Abstract
Apurinic/apyrimidinic endonuclease 1 or redox factor-1 (Ape1/Ref-1) is a pleiotropic cellular protein involved in DNA repair and, through its redox activity, enhances the binding of a select group of transcription factors to their cognate recognition sequences in DNA. Thus, we were intrigued when we identified Ape1/Ref-1 and a number of DNA repair and oxidative stress proteins in a complex associated with the DNA-bound estrogen receptor α (ERα). Because Ape1/Ref-1 interacts with a number of transcription factors and influences their activity, we determined whether it might also influence ERα activity. We found that endogenously expressed Ape1/Ref-1 and ERα from MCF-7 human breast cancer cells interact and that Ape1/Ref-1 enhances the interaction of ERα with estrogen-response elements (EREs) in DNA. More importantly, Ape1/Ref-1 alters expression of the endogenous, estrogen-responsive progesterone receptor and pS2 genes in MCF-7 cells and associates with ERE-containing regions of these genes in native chromatin. Interestingly, knocking down Ape1/Ref-1 expression or inhibiting its redox activity with the small molecule inhibitor E3330 enhances estrogen responsiveness of the progesterone receptor and pS2 genes but does not alter the expression of the constitutively active 36B4 gene. Additionally, the reduced form of Ape1/Ref-1 increases and E3330 limits ERα-ERE complex formation in vitro and in native chromatin. Our studies demonstrate that Ape1/Ref-1 mediates its gene-specific effects, in part, by associating with endogenous, estrogen-responsive genes and that the redox activity of Ape1/Ref-1 is instrumental in altering estrogen-responsive gene expression.
Redox factor 1/apurinic endonuclease-1 (Ape1/Ref-1) interacts with ERα, associates with endogenous gene regions involved in conferring estrogen responsiveness, and influences estrogen-responsive gene expression.
During the course of normal cellular metabolism, oxygen is consumed and reactive oxygen species (ROS) are produced. If not effectively dissipated, ROS can accumulate and cause damage to resident proteins, lipids, and DNA. To avoid ROS-induced damage to cellular macromolecules, cells rely on a diverse array of proteins to decrease oxidative stress and, if damage does occur, to repair the ROS-induced damage.
Zinc finger proteins, which include numerous transcription factors, are particularly vulnerable to ROS accumulation. Oxidation of their zinc fingers alters the secondary structure of these proteins and inhibits their ability to bind to DNA (1). Enzymes involved in redox regulation, such as apurinic/apyrimidinic endonuclease 1/redox factor-1 (Ape1/Ref-1), which reduces specific oxidized transcription factors such as Fos, Jun, nuclear factor-κB (NFκB), and p53 (1,2,3,4,5), are instrumental in ensuring that the transcriptional activity mediated by these proteins is maintained.
Estrogen receptor α (ERα) is a ligand-inducible transcription factor that plays a critical role in development and maintenance of the mammary gland and the reproductive tract but also influences the function of the skeletal, cardiovascular, and nervous systems (6,7,8,9,10,11). Interaction of ERα with its hormonal ligand, 17β-estradiol (E2), facilitates the interaction of the receptor with estrogen-response elements (EREs) in target genes and alters recruitment of coregulatory proteins to the DNA-bound receptor, thereby initiating changes in gene expression. In this manner, ERα modulates the expression of an array of estrogen-responsive genes (12,13) including the pS2 gene (14,15,16) and the progesterone receptor (PR) gene, which encodes two functionally distinct proteins, the 120-kDa PR-B and the 94-kDa PR-A (17,18). The capacity of ERα to initiate changes in gene expression relies on the structural integrity of its centrally located DNA binding domain, which is comprised of two zinc fingers and is required for sequence-specific DNA binding. Oxidation of ERα precludes the ability of the receptor to interact with DNA and alters estrogen-responsive gene expression (19,20).
To better understand how ERα regulates transcription of estrogen-responsive genes, we used agarose-based gel mobility shift assays to isolate large complexes of proteins associated with the ERE-bound ERα (21,22). Mass spectrometry analysis identified Ape1/Ref-1 as a component of these large protein-ERα-DNA complexes. In the current study, we demonstrate that Ape1/Ref-1 interacts with ERα, promotes the ERα-ERE interaction, influences ERα-mediated transactivation, and selectively associates with endogenous, estrogen-responsive genes in MCF-7 cells. Our findings suggest that Ape1/Ref-1 is instrumental in modulating expression of estrogen-responsive genes in this human breast cancer cell line.
Results
To better understand how estrogen-responsive genes are regulated, we developed an agarose-based gel mobility shift assay, which, unlike acrylamide-based gel mobility shift assays, can be used to isolate large, multiprotein complexes associated with the ERE-bound ERα (21,22). Mass spectrometry analysis revealed that a number of proteins associate with the DNA-bound ERα, and subsequent analysis showed that these proteins influence the activity of ERα (20,21,22,23,24,25,26,27,28,29). Interestingly, the multifunctional protein Ape1/Ref-1 was a component of these large protein-ERα-ERE complexes.
Endogenously expressed ERα and Ape1/Ref-1 interact
We first examined the level of Ape1/Ref-1 expressed in a number of cultured cell lines that have been used to study estrogen responsiveness (24,30,31,32). MCF-7 breast cancer and U2 osteosarcoma (U2OS) cells expressed significantly higher levels of Ape1/Ref-1 than either HeLa cervical cancer or MDA-MB-231 breast cancer cells, but only MCF-7 cells expressed ERα (Fig. 1A). The level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was used as a loading control, was similar in each of the cell lines tested.
Figure 1.
Expression and Interaction of ERα and Ape1/Ref-1. A, Nuclear extracts (20 μg) from MCF-7, U2OS, HeLa, or MDA-MB-231 cells were separated on a denaturing acrylamide gel and transferred to nitrocellulose. Proteins were detected with Ape1/Ref-1-, ERα-, and GAPDH-specific antibodies. GAPDH was used as a loading control. B, MCF-7 cells were treated with ethanol or 10 nm E2, cell lysates were prepared, and proteins were immunoprecipitated with an antibody directed against Ape1/Ref-1. The immunoprecipitated proteins were subjected to Western blot analysis with ERα- and Ape1/Ref-1-specific antibodies. Lanes 1 and 2 contain 1% input. IP, Immunoprecipitation.
To determine whether ERα interacted with Ape1/Ref-1 in a cell line in which these proteins are endogenously expressed, MCF-7 breast cancer cells were treated with ethanol or 10 nm E2 and immunoprecipitation assays were carried out. When an Ape1/Ref-1-specific antibody was used, both Ape1/Ref-1 and ERα were immunoprecipitated in the absence and in the presence of hormone (Fig. 1B, lanes 5 and 6). In contrast, when a control antibody was used, neither Ape1/Ref-1 nor ERα was immunoprecipitated (lanes 3 and 4).
Ape1/Ref-1 alters expression of endogenous estrogen-responsive genes
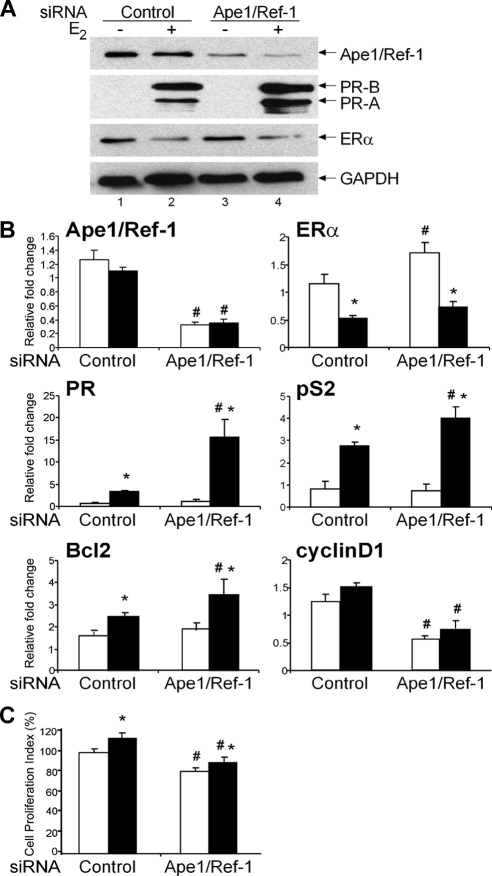
Because Ape1/Ref-1 was associated with the receptor-DNA complex (21,22) and interacted with ERα from MCF-7 cells (Fig. 1B), we investigated whether this interaction had a functional consequence and could alter ERα-mediated transcription. MCF-7 cells, which endogenously express both Ape1/Ref-1 and ERα (Fig. 1A), were transfected with a control or Ape1/Ref-1-specific small interfering RNA (siRNA). Western blot analysis confirmed that Ape1/Ref-1 expression was dramatically reduced when the Ape1/Ref-1-specific small interfering RNA (siRNA) was used (Fig. 2A, lanes 3 and 4) but that the control siRNA did not affect Ape1/Ref-1 expression (lanes 1 and 2 and data not shown). PR expression increased upon E2 treatment in the presence of control siRNA (lanes 1 and 2) as has been observed in untransfected E2-treated MCF-7 cells (33). However, when endogenous Ape1/Ref-1 was decreased, PR-B and PR-A protein expression increased 2- and 4-fold, respectively, in the presence of E2 compared with control siRNA (compare lanes 2 and 4). In contrast, neither ERα nor GAPDH, which was used as an internal control, was affected by control or Ape1/Ref-1-specific siRNA.
Figure 2.
Effect of knocking down Ape1/Ref-1 on endogenous estrogen-responsive gene expression. MCF-7 cells were transfected with control or Ape1/Ref-1-specific siRNA and then treated with ethanol (−E2 and white bars) or 10 nm E2 (+E2 and black bars) for 24 h. A, Protein samples were subjected to Western blot analysis with an antibody that recognizes Ape1/Ref-1, PR-A and PR-B, ERα, or GAPDH. B, RNA was isolated, cDNA was synthesized, and real-time PCR was carried out with gene-specific primers. The relative fold change was determined using the comparative Ct method with the housekeeping gene, 36B4, serving as the internal control. Data from four independent experiments, which had been carried out in duplicate, were combined and are presented as the mean ± sem. C, MTT assays were performed, and absorbance measurements were collected. Cell proliferation index (%) was determined by comparing the mean absorbance of Ape1/Ref-1 siRNA samples with control siRNA. Data from three independent experiments, which were carried out in duplicate, were combined and are presented as the mean ± sem. ANOVA with a post hoc Student’s t test was used to detect significant differences in mRNA levels and cell proliferation in response to E2 (*, P < 0.05) or Ape1/Ref-1-specific siRNA (#, P < 0.05).
To determine whether decreased Ape1/Ref-1 expression would influence endogenous, estrogen-responsive gene expression at the mRNA level, total RNA was isolated from MCF-7 cells that had been transfected with control or Ape1/Ref-1-specific siRNA, and cDNA was synthesized. Real-time quantitative PCR analysis revealed that Ape1/Ref-1 mRNA expression was significantly decreased with the Ape1/Ref-1-specific siRNA in the absence and in the presence of E2 (Fig. 2B). As expected, PR mRNA levels were increased when cells were treated with E2 and control siRNA was used. When Ape1/Ref-1 expression was knocked down, the level of PR mRNA was dramatically enhanced with E2 treatment compared with control siRNA. Thus, the increased PR mRNA level we observed when Ape1/Ref-1 expression was knocked down corresponded well with the increased level of PR protein (Fig. 2A). Similarly, pS2 and Bcl2 mRNA levels were increased in the presence of E2 when Ape1/Ref-1 was knocked down. However, knocking down Ape1/Ref-1 expression decreased cyclin D1 mRNA levels in the absence and presence of E2. Therefore, Ape1/Ref-1 acts to limit gene expression of the endogenous, estrogen-responsive PR, pS2, and Bcl2 genes and enhances cyclin D1 gene expression in MCF-7 cells. Although ERα mRNA levels increased in the absence of E2 when Ape1/Ref-1 was knocked down, the level of ERα protein was unaltered. These combined studies demonstrate that the effects of Ape1/Ref-1 on estrogen responsiveness are gene specific.
Previous studies in ovarian, osteosarcoma, and embryonic stem cells have shown that the loss of Ape1/Ref-1 expression results in an extended G1/S transition and decreased proliferation (34,35,36,37). To determine whether knocking down Ape1/Ref-1 in MCF-7 cells influenced proliferation, 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT) assays were performed after cells had been transfected with control or Ape1/Ref-1-specific siRNA and treated with ethanol or 10 nm E2. In fact, knocking down Ape1/Ref-1 expression resulted in an approximately 20% decrease in proliferation compared with control siRNA (Fig. 2C).
Ape1/Ref-1 associates with the endogenous estrogen-responsive PR and pS2 genes
The ability of Ape1/Ref-1 to interact with ERα and influence ERα-mediated transcription suggested that Ape1/Ref-1 might associate with regulatory regions of estrogen-responsive genes to influence transcription. To determine whether this was the case, MCF-7 cells were treated with ethanol or E2 for 20 min, 2 h, 6 h, or 24 h, and chromatin immunoprecipitation (ChIP) assays were performed. Hormone treatment elicited a dramatic increase in the association of ERα with the ERE-containing region of the pS2 gene and with two regions of the PR gene located 205 (PR205) and 221 (PR221) kb upstream of the PR-B transcription start site (Fig. 3A). PR205 and PR221 contain two and one imperfect EREs, respectively, that bind ERα and function as estrogen-responsive enhancers (Boney-Montoya, J. L., Y. S. Ziegler, C. D. Curtis, J. A. Montoya, and A. M. Nardulli, manuscript submitted). Similarly, Ape1/Ref-1 association with the pS2 gene, PR205, and PR221 was increased in the presence of E2. The capacity of Ape1/Ref-1 to associate with the PR and pS2 genes is consistent with its ability to alter the expression of these genes (Fig. 2) and suggests that Ape1/Ref-1 influences gene expression in part by associating with gene regions involved in conferring estrogen responsiveness.
Figure 3.
Ape1/Ref-1 association with endogenous estrogen-responsive genes. MCF-7 cells were treated with ethanol or 10 nm E2 for 20 min, 2 h, 6 h, or 24 h. Chromatin was prepared and immunoprecipitated with an antibody directed against ERα (A) or Ape1/Ref-1 (B). DNA was isolated and amplified by real-time quantitative PCR to determine whether ERα and Ape1/Ref-1 were associated with the ERE-containing region of the pS2 gene or regions of the PR gene located 205 kb (PR205) or 221 kb (PR221) upstream of the PR-B transcription start site, which contain two and one imperfect EREs, respectively. Four independent experiments, which were each carried out in duplicate, were combined and are presented as the mean number of copies of each estrogen-responsive region pulled down relative to the number of copies of the 36B4 gene region pulled down (Occupancy) ± sem. ANOVA with a post hoc Student’s t test was used to detect statistical increases in the association of ERα and Ape1/Ref-1 with these gene regions in the presence of E2 (*, P < 0.05).
Ape1/Ref-1 promotes the ERα-ERE interaction
Thus far, we had shown that Ape1/Ref-1 interacted with ERα, influenced endogenous, estrogen-responsive gene expression, and associated with gene regions involved in conferring estrogen responsiveness. To determine whether Ape1/Ref-1 might also influence receptor-DNA complex formation, gel mobility shift assays were performed on nondenaturing acrylamide gels. In the absence of Ape1/Ref-1, ERα bound to the ERE-containing oligos (Fig. 4A, lane 2). As increasing amounts of Ape1/Ref-1 were added, the ability of ERα to bind to ERE-containing oligos increased in a dose-dependent manner (Fig. 4A, lanes 3–7, and Fig. 4B). This increased receptor-DNA complex formation was not due to increased protein concentrations, because the total protein concentration in each reaction was held constant by addition of BSA, but was due to the presence of Ape1/Ref-1. Addition of an ERα-specific antibody (lane 8) resulted in a supershift of the protein-DNA complex, indicating that the receptor was present. However, an Ape1/Ref-1-specific antibody (lane 9) was unable to supershift the protein-DNA complex. Thus, although Ape1/Ref-1 was not present in the protein-DNA complex, it did significantly increase ERα-ERE complex formation. These studies, which were performed with purified ERα and Ape1/Ref-1 and fractionated on an acrylamide gel, do not necessarily contradict our previous studies (21,22), which were performed with HeLa nuclear extracts and purified ERα and fractionated on an agarose gel, but do suggest that the association of Ape1/Ref-1 with the ERE-bound ERα may require the participation of other nuclear proteins. These findings are reminiscent of a previous report, which demonstrated that Ape1/Ref-1 stimulated binding of Fos and Jun to AP-1-containing DNA but was not associated with the protein-DNA complex and did not alter the DNA region protected in deoxyribonuclease I footprinting or methylation interference assays (39).
Figure 4.
Effect of Ape1/Ref-1 on the ERα-ERE interaction. A, 32P-labeled oligos containing a consensus ERE were run alone (lane 1) or combined with 30 fmol baculovirus-expressed, purified ERα (lanes 2–9) and increasing (lanes 3–7) or constant (lanes 7–10) amounts of bacterially expressed, purified Ape1/Ref-1. Total protein concentrations in each lane were held constant by the addition of BSA. ERα- (lane 8) and Ape1/Ref-1- (lane 9) specific antibodies were added as indicated. Lane 10 contains purified Ape1/Ref-1 and radiolabeled ERE oligos in the absence of ERα. B, The percent of probe bound was determined from four independent experiments and is shown graphically as the mean ± sem. Statistical differences were determined using a one-way ANOVA with a post hoc Student’s t test (*, P < 0.05). Ab, Antibody.
Inhibition of Ape1/Ref-1 redox activity alters estrogen responsiveness
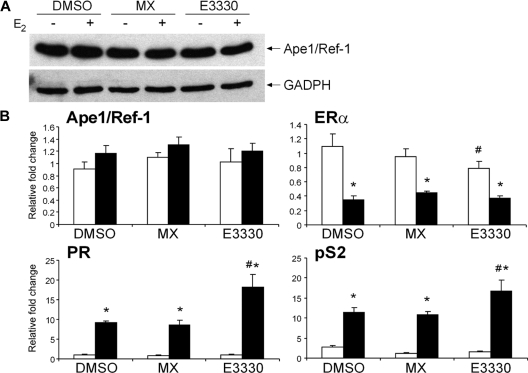
Ape1/Ref-1 has been described as a redox factor that stimulates the DNA binding activity of several transcription factors including Fos, Jun, NFκB, and p53 (2,3,4,5). Given that decreased Ape1/Ref-1 expression altered expression of estrogen-responsive genes and that Ape1/Ref-1 was able to increase ERα-ERE complex formation, we were interested in determining whether inhibiting the DNA repair or redox activity of Ape1/Ref-1 might affect endogenous, estrogen-responsive gene expression. Accordingly, MCF-7 cells were exposed to two different inhibitors of Ape1/Ref-1 activity, methoxyamine (MX) and E3330. MX binds to and occludes abasic sites in DNA and thereby inhibits Ape1/Ref-1-mediated DNA repair (36,40,41). The small molecule inhibitor, E3330, which was originally identified as a suppressor of NFκB activation, binds specifically to Ape1/Ref-1 (42,43) and blocks its redox activity, thereby inhibiting Ape1/Ref-1-induced binding of transcription factors to DNA (34,36,44,45).
Total protein and RNA were isolated from MCF-7 cells that had been exposed to MX or E3330. Western blot (Fig. 5A) and real-time quantitative PCR analysis (Fig. 5B) confirmed that neither of the inhibitors altered Ape1/Ref-1 levels. As expected, PR and pS2 mRNA levels increased when MCF-7 cells were treated with E2. When cells were exposed to MX and E2, similar changes in PR and pS2 mRNA levels were observed. However, when cells were exposed to the redox inhibitor E3330 and then treated with E2, the levels of PR and pS2 mRNA were significantly enhanced. ERα mRNA levels decreased in the absence, but not in the presence, of E2 when the redox inhibitor was present. Thus, the increased estrogen responsiveness of the PR and pS2 genes we observed when cells were exposed to E3330 was consistent with the increased PR and pS2 mRNA levels detected when Ape1/Ref-1 expression was decreased (Fig. 2). These studies suggest that the redox activity of Ape1/Ref-1, but not its DNA repair activity, is important in modulating expression of these two endogenous estrogen-responsive genes.
Figure 5.
Effect of Ape1/Ref-1 inhibitors on estrogen-responsive gene expression. MCF-7 cells were incubated with DMSO, MX, or E3330 for 1 h and then treated with ethanol (−E2 or white bars) or 10 nm E2 (+E2 or black bars) for 24 h. A, Cell lysates were prepared and separated on a denaturing acrylamide gel. The blot was subjected to Western analysis with an antibody that recognizes Ape1/Ref-1 or GAPDH. B, RNA was isolated, cDNA was synthesized, and real-time PCR was carried out with gene-specific primers. The relative fold change was determined using the comparative Ct method with the housekeeping gene, 36B4, serving as the internal control. Data from four independent experiments, which had been done in duplicate, were combined and are presented as the mean ± sem. ANOVA with a post hoc Student’s t test was used to detect significant differences in mRNA levels in response to E2 (*, P < 0.05) or the inhibitor E3330 (#, P < 0.05).
Reduced Ape1/Ref-1 facilitates ERα-ERE complex formation
Because we had shown that the redox activity of Ape1/Ref-1 was involved in altering the expression of endogenous estrogen-responsive genes, we determined whether the redox activity of Ape1/Ref-1 was involved in the Ape1/Ref-1-mediated increase in ERα-ERE complex formation.
Generally, approximately 4 mm dithiothreitol (DTT) is used in gel mobility shift assays to reduce ERα and promote the ERα-ERE interaction. As seen in Fig. 6, when DTT was omitted from the binding reaction (lane 2) or when 0.02 mm DTT concentrations were used (lane 3), ERα bound weakly to the ERE-containing oligos. When Ape1/Ref-1 was reduced with DTT, but then diluted so that the final DTT concentration was 0.02 mm, ERα-ERE complex formation was increased 4.3-fold (Fig. 6A, compare lanes 3 and 4, and Fig. 6B). Inclusion of Ape1/Ref-1, which had not been reduced with DTT before addition to the binding reaction, failed to promote the ERα-ERE interaction to the same extent as prereduced Ape1/Ref-1 (Fig. 6A, compare lanes 8 and 4, and Fig. 6B). Increasing concentrations of the Ape1/Ref-1-specific redox inhibitor E3330 elicited a dose-dependent decrease in Ape1/Ref-1-mediated ERα-ERE complex formation (Fig. 6A, lanes 5–7, and Fig. 6B). Similarly, mutation of a critical amino acid involved in conferring Ape1/Ref-1 redox activity (C65S; Refs. 46,47,48) resulted in a 20% reduction in ERα-ERE complex formation (Fig. 6C, compare lanes 2 and 3). Thus, Ape1/Ref-1 and its redox activity are instrumental in enhancing ERα-ERE complex formation.
Figure 6.
Effect of reduced Ape1/Ref-1 on ERα-ERE complex formation. A, 32P-labeled ERE-containing oligos were run alone (lane 1) or combined with 30 fmol baculovirus-expressed, purified ERα (lanes 2–8) and bacterially expressed, purified Ape1/Ref-1 (lanes 4–8). DTT (0.02 mm; lanes 3–7) or increasing concentrations of the Ape1/Ref-1-specific redox inhibitor E3330 (lanes 5–7) were included as indicated. Total protein concentrations were held constant by addition of BSA. B, The percent of probe bound was determined from four independent experiments and is shown graphically as the mean ± sem. C, 32P-labeled ERE-containing oligos were combined with 30 fmol ERα (lanes 1–3), 0.02 mm DTT (lanes 2–3) and wild-type Ape1/Ref-1 (lane 2) or redox-deficient Ape1/Ref-1 C65S (lane 3). Total protein concentrations were held constant by addition of BSA. Wt, Wild type.
Inhibition of Ape1/Ref-1 redox activity alters association of ERα with the endogenous estrogen-responsive PR and pS2 genes
Given that only reduced Ape1/Ref-1 with an intact redox domain was able to promote the ERα-ERE interaction in gel mobility shift assays, we determined whether inhibiting the redox activity of endogenously expressed Ape1/Ref-1 might also affect the association of ERα with regulatory regions of the PR and pS2 genes in native chromatin. Thus, MCF-7 cells were exposed to the Ape1/Ref-1 redox inhibitor E3330, treated with ethanol or 10 nm E2, and chromatin immunoprecipitation assays were performed. As expected, hormone treatment increased the association of ERα (Fig. 7A) and Ape1/Ref-1 (Fig. 7B) with the ERE-containing region of the pS2 gene and with two regions of the PR gene (PR205 and PR221). Addition of E3330 significantly diminished the E2-induced association of ERα and Ape1/Ref-1 with these native gene regions.
Figure 7.
Effect of Ape1/Ref-1 redox inhibitor on ERα association with endogenous estrogen-responsive genes. MCF-7 cells were exposed to DMSO or the Ape1/Ref-1 redox inhibitor E3330 for 1 h and then treated with ethanol or 10 nm E2 for 20 min. Chromatin was prepared and immunoprecipitated with an antibody directed against ERα (A) or Ape1/Ref-1 (B). DNA was isolated and amplified by real-time quantitative PCR to determine whether ERα and Ape1/Ref-1 were associated with the ERE-containing region of the pS2 gene or two upstream regulatory regions of the PR gene (PR205 or PR221). Three independent experiments, which were each carried out in duplicate, were combined and are presented as the mean number of copies of each estrogen-responsive region pulled down relative to the number of copies of the 36B4 gene region pulled down (occupancy) ± sem. ANOVA with a post hoc Student’s t test was used to detect statistical differences in the association of ERα and Ape1/Ref-1 with these gene regions in the presence of E2 (*, P < 0.05) or the inhibitor E3330 (#, P < 0.05).
Expression of Ape1/Ref-1 in human mammary tissue
Although examining the expression of Ape1/Ref-1 in MCF-7 breast cancer cells was quite informative, we were interested in determining whether Ape1/Ref-1 was expressed in normal mammary cells. Thus, paraffin-embedded, human mammary tissue was subjected to immunohistochemistry. As seen in Fig. 8, A and B, at ×10 magnification, Ape1/Ref-1 and ERα were predominantly present in the epithelial cells. When the primary antibody was omitted, no staining was observed (insets in Fig. 8, A and B). When cells were observed at a higher magnification (×40), it was apparent that both Ape1/Ref-1 and ERα were localized in the epithelial cell nuclei (Fig. 8, C and D).
Figure 8.
Expression of Ape1/Ref-1 and ERα in normal human breast tissue. Normal breast tissue was subjected to immunohistochemistry using Ape1/Ref-1- (A and C) or ERα-specific (B and D) antibody. A representative slide is shown at ×10 (A and B) and ×40 (C and D) magnification. Control slides, which were not exposed to Ape1/Ref-1 or ERα antibody, are shown in the insets in A and B. Stromal (S) and epithelial (E) cells are indicated. Scale bars, 100 μm.
Discussion
Ape1/Ref-1 has been identified as a protein involved in DNA repair and redox regulation (reviewed in Ref. 4). We have now shown that Ape1/Ref-1 interacts with ERα, associates with endogenous gene regions involved in conferring estrogen responsiveness, and influences estrogen-responsive gene expression and that the redox activity of Ape1/Ref-1, but not its DNA repair activity, is involved in mediating these effects.
Effect of Ape1/Ref-1 on transcription factor binding to DNA
ERα has two centrally located zinc fingers in its DNA-binding domain. Although both zinc fingers are susceptible to oxidative stress, the second zinc finger is particularly vulnerable (1,49). Oxidation of ERα decreases the capacity of the receptor to dimerize, diminishes DNA binding, and results in a loss of transcriptional activity (1,49). Our studies show that, just as Ape1/Ref-1 enhances the binding of Fos, Jun, NFκB, HIF1α, and p53 to their cognate binding sequences (1,3,4,39,50,51), it also enhances the binding of ERα to ERE-containing DNA, suggesting that Ape1/Ref-1 acts to maintain the reduced and active form of ERα, which in turn promotes the association of ERα with gene regions involved in conferring estrogen responsiveness.
Effect of Ape1/Ref-1 on transactivation
It is generally assumed that an increase in ERα-ERE complex formation leads to an increase in ERα-mediated gene expression. Although Ape1/Ref-1 was effective in increasing ERα-ERE complex formation and cyclin D1 gene expression, its role at the estrogen-responsive PR, pS2, and Bcl2 genes was to limit transcription. Furthermore, when Ape1/Ref-1 expression was knocked down with siRNA or when its redox activity was inhibited by the small molecule E3330, the expression of the native PR and pS2 genes in MCF-7 cells was increased (Figs. 2 and 5). Thus, the redox activity of Ape1/Ref-1 plays a critical role in estrogen-responsive gene expression. It is, however, clear that Ape1/Ref-1 is only one of many proteins involved in modulating ERα activity and that the transcriptional readout of a gene is not dependent on the activity of a single protein, but rather on the combined effects of multiple ERα-associated proteins.
The effect of knocking down Ape1/Ref-1 or inhibiting its redox activity on PR gene expression is particularly striking and may result from the combined effects of Ape1/Ref-1 on ERα as well as Fos and Jun, which are also intimately involved in regulating PR gene expression (52,53,54). Ape1/Ref-1 enhances the binding of the Fos-Jun heterodimer to its recognition sequence (39,50). Although our studies demonstrate that Ape1/Ref-1 and its redox activity play critical roles in modulating estrogen responsiveness, they did not affect expression of the constitutively active 36B4 gene. Considering the highly conserved nature of the nuclear hormone receptors, it seems possible that Ape1/Ref-1 could be involved in modulating the expression of a broad spectrum of genes regulated by other nuclear receptors.
Association of oxidative stress proteins with estrogen-responsive genes
Ape1/Ref-1 was one of a number of oxidative stress proteins we found associated with the DNA-bound ERα. Protein disulfide isomerase (PDI), Cu/Zn superoxide dismutase (SOD1), thioredoxin (Trx), and thioredoxin reductase (TrxR) were also present (20,28,55). We have shown that each of these oxidative stress proteins associates with the DNA-bound ERα and influences estrogen-responsive gene expression (20,28,55), but that each protein has unique properties as well. Through its dismutation of superoxide, SOD1 helps to regulate reactive oxygen species and limits damage to proteins, lipids, and DNA (56). PDI interacts with and acts to maintain the functional capacity of SOD1 by preventing its aggregation (57), and TrxR by facilitating the correctly folded conformation (38,58). In turn, TrxR reduces and maintains the activity of Trx (1,59,60,61), and Trx then reduces/activates a number of proteins including Ape1/Ref-1 and ERα (2,55,76,77,78,79). PDI functions as a molecular chaperone for a number of transcription factors (62,63,64,65,66), is instrumental in maintaining the structural integrity of ERα (20), and interacts with SOD1 and TrxR (57,58). Thus, multiple oxidative stress proteins including Ape1/Ref-1 collaborate to maintain ERα structure/function as well as other factors involved in regulating transcription (Fig. 9). Interestingly, the effects of these proteins on estrogen-responsive gene expression are sometimes complementary and sometimes work in opposition (20,28,55). We are intrigued by the fact that a suite of oxidative stress proteins with complementary and opposing effects influence estrogen responsiveness.
Figure 9.

An interconnected network of oxidative stress proteins interacts with the DNA bound-ERα (A and B). Ape1/Ref-1, Trx, TrxR, PDI, and SOD1 form an interconnected network of oxidative stress proteins associated with the DNA-bound ERα.
Association of DNA repair proteins with estrogen-responsive genes
A number of laboratories have shown that, in addition to its role in the oxidative stress response, Ape1/Ref-1 plays an essential role in base excision repair (4,67,68). Given that Ape1/Ref-1 is associated with ERα and was recruited to estrogen-responsive genes, it seems plausible that Ape1/Ref-1 may also play a role in fostering repair of these genes. We have, in fact, identified other proteins involved in base excision repair including 3-methyladenine DNA glycosylase, proliferating cell nuclear antigen, and flap endonuclease, which associate with ERα and influence estrogen-responsive gene expression (25,26,69). It has been suggested that DNA repair proteins may help to stabilize the transcription complex so that DNA repair and transcription can occur simultaneously. The capacity of Ape1/Ref-1 to foster ERα-ERE complex formation may contribute to this stabilization. Coupling these two cellular processes would appear to be beneficial because chromatin remodeling is essential for both transcription and DNA repair (70). The fact that multiple DNA repair proteins were found together in a complex associated with the ERE-bound ERα suggests that these proteins may act cooperatively to ensure that the integrity of transcriptionally active genes is preserved and may help to explain the preferential repair of the transcribed DNA strand (71,72). It should be noted that although Ape1/Ref-1 may be involved in helping to maintain genomic integrity, its DNA repair function is not involved in influencing expression of the estrogen-responsive PR and pS2 genes. Other DNA repair proteins including O6-methylguanine-DNA methyltransferase, poly(ADP-ribose) polymerase, and DNA-dependent protein kinase also play dual roles in regulating ERα-mediated transcription and DNA repair (73,74).
In summary, our studies show that Ape1/Ref-1 is a component of a network of proteins that act collectively to influence ERα activity. Together they ensure that the receptor is transcriptionally competent so that estrogen-responsive genes are appropriately regulated.
Materials and Methods
Identification of Ape1/Ref-1
Novel ERα-associated proteins were isolated in agarose gel mobility shift experiments in the presence of E2 and identified by mass spectrometry analysis as described elsewhere (20,21,22) using baculovirus-expressed, purified ERα and HeLa nuclear extracts. Three peptides, LPAELQELPGLSHQYWSAPSDK, QGFGELLQAVPLADSFR, and LDYFLLSHSLLPALCDSK, with an amino acid sequence identical to that found in Ape1/Ref-1, were identified.
Western blot analysis
Nuclear extracts from MCF-7 breast cancer, U2OS, HeLa cervical cancer, and MDA-MB-231 breast cancer cells were prepared as described previously (32). Nuclear extracts (20 μg) were fractionated on a 10% sodium dodecyl sulfate (SDS) polyacrylamide gel and transferred to a nitrocellulose membrane. Proteins were detected by Western blot analysis using an antibody that recognizes Ape1/Ref-1 (sc-17774; Santa Cruz Biotechnology, Inc., Santa Cruz CA), ERα (sc-543, Santa Cruz Biotechnology), or GAPDH (TAB1001; Open Biosystems, Huntsville AL). Blots were probed with horseradish peroxidase-conjugated secondary antibody, and the Supersignal West Femto Maximum Sensitivity Substrate chemiluminescent detection kit (Pierce Chemical Co., Rockford, IL) was used to visualize the proteins according to the manufacturer’s instructions.
Subcloning, expression, and purification of His-tagged Ape1/Ref-1 protein
The EcoRI fragment from pcDNA3.1-APEX (kindly provided by Takashi Kohno, National Cancer Center, Tokyo, Japan) was subcloned into the dual-tagged (His and T7) pET-28a (+) vector (Novagen, La Jolla CA) for protein expression. The QuikChangeII Site-Directed Mutagenesis kit (Stratagene, La Jolla CA) was used to convert the 5′-AAGATCTGCTCTTGG-3′ sequence to 5′-AAGATCTCCTCTTGG-3′ within the pET-28a(+)-APEX plasmid to construct a plasmid encoding the mutated Ape1/Ref-1 C65S. The wild-type and mutant plasmids were purified and used to transform BL21(DE3)pLysS Escherichia coli. Transformed bacteria were induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside for 4 h at 37 C, chilled on ice, and pelleted at 3000 × g for 5 min. Pellets were resuspended in Ni-NTA lysis buffer (50 mm NaH2PO4, 300 mm NaCl, and 10 mm imidazole) with 1× protease inhibitors (Sigma, St. Louis MO), homogenized on ice, and centrifuged at 142,000 × g for 30 min at 4 C. The supernatant was incubated with Ni-NTA agarose beads (QIAGEN, Valencia CA) for 1 h at 4 C. After three washes with Ni-NTA wash buffer (50 mm NaH2PO4, 500 mm NaCl, 20 mm imidazole, 0.5% Triton X-100, and 1× protease inhibitors), the His-tagged proteins were eluted with Ni-NTA elution buffer (50 mm NaH2PO4, 300 mm NaCl, and 250 mm imidazole). The purity of wild-type and mutant His-Ape1/Ref-1 were monitored by Coomassie staining, and the Bio-Rad protein assay was used to determine protein concentrations (Bio-Rad Laboratories, Inc., Hercules CA) according to the manufacturer’s instructions.
Coimmunoprecipitation assay
MCF-7 cells were maintained on phenol red containing MEM with 5% calf serum (CS). Three days before harvest, cells were transferred to phenol red-free MEM containing 5% charcoal dextran-treated CS (CDCS). Medium was replaced 6 h before treatment with phenol red-free MEM containing 0.4% charcoal dextran-treated fetal bovine serum. Cells (∼107) were treated with ethanol or 10 nm E2 for 2 h, washed twice with chilled PBS, and harvested in 1 ml immunoprecipitation lysis buffer [20 mm Tris (pH 7.4), 10 mm EDTA, 400 mm NaCl, 0.5% Nonidet P-40 (NP-40), 1 mm Na3VO4, 50 mm NaF, and 1× protease inhibitors]. The cell lysate was distributed equally and an Ape1/Ref-1 (sc-5572 or sc-334 from Santa Cruz Biotechnology)-specific or His control antibody (sc-802, Santa Cruz Biotechnology) was added and incubated overnight at 4 C with rotation. Protein A Sepharose 4FastFlow (GE Healthcare, Piscataway, NJ) was added, and the samples were incubated for 1 h at 4 C with rotation. The resin was washed three times with wash buffer (20 mm Tris, pH 7.4; 10 mm EDTA; 150 mm NaCl; 0.1% NP-40; 1 mm Na3VO4; 50 mm NaF; and 1× protease inhibitors), and bound proteins were eluted with 2× SDS sample buffer (125 mm Tris-HCl, pH 6.8; 150 mm SDS; 20% glycerol; 1% β-mercaptoethanol; and 0.01% bromophenol blue). Proteins were fractionated on a 10% SDS polyacrylamide gel and transferred to a nitrocellulose membrane for Western blot analysis with an antibody directed against Ape1/Ref-1 or ERα (sc-17774 or sc-8002, respectively; Santa Cruz Biotechnology).
Cell culture and transfections
U2OS cells were transfected using Lipofectin (Invitrogen, Carlsbad CA) as described elsewhere (28) with 1 ng ptk-Renilla expression vector (Promega Corp., Madison WI), 1 μg 2EREtkLUC, and 5 ng CMV5-hERα (both kindly provided by Benita Katzenellenbogen, University of Illinois, Urbana, IL). Increasing concentrations of the pcDNA3.1-APEX expression vector (kindly provided by Takashi Kohno) were added as indicated. The parental expression vector pcDNA3.1 (Invitrogen) was included to maintain the total DNA concentration at 1 μg in each well. After a 6-h incubation at 37 C, cells were treated with ethanol or 10 nm E2 for 24 h. Luciferase assays were carried out using the Dual Luciferase assay system (Promega).
Gel mobility shift assay
Baculovirus-expressed purified ERα (30 fmol), which had been isolated as described elsewhere (16), was incubated with 32P-labeled, 50 bp ERE-containing oligos (20,000 cpm) and increasing amounts of His-purified Ape1/Ref-1 in binding reaction buffer (15 mm Tris, pH 7.9; 60 mm KCl; 0.2 mm EDTA; 4 mm DTT; 50 nm E2; 10% glycerol; and 50 ng deoxyinosinic-deoxycytidylic acid) for 10 min at 25 C. Protein concentrations were held constant at 2 μg by the addition of BSA. ERα- or Ape1/Ref-1-specific antibody (sc-8002 or sc-17774, respectively) was added and incubated at 25 C for an additional 10 min. Samples were fractioned on a 6% nondenaturing polyacrylamide gel in low ionic strength buffer at 4 C with buffer recirculation. The levels of bound and free 32P-labeled DNA were quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and ImageQuant 5.0 software (GE Healthcare).
Redox activity assay
Bacterially expressed, His-purified Ape1/Ref-1 (1.25 μg/μl) was incubated with or without 1 mm DTT for 10 min at 37 C. After a 5-fold dilution in sterile water, 2 μl (500 ng) Ape1/Ref-1 was incubated with 30 fmol baculovirus-expressed purified ERα and increasing concentrations of the Ape1/Ref-1 redox inhibitor (2E)-3-[5-(2,3-dimethoxy-6-methyl-1,4-benzoquinoyl)]-2-nonyl-2-propenoic acid [E3330, kindly provided by Mark Kelley, Indiana University, Indianapolis, IN (34,36,44,45] in reaction buffer (15 mm Tris, pH 7.9; 60 mm KCl; 1 mm MgCl2; 0.2 mm EDTA; 50 nm E2; 10% glycerol; and 50 ng deoxyinosinic-deoxycytidylic acid) for 30 min at 25 C. BSA was included to maintain protein concentrations at 1 μg. 32P-labeled, 50 bp ERE-containing oligos (20,000 cpm) were added and incubated for 30 min at 25 C. Samples were fractioned on a 6% nondenaturing polyacrylamide gel in low ionic strength buffer at 4 C with buffer recirculation. The levels of bound and free 32P-labeled DNA were quantitated using a PhosphorImager (Molecular Dynamics) and ImageQuant 5.0 software (GE Healthcare).
RNA interference
MCF-7 cells were maintained and transfected using SiLentfect (Bio-Rad) as described elsewhere (75) with 100 pmol Ape1/Ref-1 specific or control Renilla luciferase siRNA (catalog nos. 51320 and 4630, respectively; Ambion, Inc., Austin TX). After a 24-h incubation, plated cells were treated with ethanol or 10 nm E2 for 24 h. Cells were harvested in TNE (40 mm Tris-HCl, pH 7.5; 140 mm NaCl; and 1.5 mm EDTA), resuspended in lysis buffer (20 mm Tris-HCl, pH 8.0; 200 mm NaCl, 1 mm EDTA; and 0.2% NP-40) and subjected to two freeze-thaw cycles. Whole-cell extracts (20 μg) were fractionated on a 10% SDS polyacrylamide gel and transferred to a nitrocellulose membrane for Western blot analysis with antibodies to Ape1/Ref-1 (sc-17774), ERα (sc-543), PR-A and PR-B (RM-9102-S0, Labvision, Fremont CA), and GAPDH (TAB1001). RNA isolation, cDNA preparation, and quantitative RT-PCR were carried out as described previously (75). The relative fold change in mRNA expression in the absence and presence of E2 was determined using the comparative Ct method and the housekeeping gene, 36B4, as the internal control.
Proliferation Assay
MCF-7 cells were maintained and transfected using SiLentfect as described with 100 pmol Ape1/Ref-1 specific or control Renilla luciferase siRNA. After 24 h incubation, plated cells were treated with ethanol or 10 nm E2 for 24 h. The following day, 1.2 mm MTT (Research Products International, Mt. Prospect, IL) was added, and cells were incubated an additional 3.5 h at 37 C. An equal volume of stop mix solution (20% SDS and 50% dimethylformaldehyde) was added, and the plate was rocked for 1 h at 25 C to dissolve the formazan precipitate. Absorbance measurements were taken at 550 nm using a plate reader, and the cell proliferation index was determined.
Inhibitors
MCF-7 cells were maintained on phenol red containing MEM with 5% CS. Cells were transferred 2 d before plating to phenol red-free MEM with 5% CDCS. Cells were resuspended in antibiotic-free, phenol red-free MEM containing 5% CDCS and seeded into 12-well plates. The following day, cells were treated with dimethylsulfoxide (DMSO), 10 mm methoxyamine hydrochloride [MX, MP Biomedicals, Solon, OH (36,40,41)] or 100 μm E3330 (34,36,44,45). After 1 h, ethanol or 10 nm E2 was added, and cells were incubated an additional 24 h at 37 C. RNA isolation, cDNA preparation, and quantitative RT-PCR were carried out as described (75). The relative fold change in mRNA expression in the absence and presence of E2 was determined using the comparative Ct method and the housekeeping gene, 36B4, as the internal control.
ChIP
MCF-7 cells were maintained on phenol red-containing MEM with 5% CS. Cells were transferred 3 d before treatment to phenol red-free MEM with 5% CDCS. Cells were treated with ethanol or 10 nm E2 for 20 min, 2 h, 6 h, or 24 h. For ChIP assays using the redox inhibitor, MCF-7 cells were treated with DMSO or 100 μm E3330 for 1 h, and ethanol or 10 nm E2 was added for the indicated times. ChIP assays were carried out essentially as recommended by Millipore Corp. (Billerica, MA) except that cell lysates were diluted in micrococcal nuclease buffer (10 mm Tris, pH 7.5; 10 mm NaCl; 3 mm MgCl2; 1 mm CaCl2; and 4% NP-40) and treated with 50 U micrococcal nuclease (United States Biochemical Corp., Cleveland OH) at 37 C for 10 min before sonication. nProtein-A sepharose (GE Healthcare) and a nonspecific fluorescein antibody (Immunological Resource Center, University of Illinois) were added to preclear the chromatin overnight at 4 C. An ERα- (sc-8002) or a mixture of Ape1/Ref-1-specific (sc-17774 and sc-334) antibodies was used for immunoprecipitation of protein-DNA complexes. PCR primers flanking the pS2 ERE, two gene regions 205 or 221 kb (PR205 and PR221, respectively) upstream of the PR-B transcription start site, or a nonspecific sequence in the 36B4 gene were used for real-time, quantitative PCR using iQ SyBr Green Supermix and the iCycler PCR thermocycler according to manufacturer’s directions (Bio-Rad). Standard curves using 1000, 5000, 10,000, 50,000, and 100,000 copies of each gene were run for each primer set during each experiment.
Immunohistochemistry of human mammary tissue
Paraffin-embedded blocks of normal human mammary tissue were obtained from Carle Foundation Hospital (Urbana, IL). The tissue was from an 81-yr-old woman who underwent elective prophylactic mastectomy in 2008. The identity of the patient is the sole property of Carle Clinic and has not been or will be in the future shared with the investigators. This study was approved by the Institutional Review Boards of the University of Illinois at Urbana-Champaign (06171) and Carle Foundation Hospital (05-44).
The paraffin-embedded blocks were sectioned and mounted on frost-free slides. The 10-μm sections were deparaffinized in xylene and rehydrated through a series of graded alcohols. Slides were washed with 1× PBS, and endogenous peroxidases were blocked with 1.5% hydrogen peroxide in 1× PBS for 20 min at 25 C. After three 5-min washes in 1× PBS, slides were incubated in blocking solution (1× PBS, 0.1% Triton X-100, and 3% BSA) with 5% normal donkey serum for 10 min at 25 C. Control (no primary antibody) and experimental slides were incubated overnight at 4 C in blocking solution without or with Ape1/Ref-1 (1:400, sc-17774) or ERα (1:400, sc-543) antibody. Slides were washed with 1× PBS between each of the following steps. Biotin-conjugated secondary antibody (1:200; Jackson ImmunoResearch, West Grove, PA) was added, and slides were incubated for 30 min at 25 C. The ABC Peroxidase Staining kit (1:100 dilution of each Reagent A and B in 1× PBS, 32020; Thermo Scientific, Rockford, IL) was applied for 30 min at 25 C. Staining was visualized with peroxidase-sensitive Sigmafast 3,3′-diaminobenzidine tablets (Sigma). Slides were counterstained with 0.1% methyl green (Sigma) for 3 min at 60 C, dehydrated in ethanol, cleared in xylene, and mounted with Permount (Fisher Scientific, Pittsburgh, PA). Images were obtained with a Leica DMI4000B confocal microscope (Leica Corp., Deerfield, IL) and a Retiga 2000R digital camera (W. Nushbaum, McHenry, IL).
Acknowledgments
We thank B. Hall and M. Frank (Carle Foundation Hospital, Urbana, IL) for assistance in procuring the human mammary tissue. We also thank J. Yokota and T. Kohno (National Cancer Center Research, Tokyo, Japan), and B. Katzenellenbogen (UIUC, Urbana, IL) for expression vectors; M. Kelley (Indiana University, Indianapolis, IN) for E3330, plasmids, and valuable advice; L. Kraus (Cornell University, Ithaca, NY) and J. Kadonaga (University of California, San Diego, CA) for viral stock used in ERα production; and J. Boney-Montoya, A. Rao, and G. Freund (UIUC, Urbana, IL) for helpful discussions.
Footnotes
This work was supported by National Institutes of Health Grants R01 and R56 DK 53884 (to A.M.N.) and P41 RR011823 (to J.R.Y.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online May 21, 2009
Abbreviations: Ape1, Apurinic/apyrimidinic endonuclease 1; CDCS, charcoal dextran-treated CS; ChIP, Chromatin immunoprecipitation; CS, calf serum; DTT, dithiothreitol; E2, 17β-estradiol; ER, estrogen receptor; ERE, estrogen-response element; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MTT, 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide; MX, methoxyamine; NFκB, nuclear factor-κB; NP-40, Nonidet P-40; PDI, protein disulfide isomerase; PR, progesterone receptor; Ref-1, redox factor-1; ROS, reactive oxygen species; SDS, sodium dodecyl sulfate; siRNA, small interfering RNA; SOD1, superoxide dismutase; Trx, thioredoxin; TrxR, thioredoxin reductase; U2OS, U2 osteosarcoma.
References
- Webster KA, Prentice H, Bishopric NH 2001 Oxidation of zinc finger transcription factors: physiological consequences. Antioxid Redox Signal 3:535–548 [DOI] [PubMed] [Google Scholar]
- Hirota K, Matsui M, Iwata S, Nishiyama A, Mori K, Yodoi J 1997 AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci USA 94:3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C 1997 Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev 11:558–570 [DOI] [PubMed] [Google Scholar]
- Evans AR, Limp-Foster M, Kelley MR 2000 Going APE over ref-1. Mutat Res 461:83–108 [DOI] [PubMed] [Google Scholar]
- Sweasy JB, Lang T, DiMaio D 2006 Is base excision repair a tumor suppressor mechanism? Cell Cycle 5:250–259 [DOI] [PubMed] [Google Scholar]
- Kumar V, Chambon P 1988 The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell 55:145–156 [DOI] [PubMed] [Google Scholar]
- Klein-Hitpass L, Ryffel GU, Heitlinger E, Cato ACB 1988 A 13 bp palindrome is a functional estrogen responsive element and interacts specifically with estrogen receptor. Nucleic Acids Res 16:647–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couse JF, Korach KS 1999 Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev 20:358–417 [DOI] [PubMed] [Google Scholar]
- Korach KS 1994 Insights from the study of animals lacking functional estrogen receptor. Science 266:1524–1527 [DOI] [PubMed] [Google Scholar]
- Subbiah MTR 1998 Mechanisms of cardioprotection by estrogens. Proc Soc Exp Biol Med 217:23–29 [DOI] [PubMed] [Google Scholar]
- Toran-Allerand CD 1996 Mechanisms of estrogen action during neural development: mediation by interactions with the neurotophins and their receptors? J Steroid Biochem 56:169–178 [DOI] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS 2003 Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144:4562–4574 [DOI] [PubMed] [Google Scholar]
- Stossi F, Barnett DH, Frasor J, Komm B, Lyttle CR, Katzenellenbogen BS 2004 Transcriptional profiling of estrogen-regulated gene expression via estrogen receptor (ER) α or ERβ in human osteosarcoma cells: distinct and common target genes for these receptors. Endocrinology 145:3473–3486 [DOI] [PubMed] [Google Scholar]
- Jakowlew SB, Breathnach R, Jeltsch JM, Masiakowski P, Chambon P 1984 Sequence of the pS2 mRNA induced by estrogen in the human breast cancer cell line MCF-7. Nucleic Acids Res 12:2861–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio MC, Bellocq JP, Gairard B, Rasmussen UB, Krust A, Koehl C, Calderoli H, Schiff V, Renaud R, Chambon P 1987 Specific expression of the pS2 gene in subclasses of breast cancers in comparison with expression of the estrogen and progesterone receptors and the oncogene ERBB2. Proc Natl Acad Sci USA 84:9243–9247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Petz LN, Ziegler YS, Wood JR, Potthoff SJ, Nardulli AM 2000 Regulation of the estrogen-responsive pS2 gene in MCF-7 human breast cancer cells. J Steroid Biochem 74:157–168 [DOI] [PubMed] [Google Scholar]
- Giangrande PH, Kimbrel EA, Edwards DP, McDonnell DP 2000 The opposing transcriptional activities of the two isoforms of the human progesterone receptor are due to differential cofactor binding. Mol Cell Biol 20:3102–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P 1990 Two distinct estrogen-regulated promoters generate transcripts encoding two functionally different human progesterone receptor forms A and B. EMBO J 9:1603–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Hajiro-Nakanishi K, Makino Y, Eguchi H, Yodoi J, Tanaka H 1997 Functional modulation of estrogen receptor by redox state with reference to thioredoxin as a mediator. Nucleic Acids Res 25:4035–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz-Norton JR, McDonald WH, Yates JR, Nardulli AM 2006 Protein disulfide isomerase serves as a molecular chaperone to maintain estrogen receptor α structure and function. Mol Endocrinol 20:1982–1995 [DOI] [PubMed] [Google Scholar]
- Schultz-Norton JR, Ziegler YS, Likhite VS, Nardulli AM 2009 Isolation of proteins associated with the DNA-bound estrogen receptor α. In: Park Sarge O-K, Curry T, eds. Molecular endocrinology: a comprehensive guide to current methodologies. Totowa, NJ: Humana Press [Google Scholar]
- Schultz-Norton JR, Ziegler YS, Likhite VS, Yates JR, Nardulli AM 2008 Isolation of novel coregulatory protein networks associated with DNA-bound estrogen receptor α. BMC Mol Biol 9:97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven MA, Davis RE, Curtis CD, Muster N, Yates JR, Nardulli AM 2004 A novel estrogen receptor α-associated protein alters receptor-deoxyribonucleic acid interactions and represses receptor-mediated transcription. Mol Endocrinol 18:2649–2659 [DOI] [PubMed] [Google Scholar]
- Loven MA, Muster N, Yates JR, Nardulli AM 2003 A novel estrogen receptor α associated protein, template activating factor I β, inhibits acetylation and transactivation. Mol Endocrinol 17:67–78 [DOI] [PubMed] [Google Scholar]
- Schultz-Norton JR, Walt KA, Ziegler YS, McLeod IX, Yates JR, Raetzman LT, Nardulli AM 2007 The deoxyribonucleic acid repair protein flap endonuclease-1 modulates estrogen-responsive gene expression. Mol Endocrinol 21:1569–1580 [DOI] [PubMed] [Google Scholar]
- Schultz-Norton JR, Gabisi VA, Ziegler YS, McLeod IX, Yates JR, Nardulli AM 2007 Interaction of estrogen receptor α with proliferating cell nuclear antigen. Nucleic Acids Res 35:5028–5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Marzouk S, Schultz-Norton JR, Likhite VS, McLeod IX, Yates JR, Nardulli AM 2007 Rho GDP dissociation inhibitor α interacts with estrogen receptor α and influences estrogen responsiveness. J Mol Endocrinol 39:249–259 [DOI] [PubMed] [Google Scholar]
- Rao AK, Ziegler YS, McLeod IX, Yates JR, Nardulli AM 2008 Effects of Cu/Zn superoxide dismutase on estrogen responsiveness and oxidative stress in human breast cancer cells. Mol Endocrinol 22:1113–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creekmore AL, Walt KA, Schultz-Norton JR, Ziegler YS, McLeod IX, Yates JR, Nardulli AM 2008 The role of retinoblastoma associated proteins 46 and 48 in estrogen receptor α mediated gene expression. Mol Cell Endocrinol 291:79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henttu PM, Kalkhoven E, Parker MG 1997 AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol Cell Biol 17:1832–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaidarun SS, Swearingen B, Alexander JM 1998 Differential expression of estrogen receptor-β (ER β) in human pituitary tumors: functional interactions with ER α and a tumor-specific splice variant. J Clin Endocrinol Metab 83:3308–3315 [DOI] [PubMed] [Google Scholar]
- Wood JR, Likhite VS, Loven MA, Nardulli AM 2001 Allosteric modulation of estrogen receptor conformation by different estrogen response elements. Mol Endocrinol 15:1114–1126 [DOI] [PubMed] [Google Scholar]
- Nardulli AM, Greene GL, O'Malley BW, Katzenellenbogen BS 1988 Regulation of progesterone receptor messenger ribonucleic acid and protein levels in MCF-7 cells by estradiol: analysis of estrogen’s effect on progesterone receptor synthesis and degradation. Endocrinology 122:935–944 [DOI] [PubMed] [Google Scholar]
- Zou GM, Luo MH, Reed A, Kelley MR, Yoder MC 2007 Ape1 regulates hematopoietic differentiation of embryonic stem cells through its redox functional domain. Blood 109:1917–1922 [DOI] [PubMed] [Google Scholar]
- Demple B, Sung JS 2005 Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair 4:1442–1449 [DOI] [PubMed] [Google Scholar]
- Fishel ML, Kelley MR 2007 The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med 28:375–395 [DOI] [PubMed] [Google Scholar]
- Fishel ML, He Y, Reed AM, Chin-Sinex H, Hutchins GD, Mendonca MS, Kelley MR 2008 Knockdown of the DNA repair and redox signaling protein Ape1/Ref-1 blocks ovarian cancer cell and tumor growth. DNA Repair 7:177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundström J, Holmgren A 1990 Protein disulfide-isomerase is a substrate for thioredoxin reductase and has thioredoxin-like activity. J Biol Chem 265:9114–9120 [PubMed] [Google Scholar]
- Xanthoudakis S, Curran T 1992 Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J 11:653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S, Fortini P, Karran P, Bignami M, Dogliotti E 1991 Processing in vitro of an abasic site reacted with methoxyamine: a new assay for the detection of abasic sites formed in vivo. Nucleic Acids Res 19:5569–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Gerson SL 2004 Therapeutic impact of methoxyamine: blocking repair of abasic sites in the base excision repair pathway. Curr Opin Investig Drugs 5:623–627 [PubMed] [Google Scholar]
- Hiramoto M, Shimizu N, Sugimoto K, Tang J, Kawakami Y, Ito M, Aizawa S, Tanaka H, Makino I, Handa H 1998 Nuclear targeted suppression of NF-κ B activity by the novel quinone derivative E3330. J Immunol 160:810–819 [PubMed] [Google Scholar]
- Shimizu N, Sugimoto K, Tang J, Nishi T, Sato I, Hiramoto M, Aizawa S, Hatakeyama M, Ohba R, Hatori H, Yoshikawa T, Suzuki F, Oomori A, Tanaka H, Kawaguchi H, Watanabe H, Handa H 2000 High-performance affinity beads for identifying drug receptors. Nat Biotechnol 18:877–881 [DOI] [PubMed] [Google Scholar]
- Zou GM, Maitra A 2008 Small-molecule inhibitor of the AP endonuclease 1/REF-1 E3330 inhibits pancreatic cancer cell growth and migration. Mol Cancer Ther 7:2012–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Delaplane S, Jiang A, Reed A, He Y, Fishel M, Nyland 2nd RL, Borch RF, Qiao X, Georgiadis MM, Kelley MR 2008 Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: small-molecule Inhibition of the redox function of Ape1. Antioxid Redox Signal 10:1853–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LJ, Robson CN, Black E, Gillespie D, Hickson ID 1993 Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol 13:5370–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S 2005 Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci USA 102:5739–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, Kelley MR 2008 Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res 643:54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittal RM, Benz CC, Scott G, Semyonov J, Burlingame AL, Baldwin MA 2000 mol DNA-binding domain prevents dimerization and, hence, DNA binding. Biochemistry 39:8406–8417 [DOI] [PubMed] [Google Scholar]
- Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T 1992 Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J 11:3323–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D, Pongratz I, Poellinger L, Whitelaw ML 2000 A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1α and the HIF-like factor. J Biol Chem 275:4618–4627 [DOI] [PubMed] [Google Scholar]
- Petz LN, Ziegler YS, Loven MA, Nardulli AM 2002 Estrogen receptor α and activating protein-1 mediate estrogen responsiveness of the progesterone receptor gene in MCF-7 breast cancer cells. Endocrinology 143:4583–4591 [DOI] [PubMed] [Google Scholar]
- Petz LN, Ziegler YS, Schultz JR, Nardulli AM 2004 Fos and Jun inhibit estrogen-induced transcription of the human progesterone receptor gene through an activator protein-1 site. Mol Endocrinol 18:521–532 [DOI] [PubMed] [Google Scholar]
- Schultz JR, Petz LN, Nardulli AM 2005 Cell- and ligand-specific regulation of promoters containing activator protein-1 and Sp1 sites by estrogen receptors α and β. J Biol Chem 280:347–354 [DOI] [PubMed] [Google Scholar]
- Rao AK, Ziegler YS, McLeod IX, Yates JR, Nardulli AM 20 July 2009 Thioredoxin and thioredoxin reductase influence estrogen receptor α mediated gene expression in human breast cancer cells. J Mol Endocrinol 10.1677/JME-09-0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich I 1995 Superoxide radical and superoxide dismutases. Annu Rev Biochem 64:97–112 [DOI] [PubMed] [Google Scholar]
- Atkin JD, Farg MA, Turner BJ, Tomas D, Lysaght JA, Nunan J, Rembach A, Nagley P, Beart PM, Cheema SS, Horne MK 2006 Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J Biol Chem 281:30152–30165 [DOI] [PubMed] [Google Scholar]
- Cheung PY, Churchich JE, Lee KS 1999 Refolding of thioredoxin reductase assisted by groEL and PDI. Biochem Biophys Res Commun 255:17–22 [DOI] [PubMed] [Google Scholar]
- Holmgren A 1985 Thioredoxin. Annu Rev Biochem 54:237–271 [DOI] [PubMed] [Google Scholar]
- Holmgren A, Björnstedt M 1995 Thioredoxin and thioredoxin reductase. Methods Enzymol 252:199–208 [DOI] [PubMed] [Google Scholar]
- Mustacich D, Powis G 2000 Thioredoxin reductase. Biochem J 346:1–8 [PMC free article] [PubMed] [Google Scholar]
- Lyles MM, Gilbert HF 1991 Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: pre-steady-state kinetics and the utilization of the oxidizing equivalents of the isomerase. Biochemistry 30:619–625 [DOI] [PubMed] [Google Scholar]
- Noiva R, Freedman RB, Lennarz WJ 1993 Peptide binding to protein disulfide isomerase occurs at a site distinct from the active sites. J Biol Chem 268:19210–19217 [PubMed] [Google Scholar]
- Puig A, Lyles MM, Noiva R, Gilbert HF 1994 The role of the thiol/disulfide centers and peptide binding site in the chaperone and anti-chaperone activities of protein disulfide isomerase. J Biol Chem 269:19128–19135 [PubMed] [Google Scholar]
- Wang CC, Tsou CL 1993 Protein disulfide isomerase is both an enzyme and a chaperone. FASEB J 7:1515–1517 [DOI] [PubMed] [Google Scholar]
- Quan H, Fan G, Wang CC 1995 Independence of the chaperone activity of protein disulfide isomerase from its thioredoxin-like active site. J Biol Chem 270:17078–17080 [DOI] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer JA 2000 DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination Nature. [Erratum (2000) 404:6777] Nature 403:451–456 [DOI] [PubMed] [Google Scholar]
- Wilson SH, Kunkel TA 2000 Passing the baton in base excision repair. Nat Struct Biol 7:176–178 [DOI] [PubMed] [Google Scholar]
- Likhite VS, Cass EI, Anderson SD, Yates JR, Nardulli AM 2004 Interaction of estrogen receptor α with 3-methyladenine DNA glycosylase modulates transcription and DNA repair. J Biol Chem 279:16875–16882 [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Shen X 2006 Chromatin modifications in DNA repair. Results Probl Cell Differ 41:109–125 [DOI] [PubMed] [Google Scholar]
- Mellon I, Bohr VA, Smith CA, Hanawalt PC 1986 Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci USA 83:8878–8882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I, Spivak G, Hanawalt PC 1987 Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51:241–249 [DOI] [PubMed] [Google Scholar]
- Teo AK, Oh HK, Ali RB, Li BF 2001 The modified human DNA repair enzyme O(6)-methylguanine-DNA methyltransferase is a negative regulator of estrogen receptor-mediated transcription upon alkylation DNA damage. Mol Cell Biol 21:7105–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG 2006 A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312:1798–1802 [DOI] [PubMed] [Google Scholar]
- Curtis CD, Likhite VS, McLeod IX, Yates JR, Nardulli AM 2007 Interaction of the tumor metastasis suppressor nonmetastatic protein 23 homologue H1 and estrogen receptor α alters estrogen-responsive gene expression. Cancer Res 67:10600–10607 [DOI] [PubMed] [Google Scholar]
- Xanthoudakis S, Miao GG, Curran T 1994 The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc Natl Acad Sci USA 91:23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Clore GM, Kennedy WP, Kuszewski J, Gronenborn AM 1996 The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal. Structure 4:613–620 [DOI] [PubMed] [Google Scholar]
- Ando K, Hirao S, Kabe Y, Ogura Y, Sato I, Yamaguchi Y, Wada T, Handa H 2008 A new APE1/Ref-1-dependent pathway leading to reduction of NF-κB and AP-1, and activation of their DNA-binding activity. Nucleic Acids Res 36:4327–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, Botero A, Hirota K, Bradbury CM, Markovina S, Laszlo A, Spitz DR, Goswami PC, Yodoi J, Gius D 2000 Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res 60:6688–6695 [PubMed] [Google Scholar]