mechanism, wherein androgens increase the expression of SGK3 and CXCR4, and upon binding CXCL12, SGK3 is also activated via PI3K stimulation. This, in turn, would lead to stabilization of CXCR4 protein and enhanced ability to respond to increasing concentrations of CXCL12, facilitating chemotaxis along a gradient of CXCL12. CXCR4 could then also be elevated when PI3K is constitutively activated after loss of phosphatase and tensin homologue activity (which suppresses PI3K) as is found in most prostate cancers (45).
The CXCR4-CXCL12 signaling axis is used by tumors to metastasize to sites, such as bone, in which CXCL12 is elevated. Not surprisingly, therefore, it has been demonstrated that the expression of CXCR4 is significantly elevated in more aggressive primary tumors and in those that have metastasized to bone (9,46). CXCL12 signaling through CXCR4 enhances the adhesion of prostate cancer cells to bone marrow endothelial cells, possibly by activating αVβ3 integrins, and stimulates prostate cancer migration toward a CXCL12 gradient (11,47,48). Furthermore, activation of CXCR4 by CXCL12 stimulates the production of multiple matrix metalloproteinases (MMPs), including MMP-1, MMP-13, MMP-2, and MMP-10, facilitating invasion (8,49). However, in our hands, and in the model systems we examined, CXCL12 alone did not increase the invasiveness of androgen-treated prostate cancer cells. Importantly, however, others have demonstrated that neutralizing anti-CXCR4 antibodies significantly inhibited PC3 prostate cancer metastasis and growth in osseous sites in an in vivo animal model (10).
Although androgens facilitated an enhanced migration of prostate cancer cells toward a serum gradient (Fig. 3), this activity was not blocked by the CXCR4 antagonist AMD3100 (data not shown). Thus, it is likely that serum contains additional chemokines that function independently of CXCR4. For example, IL-8 has previously been shown to enhance R1881-mediated LNCaP migration (50). If this is indeed the case, future treatment regimens may require a cocktail of drugs that can block multiple overlapping chemokine-signaling pathways.
In addition to uncovering a link between androgens and the migratory potential of prostate cancer cells, we also defined a novel mechanism by which androgens indirectly regulate their target genes. Specifically, we observed that androgens increased CXCR4 expression, in part, through the direct up-regulation of the transcription factor KLF5 (Figs. 4–7). Although KLF5 is necessary and sufficient for maximal CXCR4 expression and migration, it is still unclear how this is accomplished. We have not as yet been able to define the KLF5-responsive enhancer within the CXCR4 promoter. At this time, we cannot rule out the possibility that KLF5 may indirectly regulate CXCR4 through the expression of yet another factor. Additionally, there may be other factors independent of KLF5 that contribute to androgen-mediated CXCR4 regulation. For example, activator protein-1 has been implicated in the regulation of a number of androgen-regulated genes where no canonical ARE has been identified (51,52). KLF5 could increase the expression of a Jun or Fos family member, and thereby expression of activator protein-1-regulated genes. Further, the ability of AR to form large transcriptional complexes with other factors (39) opens the possibility that androgen regulation of CXCR4 may utilize both direct and indirect AR actions.
Although we have focused here on the ability of KLF5 to regulate a receptor involved in migration, we acknowledge the potentially pleiotropic effects of this transcription factor due to its ability to regulate transcription through relatively common GC-rich elements. Although KLF5 levels are not strongly correlated with overall prostate cancer progression, it is possible that KLF5 is important for a subset of cancers, particularly considering previous reports that suggest KLF5 expression impacts cell proliferation (31,32,33,53). KLF5 belongs to the large Sp family of transcription factors that all bind GC-rich sequences. Interestingly, other members of this family, such as the closely related KLF4, appear to oppose KLF5 actions, whereas yet other members may be functionally redundant to KLF5 (31,53,54). Hence, although KLF5 may not correlate with a particular cancer, it remains to be seen if another Sp family member may exhibit a similar activity. For example, more than 75% of prostate cancers were found to contain mutations in the KLF6 gene (55,56). KLF6 is thought to be a tumor suppressor, and thus inactivation of KLF6 may subsequently allow a balance in the Sp/KLF family to tilt toward a more tumorigenic role. Certainly, more studies are needed to better understand how changes in Sp/KLF activities impact overall biological processes.
Collectively, these data demonstrate the indirect androgen-mediated regulation of a gene involved in cancer cell migration. Although it has become apparent that the CXCL12/CXCR4 signaling axis plays a critical role in a number of cancer metastases, this is the first report that links it to androgen signaling. Given the recent findings that AR remains active and necessary for the growth and survival of advanced prostate cancers, there is renewed interest in the pharmacological exploitation of downstream AR-signaling pathways for the development of novel therapeutic approaches. These studies indicate the KLF5-CXCR4 pathway may be a suitable target for such future endeavors.
Materials and Methods
Chemicals
Methyltrienolone (R1881) was purchased from PerkinElmer (Waltham, MA) and bicalutamide (Casodex) was provided as a gift from P. Turnbull (GlaxoSmithKline, Research Triangle Park, NC). Cycloheximide, mithramycin A, and AMD3100 were obtained from Sigma (St. Louis, MO). CXCL12/stromal-cell derived factor-1α was purchased from R&D Systems (Minneapolis, MN).
Plasmids
The CMV-βgal plasmid was obtained from CLONTECH Laboratories, Inc. (Palo Alto, CA), the pBSII vector was purchased from Stratagene (La Jolla, CA), pGL4.26 was obtained from Promega Corp. (Madison, WI) and pcDNA3.1 was obtained from Invitrogen (Carlsbad, CA). The creation of pcDNA3.1-ARwt has previously been described (57). MSCV-GWb-GAL4(DBD)-IRES-EGFP and MSCV-GWb-KLF5-IRES-EGFP were created using the Invitrogen Gateway recombinase subcloning system according to the manufacturer’s instructions. To do this, GAL4(DBD) or KLF5 was shuttled from pENTR-GAL4(DBD) or pOTB7-KLF5 (Invitrogen) to MSCV-IRES-EGFP that was converted to a Gateway destination vector. pGL4.26-KLF5enh (2.1 kb) was created by PCR amplifying a 2.1-kb genomic sequence within intron 1 of the KLF5 gene that encompassed the potential AR binding site identified using ChIP on Chip [previously described (39)]. This fragment was then cloned into the pGL4.26 vector using NheI and HindIII restriction sites. Subsequent deletion constructs were created by PCR amplifying smaller fragments that were again cloned into pGL4.26 using NheI and HindIII restriction sites. Finally, the pGL4.26-KLF5enh(1.3kb)-ARE deletion construct was created from the pGL4.26-KLF5enh (1.3 kb) construct using the ExSite PCR-Based Site-Directed Mutagenesis Kit (Stratagene). All primers used for the construct creation are listed in supplemental Table 1. All sequences were confirmed using restriction digests and sequencing.
Antibodies
The CXCR4 antibody (clone 44717)-PE conjugate, its control, mouse IgG2B, and the CXCR4 antibody (clone 12G5) used for Western blot were obtained from R&D Systems. The rabbit KLF5 (sc-22797) and goat polyclonal glyceraldehyde-3-phosphate dehydrogenase (GAPDH) V-18 antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and the mouse monoclonal AR441 antibody against AR was received as a gift from D. Edwards (Baylor College of Medicine, Houston, TX). The secondary horseradish peroxidase antibody conjugates were purchased from Amersham Biosciences (Buckinghamshire, UK).
Cell culture
The human prostate carcinoma cell line LNCaP and the human hepatoma cell line HepG2 were obtained from American Type Culture Collection (Manassas, VA). The LNCaP cells were maintained in RPMI Medium 1640 (Invitrogen) supplemented with 8% fetal bovine serum (FBS) (Sigma), 0.1 mm nonessential amino acids (NEAA), and 1 mm sodium pyruvate (NaPyr) (Invitrogen). The HepG2 cells were maintained in MEM (Invitrogen) supplemented with 8% FBS, 0.1 mm NEAA, and 1 mm NaPyr.
Transient transfections and reporter gene assays
For transient transfections, cells were split into phenol red-free (PRF) medium, supplemented with charcoal-stripped fetal bovine serum (CS-FBS) (Hyclone Laboratories, Inc., Logan, UT), 0.1 mm NEAA, and 1 mm NaPyr in 24-well plates 24 h before transfection. Lipofectin (Invitrogen)-mediated transfection has been previously described in detail (58). Cells were treated with hormones approximately 16 h before the assay. Luciferase and β-galactosidase activities were measured as previously described (58). Each treatment was performed in triplicate, and results are expressed as mean ± se. Each experiment was repeated at least three times, with a representative experiment shown.
Creation of LNCaP stable cell lines
To create LNCaP-SRα and LNCaP-SRαAR cells, cells were infected with empty SRα or SRα retrovirus expressing wild-type AR (gift of C.L. Sawyers, Memorial Sloan-Kettering Cancer Center, New York, NY). Infected cells were selected by growth in RPMI 1640 supplemented with 8% FBS, 0.1 mm NEAA, 1 mm NaPyr, and 400 μg/ml G418 (Invitrogen). AR expression levels were confirmed by qPCR and Western analysis (supplemental Fig. S5). To create LNCaP-Vec and LNCaP-KLF5 cells, parental cells were infected with retrovirus expressing either MSCV-GWb-GAL4(DBD)-IRES-EGFP (negative control) or MSCV-GWb-KLF5-IRES-EGFP. Enhanced green fluorescent protein (EGFP)-positive cells were then selected through two rounds of cell sorting using flow cytometry, and KLF5 expression levels were confirmed by Western blot (Fig. 5E, inset).
siRNA transfection of cultured human prostate cell lines
Stealth siRNA (Invitrogen) transfections were previously described (30). A Stealth siRNA targeting a scrambled sequence (siScramble) was used as a negative control. All siRNAs were used at a final concentration of 100 nm.
RNA Isolation, cDNA preparation, and quantitative RT-PCR
RNA isolation, cDNA preparation and qPCR were performed as previously described (59). Each cDNA reflecting each biological sample was analyzed in triplicate and expressed as an average ± se. Each experiment was performed at least three times, with a representative experiment shown. All qPCR primers used in this study are listed in supplemental Table 1.
Western blot analysis
Western blots were performed as previously described (59). Of note, although a number of groups have suggested various technical modifications are necessary for isolating cell extracts suitable for CXCR4 (clone 12G5) Western blots, we found that altering the protein sample preparation at a variety of points (e.g. lysis buffer, lysis technique, boiling, cell debris spin-downs, etc.) continually generated similar results.
Flow cytometry
LNCaP cells were plated in six-well tissue culture-treated plates (Corning, Inc., Corning, NY) at a density of approximately 3 × 105 cells/ml in 2 ml of PRF-RPMI 1640 supplemented with 8% CS-FBS, 0.1 mm NEAA, and 1 mm NaPyr in a 37 C incubator with 5% CO2. After 2 d, media was replaced with PRF-RPMI 1640 with 0.1% fatty acid free BSA (Invitrogen) and 10 mm HEPES (Invitrogen) with ethanol (vehicle control) or R1881. After 24 h or the time points indicated in Fig. 2B, cells were dissociated with Accutase dissociation buffer (Millipore Corp., Billerica, MA). Cells were resuspended in cold 1× PBS with 1% BSA to a concentration of 1000 cells/μl. Cells (100 μl) were stained with 10 μl anti-CXCR4-PE or the isotype control IgG2B-PE conjugate at 4 C in the dark for 30–45 min. Cells were washed two times with 4 ml cold 1× PBS with 1% BSA and resuspended in 1× PBS with 1% BSA for flow cytometry analysis with a FACSCaliber flow cytometry machine. Cells were gated for live cells based on light scatter, and the percentage of stained cells was quantitated.
Migration assays
LNCaP, LNCaP-SRα, LNCaP-SRαAR, LNCaP-Vec, or LNCaP-KLF5 cells were plated in six-well tissue culture treated plates at a density of approximately 5 × 105 cells/ml in 2 ml of PRF-RPMI 1640 supplemented with 8% CS-FBS, 0.1 mm NEAA, and 1 mm NaPyr in a 37 C incubator with 5% CO2. After 2 d, media were replaced with PRF-RPMI 1640 with 0.1% fatty acid free BSA and 10 mm HEPES with ethanol (vehicle control) or R1881 (10 nm) in conjunction with sterile water (vehicle control) or the selective CXCR4 antagonist AMD3100. After 24 h, cells were dissociated with Accutase. Cells were counted with a hemacytometer, and 50,000 cells were seeded in PRF-RPMI + 0.1% fatty acid free BSA + 10 mm HEPES with treatments in the top chamber of a Biocoat control insert (BD Biosciences, Palo Alto, CA) with an 8 μm pore size for 24-well Boyden dual chamber migration assays. Before cells were added, the top and bottom of the membrane in the control insert were coated with 5 μg/ml of fibronectin in PBS at 37 C for 2 h, followed by washing with 1× PBS and subsequent drying of the coated membrane at 25 C. The bottom chamber contained PRF-RPMI + 0.1% fatty acid free BSA + 10 mm HEPES ± R1881 ± AMD3100 with 0–400 ng/ml of CXCL12 or 5% FBS. The migration assay was allowed to proceed at 37 C, 5% CO2 for 18–24 h, at which point the cells were fixed with 1.85% formaldehyde and 0.1% glutaraldehyde in PBS for 16 h at 4 C. The inserts were washed three times with 1× PBS, followed by staining with 0.5% crystal violet (Sigma) in 20% methanol for 1 h at 25 C and then washed three more times with 1×PBS. The inserts were visualized under a light microscope, pictures were taken, and the cells were quantitated by counting the number of cells in three randomly selected microscopic fields at ×100 magnification. The number of cells in all three fields were added together, and an average of three inserts ± se was determined. Light microscope photographs were taken of cells using a Coolpix 990 digital camera (Nikon, Melville, NY) on an Olympus CK2 inverted microscope (Olympus Corp., Lake Success, NY).
Cell proliferation assay
Proliferation assays were performed as previously described (59) by measuring the cellular DNA content using the FluoReporter Blue Fluorometric double-stranded DNA Quantitation Kit (Invitrogen) as per the manufacturer’s protocol.
Chromatin immunoprecipitation
LNCaP, LNCaP-SRα, or LNCaP-SRαAR cells were plated in 15-cm dishes in the presence of 20 ml of RPMI 1640 supplemented with 8% FBS, 0.1 mm NEAA, and 1 mm NaPyr until 90% confluent. Medium was changed to PRF-RPMI 1640 supplemented with 8% CS-FBS, 0.1 mm NEAA, and 1 mm NaPyr for 24 h, and cells were treated with specified ligands for 4 h. After ligand treatment, 1% formaldehyde was added to the media for 10 min and quenched with 250 mm glycine for 5 min. Cells were washed twice with PBS, pelleted, lysed in RIPA buffer [50 mm Tris, pH 7.5; 0.15 m NaCl; 1% Nonidet P-40; 0.5% Na-deoxycholate; 0.05% sodium dodecyl sulfate (SDS); 1 mm EDTA] and sonicated. Each sonicated cross-linked chromatin sample was diluted and precleared with 100 μl of 50% protein A/G agarose slurry (SC-2003, Santa Cruz) containing 20 μg sonicated salmon sperm DNA and 50 μg of BSA. Precleared chromatin was then immunoprecipitated with the following antibodies from Santa Cruz: 5 μg AR (SC-816), 10 μg Pol II (SC-899), or 5 μg control IgG for 2 h at 4 C, after which protein A/G agarose beads were added and incubated overnight at 4 C. Protein A/G beads were washed two times sequentially in low-salt buffer (50 mm HEPES, pH 7.8; 140 mm NaCl; 1 mm EDTA; 1% Triton X-100; 0.1% Na-deoxycholate; 0.1% SDS), high-salt buffer (same as low-salt with 500 mm NaCl), LiCl buffer (20 mm Tris, pH 8.0; 1 mm EDTA; 250 mm LiCl; 0.5% Nonidet P-40; 0.5% Na-deoxycholate), and TE buffer (50 mm Tris, pH 8.0; 1 mm EDTA). Protein-DNA complexes were eluted twice in elution buffer (50 mm Tris, pH 8.0; 1 mm EDTA; 1% SDS) at 65 C for 15 min. Eluted protein-DNA complexes were reverse cross-linked in the presence of NaCl overnight at 65 C and further treated with EDTA and proteinase K at 42 C for 1 h. The DNA fragments were purified and eluted in 10 mm Tris (pH 8.5) using the QIAquick PCR purification kit (QIAGEN, Valencia, CA), diluted and analyzed by qPCR.
Statistical analysis
Data were analyzed using one-way ANOVA and post hoc Dunnett’s test with GraphPad Prism, Version 4 (GraphPad Software, Inc., San Diego, CA). Statistically significant changes were determined at the P < 0.05 level.
Supplementary Material
Acknowledgments
We thank the members of the McDonnell laboratory, especially Dr. Ching-yi Chang, for critical reading of this manuscript and insightful discussions; the Duke Comprehensive Cancer Center Flow Cytometry Shared Resource for help with cell sorting; and Dr. Dean Edwards (Baylor College of Medicine, Houston, TX) for AR antibodies, Dr. Charles Sawyers (Memorial Sloan-Kettering Cancer Center, New York, NY) for AR-expressing retroviruses, and Dr. Robin Bachelder (Duke University Medical Center) for advice on CXCR4 Western blots.
Footnotes
This work was supported by National Institutes of Health Grants F32 DK072794 (to D.E.F.), F32 CA119642 (to B.M.W.), and R01 CA139818 (to D.P.M.), Department of Defense Grant DAMD17-03-1-0569 (to A.B.S.), and the FÁS Science Challenge (to A.P.T).
Disclosure Summary: The authors have nothing to disclose.
First Published Online May 21, 2009
Abbreviations: AR, Androgen receptor; ARE, androgen response element; ChIP, chromatin immunoprecipitation; CS-FBS, charcoal-stripped FBS; CXCR4, chemokine (C-X-C motif) receptor 4; DBD, DNA-binding domain; FBS, fetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; KLF5, Krüppel-like factor 5; MMP, matrix metalloproteinase; NEAA, nonessential amino acids; PE, phycoerythrin; PI3K, phosphatidylinositol 3-kinase; Pol II, RNA polymerase II; PRF, phenol red-free; qPCR, quantitative RT-PCR; SDS, sodium dodecyl sulfate; SGK1, serum and glucocorticoid-regulated kinase 1; siRNAs, small interfering RNAs.
References
- American Cancer Society 2007 Cancer facts and figures [Google Scholar]
- Isaacs JT, Isaacs WB 2004 Androgen receptor outwits prostate cancer drugs. Nat Med 10:26–27 [DOI] [PubMed] [Google Scholar]
- Lawton CA, Winter K, Murray K, Machtay M, Mesic JB, Hanks GE, Coughlin CT, Pilepich MV 2001 Updated results of the phase III radiation therapy oncology group (RTOG) trial 85-31 evaluating the potential benefit of androgen suppression following standard radiation therapy for unfavorable prognosis carcinoma of the prostate. Int J Radiat Oncol Biol Phys 49:937–946 [DOI] [PubMed] [Google Scholar]
- Bolla M, Collette L, Blank L, Warde P, Dubois JB, Mirimanoff RO, Storme G, Bernier J, Kuten A, Sternberg C, Mattelaer J, Lopez Torecilla J, Pfeffer JR, Lino Cutajar C, Zurlo A, Pierart M 2002 Long-term results with immediate androgen suppression and external irradiation in patients with locally advanced prostate cancer (an EORTC study): a phase III randomised trial. Lancet 360:103–106 [DOI] [PubMed] [Google Scholar]
- Bubendorf L, Schöpfer A, Wagner U, Sauter G, Moch H, Willi N, Gasser TC, Mihatsch MJ 2000 Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum Pathol 31:578–583 [DOI] [PubMed] [Google Scholar]
- Burger JA, Kipps TJ 2006 CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood 107:1761–1767 [DOI] [PubMed] [Google Scholar]
- Du YF, Shi Y, Xing YF, Zeng FQ 2008 Establishment of CXCR4-small interfering RNA retrovirus vector driven by human prostate-specific antigen promoter and its biological effects on prostate cancer in vitro and in vivo. J Cancer Res Clin Oncol [Erratum (2008) 134:1265] 134:1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Singh UP, Grizzle WE, Lillard Jr JW 2004 CXCL12-CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab Invest 84:1666–1676 [DOI] [PubMed] [Google Scholar]
- Sun YX, Wang J, Shelburne CE, Lopatin DE, Chinnaiyan AM, Rubin MA, Pienta KJ, Taichman RS 2003 Expression of CXCR4 and CXCL12 (SDF-1) in human prostate cancers (PCa) in vivo. J Cell Biochem 89:462–473 [DOI] [PubMed] [Google Scholar]
- Sun YX, Schneider A, Jung Y, Wang J, Dai J, Wang J, Cook K, Wang J, Osman NI, Koh-Paige AJ, Shim H, Pienta KJ, Keller ET, McCauley LK, Taichman RS 2005 Skeletal localization and neutralization of the SDF-1(CXCL12)/CXCR4 axis blocks prostate cancer metastasis and growth in osseous sites in vivo. J Bone Miner Res 20:318–329 [DOI] [PubMed] [Google Scholar]
- Taichman RS, Cooper C, Keller ET, Pienta KJ, Taichman NS, McCauley LK 2002 Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res 62:1832–1837 [PubMed] [Google Scholar]
- Vaday GG, Hua SB, Peehl DM, Pauling MH, Lin YH, Zhu L, Lawrence DM, Foda HD, Zucker S 2004 CXCR4 and CXCL12 (SDF-1) in prostate cancer: inhibitory effects of human single chain Fv antibodies. Clin Cancer Res 10:5630–5639 [DOI] [PubMed] [Google Scholar]
- Wang J, Loberg R, Taichman RS 2006 The pivotal role of CXCL12 (SDF-1)/CXCR4 axis in bone metastasis. Cancer Metastasis Rev 25:573–587 [DOI] [PubMed] [Google Scholar]
- Busillo JM, Benovic JL 2007 Regulation of CXCR4 signaling. Biochim Biophys Acta 1768:952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darash-Yahana M, Pikarsky E, Abramovitch R, Zeira E, Pal B, Karplus R, Beider K, Avniel S, Kasem S, Galun E, Peled A 2004 Role of high expression levels of CXCR4 in tumor growth, vascularization, and metastasis. FASEB J 18:1240–1242 [DOI] [PubMed] [Google Scholar]
- Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, Michalopoulos G, Becich M, Luo JH 2004 Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol 22:2790–2799 [DOI] [PubMed] [Google Scholar]
- Kazmin D, Prytkova T, Cook CE, Wolfinger R, Chu TM, Beratan D, Norris JD, Chang CY, McDonnell DP 2006 Linking ligand-induced alterations in androgen receptor structure to differential gene expression: a first step in the rational design of selective androgen receptor modulators. Mol Endocrinol 20:1201–1217 [DOI] [PubMed] [Google Scholar]
- Liao X, Thrasher JB, Pelling J, Holzbeierlein J, Sang QX, Li B 2003 Androgen stimulates matrix metalloproteinase-2 expression in human prostate cancer. Endocrinology 144:1656–1663 [DOI] [PubMed] [Google Scholar]
- Pandini G, Mineo R, Frasca F, Roberts Jr CT, Marcelli M, Vigneri R, Belfiore A 2005 Androgens up-regulate the insulin-like growth factor-I receptor in prostate cancer cells. Cancer Res 65:1849–1857 [DOI] [PubMed] [Google Scholar]
- BouHamdan M, Strayer DS, Wei D, Mukhtar M, Duan LX, Hoxie J, Pomerantz RJ 2001 Inhibition of HIV-1 infection by down-regulation of the CXCR4 co-receptor using an intracellular single chain variable fragment against CXCR4. Gene Ther 8:408–418 [DOI] [PubMed] [Google Scholar]
- Choi B, Gatti PJ, Fermin CD, Vigh S, Haislip AM, Garry RF 2008 Down-regulation of cell surface CXCR4 by HIV-1. Virol J 5:6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster R, Kremmer E, Schubel A, Breitfeld D, Kleinschmidt A, Nerl C, Bernhardt G, Lipp M 1998 Intracellular and surface expression of the HIV-1 coreceptor CXCR4/fusin on various leukocyte subsets: rapid internalization and recycling upon activation. J Immunol 160:1522–1531 [PubMed] [Google Scholar]
- Signoret N, Oldridge J, Pelchen-Matthews A, Klasse PJ, Tran T, Brass LF, Rosenkilde MM, Schwartz TW, Holmes W, Dallas W, Luther MA, Wells TN, Hoxie JA, Marsh M 1997 Phorbol esters and SDF-1 induce rapid endocytosis and down modulation of the chemokine receptor CXCR4. J Cell Biol 139:651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasova NI, Stauber RH, Michejda CJ 1998 Spontaneous and ligand-induced trafficking of CXC-chemokine receptor 4. J Biol Chem 273:15883–15886 [DOI] [PubMed] [Google Scholar]
- Hatse S, Princen K, Bridger G, De Clercq E, Schols D 2002 Chemokine receptor inhibition by AMD3100 is strictly confined to CXCR4. FEBS Lett 527:255–262 [DOI] [PubMed] [Google Scholar]
- Balkwill F 2004 The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol 14:171–179 [DOI] [PubMed] [Google Scholar]
- Begley L, Monteleon C, Shah RB, Macdonald JW, Macoska JA 2005 CXCL12 overexpression and secretion by aging fibroblasts enhance human prostate epithelial proliferation in vitro. Aging Cell 4:291–298 [DOI] [PubMed] [Google Scholar]
- Mowafi F, Cagigi A, Matskova L, Björk O, Chiodi F, Nilsson A 2008 Chemokine CXCL12 enhances proliferation in pre-B-ALL via STAT5 activation. Pediatr Blood Cancer 50:812–817 [DOI] [PubMed] [Google Scholar]
- Pattarozzi A, Gatti M, Barbieri F, Würth R, Porcile C, Lunardi G, Ratto A, Favoni R, Bajetto A, Ferrari A, Florio T 2008 17β-Estradiol promotes breast cancer cell proliferation-inducing stromal cell-derived factor-1-mediated epidermal growth factor receptor transactivation: reversal by gefitinib pretreatment. Mol Pharmacol 73:191–202 [DOI] [PubMed] [Google Scholar]
- Sherk AB, Frigo DE, Schnackenberg CG, Bray JD, Laping NJ, Hammond M, Patterson J, Thompson SK, Kazmin D, Norris JD, McDonnell DP 2008 Development of a small molecule serum and glucocorticoid-regulated kinase 1 antagonist and its evaluation as a prostate cancer therapeutic. Cancer Res 68:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW 2005 Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res 15:92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Ghaleb AM, Nandan MO, Yang VW 2007 The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays 29:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan MO, McConnell BB, Ghaleb AM, Bialkowska AB, Sheng H, Shao J, Babbin BA, Robine S, Yang VW 2008 Kruppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology 134:120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui S, Sugimoto N, Takuwa N, Sakagami S, Takata S, Kaneko S, Takuwa Y 2004 Blood lipid mediator sphingosine 1-phosphate potently stimulates platelet-derived growth factor-A and -B chain expression through S1P1-Gi-Ras-MAPK-dependent induction of Kruppel-like factor 5. J Biol Chem 279:12300–12311 [DOI] [PubMed] [Google Scholar]
- Yang Y, Tetreault MP, Yermolina YA, Goldstein BG, Katz JP 2008 Kruppel-like factor 5 controls keratinocyte migration via the integrin-linked kinase. J Biol Chem 283:18812–18820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk G, Urrutia R 2005 The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem J 392:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britschgi A, Trinh E, Rizzi M, Jenal M, Ress A, Tobler A, Fey MF, Helin K, Tschan MP 2008 DAPK2 is a novel E2F1/KLF6 target gene involved in their proapoptotic function. Oncogene 27:5706–5716 [DOI] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL 2004 Molecular determinants of resistance to antiandrogen therapy. Nat Med 10:33–39 [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M 2007 A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 27:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi L, Carloni V, Muratori M, Salvadori A, Giannini A, Carini M, Serio M, Forti G, Baldi E 2000 Androgen receptor expression in prostate carcinoma cells suppresses α6β4 integrin-mediated invasive phenotype. Endocrinology 141:3172–3182 [DOI] [PubMed] [Google Scholar]
- Heisler LE, Evangelou A, Lew AM, Trachtenberg J, Elsholtz HP, Brown TJ 1997 Androgen-dependent cell cycle arrest and apoptotic death in PC-3 prostatic cell cultures expressing a full-length human androgen receptor. Mol Cell Endocrinol 126:59–73 [DOI] [PubMed] [Google Scholar]
- Thalmann GN, Sikes RA, Wu TT, Degeorges A, Chang SM, Ozen M, Pathak S, Chung LW 2000 LNCaP progression model of human prostate cancer: androgen-independence and osseous metastasis. Prostate 44:91–103 [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Marchese A, Brech A, Stenmark H 2006 CISK attenuates degradation of the chemokine receptor CXCR4 via the ubiquitin ligase AIP4. EMBO J 25:3738–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, Benovic JL 2003 The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev Cell 5:709–722 [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R 1997 PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275:1943–1947 [DOI] [PubMed] [Google Scholar]
- Mochizuki H, Matsubara A, Teishima J, Mutaguchi K, Yasumoto H, Dahiya R, Usui T, Kamiya K 2004 Interaction of ligand-receptor system between stromal-cell-derived factor-1 and CXC chemokine receptor 4 in human prostate cancer: a possible predictor of metastasis. Biochem Biophys Res Commun 320:656–663 [DOI] [PubMed] [Google Scholar]
- Engl T, Relja B, Marian D, Blumenberg C, Müller I, Beecken WD, Jones J, Ringel EM, Bereiter-Hahn J, Jonas D, Blaheta RA 2006 CXCR4 chemokine receptor mediates prostate tumor cell adhesion through α5 and β3 integrins. Neoplasia 8:290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YX, Fang M, Wang J, Cooper CR, Pienta KJ, Taichman RS 2007 Expression and activation of α v β 3 integrins by SDF-1/CXC12 increases the aggressiveness of prostate cancer cells. Prostate 67:61–73 [DOI] [PubMed] [Google Scholar]
- Chinni SR, Sivalogan S, Dong Z, Filho JC, Deng X, Bonfil RD, Cher ML 2006 CXCL12/CXCR4 signaling activates Akt-1 and MMP-9 expression in prostate cancer cells: the role of bone microenvironment-associated CXCL12. Prostate 66:32–48 [DOI] [PubMed] [Google Scholar]
- Lee LF, Louie MC, Desai SJ, Yang J, Chen HW, Evans CP, Kung HJ 2004 Interleukin-8 confers androgen-independent growth and migration of LNCaP: differential effects of tyrosine kinases Src and FAK. Oncogene 23:2197–2205 [DOI] [PubMed] [Google Scholar]
- Church DR, Lee E, Thompson TA, Basu HS, Ripple MO, Ariazi EA, Wilding G 2005 Induction of AP-1 activity by androgen activation of the androgen receptor in LNCaP human prostate carcinoma cells. Prostate 63:155–168 [DOI] [PubMed] [Google Scholar]
- Takai H, Nakayama Y, Kim DS, Arai M, Araki S, Mezawa M, Nakajima Y, Kato N, Masunaga H, Ogata Y 2007 Androgen receptor stimulates bone sialoprotein (BSP) gene transcription via cAMP response element and activator protein 1/glucocorticoid response elements. J Cell Biochem 102:240–251 [DOI] [PubMed] [Google Scholar]
- Black AR, Black JD, Azizkhan-Clifford J 2001 Sp1 and Kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol 188:143–160 [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R 2003 Sp1- and Kruppel-like transcription factors. Genome Biol 4:206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Hyytinen ER, Sun X, Helin HJ, Koivisto PA, Frierson Jr HF, Vessella RL, Dong JT 2003 Deletion, mutation, and loss of expression of KLF6 in human prostate cancer. Am J Pathol 162:1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, Ferrari AC, Martignetti JA, Friedman SL 2001 KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294:2563–2566 [DOI] [PubMed] [Google Scholar]
- Chang CY, McDonnell DP 2002 Evaluation of ligand-dependent changes in AR structure using peptide probes. Mol Endocrinol 16:647–660 [DOI] [PubMed] [Google Scholar]
- Fan JD, Wagner BL, McDonnell DP 1996 Identification of the sequences within the human complement 3 promoter required for estrogen responsiveness provides insight into the mechanism of tamoxifen mixed agonist activity. Mol Endocrinol 10:1605–1616 [DOI] [PubMed] [Google Scholar]
- Frigo DE, McDonnell DP 2008 Differential effects of prostate cancer therapeutics on neuroendocrine transdifferentiation. Mol Cancer Ther 7:659–669 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.