Abstract
Understanding the molecular details associated with aberrant high mobility group A2 (HMGA2) gene expression is key to establishing the mechanism(s) underlying its oncogenic potential and impact on the development of therapeutic strategies. Here, we report the involvement of HMGA2 in impairing DNA-dependent protein kinase (DNA-PK) during the non-homologous end joining (NHEJ) process. We demonstrated that HMGA2-expressing cells displayed deficiency in overall and precise DNA end-joining repair and accumulated more endogenous DNA damage. Proper and timely activation of DNA-PK, consisting of Ku70, Ku80 and DNA-PKcs subunits, is essential for the repair of DNA double strand breaks (DSBs) generated endogenously or by exposure to genotoxins. In cells overexpressing HMGA2, accumulation of histone 2A variant X phosphorylation at Ser-139 (γ-H2AX) was associated with hyper-phosphorylation of DNA-PKcs at Thr-2609 and Ser-2056 before and after the induction of DSBs. Also, the steady-state complex of Ku and DNA ends was altered by HMGA2. Microirradiation and real-time imaging in living cells revealed that HMGA2 delayed the release of DNA-PKcs from DSB sites, similar to observations found in DNA-PKcs mutants. Moreover, HMGA2 alone was sufficient to induce chromosomal aberrations, a hallmark of deficiency in NHEJ-mediated DNA repair. In summary, a novel role for HMGA2 to interfere with NHEJ processes was uncovered, implicating HMGA2 in the promotion of genome instability and tumorigenesis.
Keywords: HMGA2, Ku70/80, DNA-PKcs, NHEJ, genome instability
Introduction
High mobility group A (HMGA) genes encode four chromatin binding proteins (HMGA1a, HMGA1b, HMGA1c and HMGA2), each containing several “AT-hooks”, the functional motif characteristic of HMGA family (1). HMGA2 (formerly HMGI-C) function has been studied in detail in mice, where it is expressed in pluripotent embryonic stem cells and during embryogenesis, but is absent or present at low levels in terminally differentiated tissues. The physiological function of HMGA2 seems critical for cell growth and adipogenesis as mice lacking functional HMGA2 exhibit a pygmy phenotype with greatly reduced fat tissue (2). In addition, a critical role of HMGA2 in stimulating normal cardiogenesis has been suggested from studies using mouse embryonic carcinoma cells P19CL6 (3) or promoting mouse central and peripheral neural stem cell self-renewal (4). By contrast, overexpression of HMGA2 in pituitary glands induces pituitary adenomas (5), and misexpression of HMGA2 in mesenchymal cells of HMGA2 transgenic mice produces fibroadenomas of the breast and salivary gland adenomas (6). Hence, HMGA2 is referred to as an oncofetal protein.
In humans, the HMGA2 gene is located on chromosome 12q14.3, which is frequently amplified or subjected to chromosomal rearrangement. Elevated HMGA2 protein levels are associated with more malignant neoplasia, and HMGA2 rearrangements are frequently found in tumors of mesenchymal origin (7, 8). Furthermore, recent data hints at novel roles for HMGA2 in the epithelial-mesenchymal transition during tumor metastasis (9). Several independent studies have also identified HMGA2 transcript as a target for the let-7 family of microRNAs. Notably, disruption of let-7 repression of HMGA2 enhances tumor cell proliferation and cell transformation (10, 11), further supporting the role of HMGA2 as an oncogene.
DNA double-strand breaks (DSBs) can be generated by endogenous reactive oxygen species or destabilization of stalled replication forks as well as by exposure to a variety of exogenous agents, including ionizing radiation (IR) and chemotherapeutic agents. Proper repair of DSBs plays a prominent role in cell survival, maintenance of genomic integrity, and prevention of tumorigenesis (12). Eukaryotes have evolved at least two major mechanisms for DSB repair: homologous recombination (HR) and non-homologous end joining (NHEJ). While HR is the main pathway in simple eukaryotes, NHEJ predominantly repairs DSBs in mammalian cells resulting from DNA-damaging agents (13). While NHEJ does not depend on the presence of homologous DNA sequences, it requires the DNA-dependent protein kinase (DNA-PK) complex, which is comprised of a heterodimer of 70- and 80-kDa proteins (Ku70 and Ku80) and a 470-kDa DNA-PK catalytic subunit (DNA-PKcs) (14). The current model predicts that the DNA-PKcs functions as a “gatekeeper”, protecting DNA ends from premature processing and undesirable ligation until the two ends are properly juxtaposed and DNA-PKcs autophosphorylation can then take place (14). While efficient and appropriate repair of DSBs is essential for the preservation of genome integrity, the possibility of interfering with DNA-PK and/or NHEJ function by oncoprotein(s) other than genetic mutations of NHEJ components (Ku70, Ku80, DNA-PKcs, Artemis, Xrcc4 and DNA ligase 4) per se is still unclear. Given the evidence that HMGA2 is an oncoprotein, we investigated whether HMGA2 plays a role in impairing cellular DSB repair functions.
This report is, to our knowledge, the first direct cell biological, biochemical and cytogenetic evidence to suggest a novel role for HMGA2 in modulating the NHEJ processes during the repair of DSBs, resulting in accumulation of chromosomal aberrations. We showed that HMGA2 caused DNA-PKcs hyper-phosphorylation at Ser-2956 and Thr-2609 and accumulation of γ-H2AX both prior to and after induction of DSBs. Conversely, knockdown of HMGA2 decreased the levels of both basal and Dox-induced DNA-PKcs hyper-phosphorylation. Our studies using real-time imaging in living cells demonstrated a prolonged presence of DNA-PKcs at DSB sites in HMGA2-expressing cells receiving microirradiation, a photobleaching technique that utilized a 365-nm pulsed nitrogen laser to introduce a small area of DNA damage in the cell nucleus. We also showed that HMGA2 had profound effect on the rate for γ-H2AX clearance after exposure to IR. As genomic instability is particularly important in tumor progression (15), our results suggest that HMGA2 interference of NHEJ function contributes to tumorigenesis by promoting genome instability.
Materials and Methods
Cell culture and Construct preparation
In all assays, cells were serum starved (48-72 h) using Opti-MEM (Gibco Lifetech) to achieve at least 60% synchronization in G1-phase, as confirmed by fluorescence-activated cell sorting analyses. Normal human lung fibroblast WI-38 cells (ATCC CCL-75) were maintained in DMEM (Mediatech) medium containing 10% fetal bovine serum (FBS) (Hyclone) supplemented with 0.1mM non-essential amino acids. CHO-AA8, V3, Xrs6, CL48, Hep3B, HepG2, MEF, MEF/Ku70(-/-), M059K, and M059J cells were all maintained in DMEM medium containing 10% FBS. HCC1419, Hs578T Pa-4 and Pa-4/HMGA2 cells were cultured as previously described (16). HeLa/HMGA2, and HeLa/HMGA2(4P/A) cells were generated by transfecting parental HeLa cells with HA-HMGA2- and HA-HMGA2(4P/A)-pcDNA3.1 plasmids respectively using Lipofectamine 2000 (Invitrogen). Cells were then cultured in 10% FBS/DMEM medium supplemented with 1 mg/mL and 0.3 mg/mL G418 (Invitrogen) for selection and clonal expansion, respectively. HA-HMGA2 and HA-HMGA2(4P/A), which carried a combination of P48A, P53A, P76A and P80A mutations, were generated using the previously described methods (17). HMGA2 and HMGA2(4P/A) were PCR amplified with XhoI flanking the 5′- and BamHI flanking the 3′-ends followed by XhoI/BamHI digestion. The digested products were subsequently ligated into XhoI/BamHI-linearized pIRES2-DsRed2 vector (Clontech) to generate HMGA2-IRES-DsRed and HMGA2(4P/A)-IRES-DsRed construct, respectively. All engineered constructs were verified by DNA sequencing, and expression of these constructs was verified by Western analyses.
Western analyses, Antibodies and Immunofluorescence
Western analyses were performed as previously described (16). The antibodies used in this study were as follows: anti-HA (Covance), anti-HMGA2 (Biocheck), anti-actin (Chemicon), anti-γ-H2AX (Upstate), and anti-DNA-PKcs, anti-nucleolin, and anti-H2AX (Santa Cruz), and anti-DNA-PKcs (p-T2609), anti-DNA-PKcs (p-S2056), and anti-Ku80 polyclonal antibodies were generated as previously described (18). Immunofluorescent staining was performed as previously described (19). For confocal images, the volume of each DNA-PKcs-T2609 foci was quantified by first using the scale provided by Zeiss Software to measure the diameter, and then using the formula, V = 4/3 πr3, to calculate the volume.
Viral transduction, Cytogenetic analyses and HMGA2 knockdown
Lentiviral vectors pRRLsin.hCMV-HMGA2, pΔ8.7, and pVSV-G were constructed and used for lentiviral production in HEK 293T cells as previously described (20). WI-38 cells were infected with lentiviruses encoding HMGA2 or vector alone, and the transduced WI-38 cells were maintained in culture for three passages, at 1:3 dilutions, to allow time for the genome to stabilize before further analysis was performed. For cytogenetic analysis, the HMGA2- and vector-transduced WI-38 cells were harvested according to established protocol (20). HMGA2 was knocked down by retroviruses produced using pSin-Puro-miR30-shHMGA2 vector generously provided by Dr. Scott Lowe (Cold Spring Harbor Laboratory).
Laser microirradiation, Time lapse imaging and Calculation of protein recruitment kinetics
DSBs were introduced in the nuclei of cultured cells by microirradiation and imaged using previously described methods (21). Time-lapse image acquisition began before laser microirradiation so as to obtain an image of the cell prior to irradiation. Images before and after irradiation were captured with a 400 ms exposure time. Signal intensities of accumulated YFP at the microirradiated site were calculated as previously described (21).
DNA end-binding activity of Ku80
HeLa/vector, HeLa/HMGA2 and HeLa/HMGA2(4P/A) cells were treated with Dox (5 μM) at indicated time points. Preparation of nuclear extracts and assessment of DNA end-binding activity of Ku80 were carried out using a Nuclear Extract kit and a Ku70/Ku80 DNA Repair kit, respectively (Active Motif), according to suggested protocols. Protein from each sample (2.5 μg) was used in the Ku70/80 DNA repair assay.
X-irradiation
Cells were irradiated (3 Gy/min, room temperature) using a 137Cs source (Mark II, gamma irradiator). Cells were then harvested using Laemmli sample buffer at respective time points post-irradiation followed by Western analysis.
Statistical analysis
Statistical analyses were performed using one-way ANOVA, followed by posthoc comparisons based on modified Newman-Keuls-Student procedure with p<0.05 considered significant. Where appropriate, unpaired Student's t-tests were also performed to determine the difference between two data groups.
Results
The aim of this study was to investigate the role of HMGA2 in the perturbation of DSB repair by NHEJ. In light of the fact that HR uses sister chromatids for DSB repair, and therefore occurs only in dividing cells in S- or G2-phase (22), cells enriched in G1-phase were used throughout the study.
HMGA2 stimulates basal and DSB-induced DNA-PKcs phosphorylation at Ser-2056 and Thr-2609 and reduces Ku80 DNA end-binding
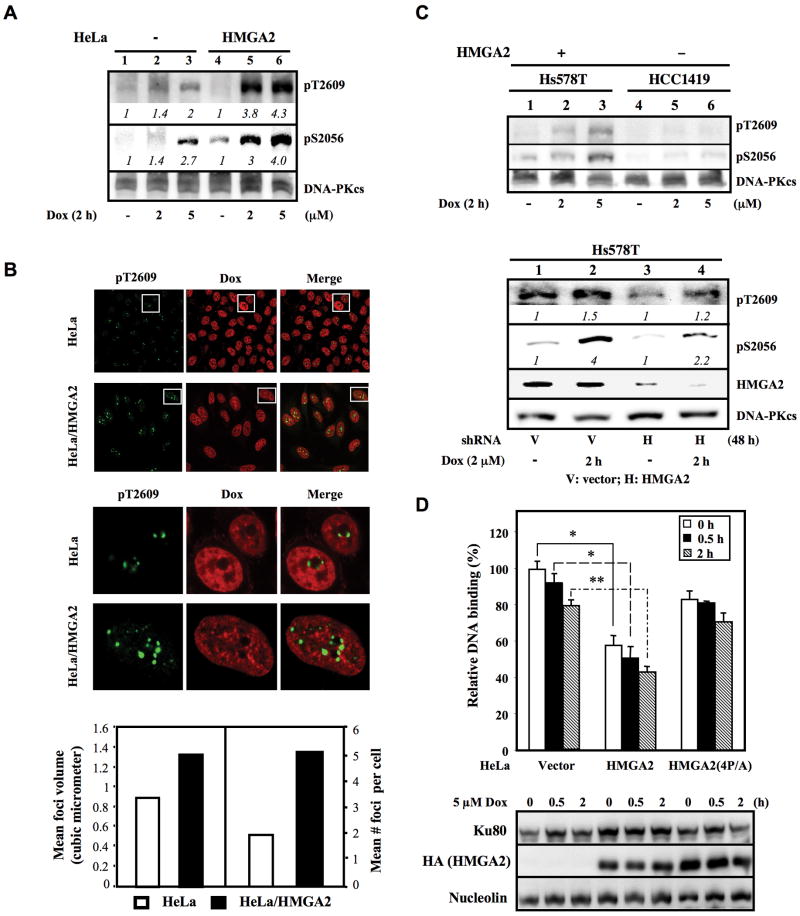
Given that HMGA2 overexpression caused accumulation of DNA lesions and dysregulation of NHEJ (Supplementary Figure S1), we investigated whether HMGA2 altered the Dox-induced DNA-PKcs activation profile by monitoring DNA-PKcs phosphorylation at both Ser-2056 and Thr-2609, which was demonstrated to be important for proper NHEJ-mediated repair of DSBs (18, 23). DNA-PKcs-Ser-2056/-Thr-2609 hyper-phosphorylation signals were detected 2 h post Dox treatment in HeLa/HMGA2 cells compared to Dox-treated HeLa cells (Figure 1A, lanes 5 and 6 versus lanes 2 and 3). Consistent with these results, immunofluorescent staining revealed there were more pT2609-DNA-PKcs foci in Dox-treated HeLa/HMGA2 cells (Figure 1B). Quantitative analyses further confirmed an increase in the average number and volume of discrete nuclear pT2609-DNA-PKcs foci in HeLa/HMGA2 cells 30 min post Dox treatment (Figure 1B). Comparable nuclear accumulation of Dox-associated red fluorescence intensity was measured (data not shown) in both cell types suggested T2609-DNA-PKcs hyper-phosphorylation in HeLa/HMGA2 cells was not due to increased Dox in the nucleus. The importance of proper DNA-PK function in DSB repairs was further confirmed in Ku70-deficient (Ku70-/-) MEF cells (Supplementary Figure S2B) and DNA-PKcs-deficient M059J cells (Supplementary Figure S2C), as seen by the marked decrease in cell viability post treatment with increasing concentrations of Dox or cisplatin compared to their wild-type (wt) counterparts. DNA-PKcs hyper-phosphorylation signals were also observed in HMGA2-expressing Hs578T, but not HMGA2-underexpressing HCC1419 breast cancer cells (Figure 1C, upper-panel). Knockdown of endogenous HMGA2 by sh-HMGA2 conferred a decrease in both basal and Dox-induced pS2056- and pT2609-DNA-PKcs signals as compared to vector shRNA in Hs578T cells (Figure 1C, lower-panel, lanes 3 and 4 versus lanes 1 and 2). Notably, the observed increase in pS2056-DNA-PKcs signal prior to Dox treatment (Figures 1A and 1C) possibly reflects the basal activation level of DNA-PKcs in response to elevated DSBs resulting from the defect in DSB repair caused by HMGA2 expression.
Figure 1. HMGA2 expression induces DNA-PKcs hyper-phosphorylation and reduces DNA end-binding of Ku80.
(A) Relative levels (numbers in italic) of Dox-induced pDNA-PKcs-T2609 and pDNA-PKcs-S2056 in HeLa and HeLa/HMGA2 cells normalized against that of DNA-PKcs are shown. (B) Confocal images of pDNA-PKcs-T2609 foci (green) and Dox (red) distribution in HeLa and HeLa/HMGA2 cells 30 min post-Dox (5μM) treatment. Middle-panel is enlarged view of the selected pDNA-PKcs-T2609 foci (upper-panel) as indicated by the white boxes. The mean foci volume (lower-left-panel) and the average number of foci per cell in 75-100 cells (lower-right-panel) are shown. (C, top-panel) Relative levels of Dox-induced pDNA-PKcs-T2609 and pDNA-PKcs-S2056 in Hs578T and HCC1419 breast cancer cells. (bottom-panel) Hs578T cells were transduced with vector or shHMGA2 for 72 h prior to Dox treatment. Relative levels of pDNA-PKcs-T2609 and pDNA-PKcs-S2056 normalized against that of DNA-PKcs are italicized. (D) Relative DNA binding by Ku in HeLa, HeLa/HMGA2, and HeLa/HMGA2(4P/A) cells following Dox treatment. *, p<0.05; **, p<0.01, from three independent experiments.
Ku is a heterodimer that has DNA end binding activity and is required for proper DSB repair by NHEJ (20). To explore the mechanisms associated with HMGA2 and DNA-PKcs hyper-phosphorylation, we utilized an in vitro Ku70/Ku80 DNA repair assay to monitor whether or not HMGA2 or HMGA2(4P/A), a mutant form that carries 4 Pro→Ala substitutions at amino acid residues 48, 52 (second AT-hook), 76 and 80 (third AT-hook), altered Ku DNA end-binding. HMGA2(4P/A) expression almost completely abrogated the wt HMGA2-induced sensitivity to DSBs (data not shown), suggesting that the HMGA2(4P/A) protein did not affect DSB repair. Expression of HMGA2, but not HMGA2(4P/A), significantly reduced the steady-state Ku80 complex in the nuclear extract and DNA ends in vitro (Figure 1D). A slightly decreased steady-state complex of Ku80 and DNA ends was noticed 2 h post Dox treatment, consistent with observations by Turchi et al. that cisplatin decreased the number of Ku molecules bound to DNA (24).
HMGA2 induces a prolonged presence of DNA-PKcs at DSB sites in response to laser damage in living cells
The effects of HMGA2 on the spatiotemporal dynamics of DNA-PKcs recruitment to DSBs were then investigated. V3/YFP-DNA-PKcs cells, a DNA-PKcs-deficient cell line that stably expresses YFP-tagged DNA-PKcs, were transiently transfected with HMGA2 or its mutant HMGA2(4P/A) in a bicistronic vector with IRES-driven DsRed (Supplementary Figure S3A). A small area of DNA damage in the nucleus was introduced with a microirradiation system and laser-induced DSBs were confirmed by immunofluorescent staining with γ-H2AX (data not shown). DNA-PKcs was localized at laser-induced DSB sites and the expression of DsRed did not interfere with the proper response of the transfected V3/YFP-DNA-PKcs cells to the laser damage (Supplementary Figure S3B). Immediately after microirradiation, there was a rapid increase (first 30 s) in YFP-DNA-PKcs at the damaged sites in YFP-DNA-PKcs-expressing V3/vector, V3/HMGA2, and V3/HMGA2(4P/A) cells (Figure 2A, left-panel). The initial kinetics of DNA-PKcs accumulation at DSB sites were comparable among vector-, HMGA2-, and HMGA2(4P/A)-transfected V3/YFP-DNA-PKcs cells (Figure 2B).
Figure 2. HMGA2 induces a continued presence of DNA-PKcs at DSB sites in response to laser damage in living cells.
(A) Short term (1 min, left-panel) and long term (2 h, right-panel) time-lapse imaging of YFP-DNA-PKcs-expressing V3/vector, V3/HMGA2, and V3/HMGA2(4P/A) cells before and after microirradiation. (B and C) Initial accumulation (B) and 2 h time-course accumulation (C) kinetics of YFP-DNA-PKcs of respective cells at DSB sites after microirradiation. Error bars, ± SD from 10 independent measurements.
Time-lapse imaging was then performed for a period of 2 h following laser irradiation. Interestingly, there were significant differences among the three cell lines in the behavior of DNA-PKcs at DSB sites (Figure 2A, right-panel). The fluorescence intensity of the accumulation area in both YFP-DNA-PKcs-expressing V3/vector and V3/HMGA2(4P/A) cells decreased to about 20% of the maximum level at the end of a 2 h period. However, the intensity of the accumulation area in V3/HMGA2 cells was at approximately 60% of the maximal level at 2 h post-treatment (Figure 2C). These results demonstrated that the expression of HMGA2, but not HMGA2(4P/A), conveyed a prolonged presence of DNA-PKcs at the DSB sites. Since DNA-PKcs mutants that are deficient in NHEJ showed prolonged stay at introduced DSB sites (21), we conclude that the sustained DNA-PKcs accumulation area in HMGA2-transfected cells likely reflected the inability to timely repair of DSBs. The specific involvement of HMGA2 in dysregulating NHEJ DNA repair was further confirmed by the expression of a vector, HMGA2 or HMGA2(4P/A) in CHO-AA8 (wild-type), V3 (DNA-PKcs-/-) and Xrs6 (Ku80-/-) cells (Supplementary Figure S4A) followed by cell viability assay in response to Dox treatment. Enhanced sensitivity to Dox treatment was observed in HMGA2-transfected AA8 cells compared to both vector- or HMGA2(4P/A)-transfectants (Supplementary Figure S4B). However, HMGA2 failed to further sensitize V3 and Xrs6 cells (Supplementary Figure S4C and S4D), suggesting that HMGA2 might be specifically perturbing NHEJ, since no additional sensitization to Dox was conferred in cells already NHEJ-deficient due to the lack of crucial NHEJ components (e.g., DNA-PKcs or Ku80).
Delayed clearance of γ-H2AX post X-irradiation in HMGA2-expressing cells
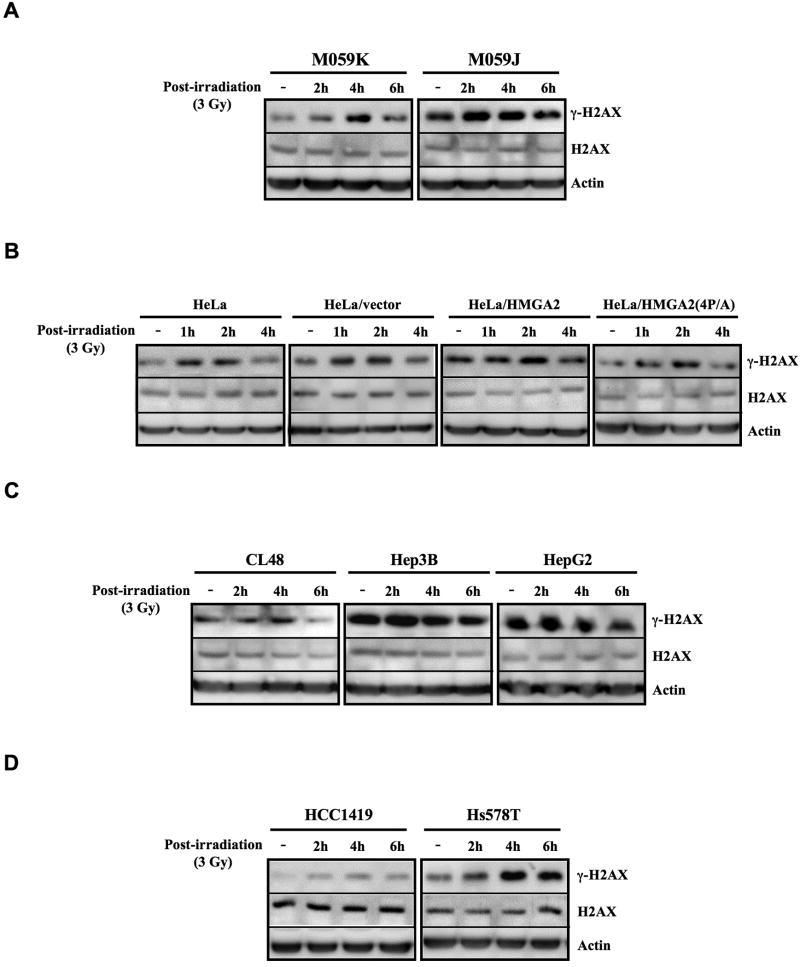
Histone H2AX rapidly undergoes phosphorylation at Ser-139 (γ-H2AX) in response to DSB. As γ-H2AX foci provide a measure of the number of DSBs within a cell, the elimination of γ-H2AX is indicative of DSB repair (25). Accumulation of γ-H2AX in DSB repair has been demonstrated as an apparent DNA repair deficit caused by DNA-PK inhibitors (26) or DNA-PK-deficiency following irradiation (27). Accordingly, we asked whether HMGA2 also modulated γ-H2AX clearance rates at various time points post X-ray exposure. DNA-PKcs-deficient M059J cells exhibited marked increases in the level of γ-H2AX both prior to and post irradiation as compared to their DNA-PKcs-proficient counterparts, M059K cells (Figure 3A). Although M059K and M059J cells are isolated from the same tumor specimen, DNA-PK-proficient M059K cells express higher levels of ATM than M059J cells (28). To rule out the effect of ATM on γ-H2AX clearance, we monitored the disappearance of γ-H2AX in HeLa cells with distinct HMGA2 contexts. Stable expression of HMGA2, but not HMGA2(4P/A), rendered enhanced and prolonged phosphorylation of H2AX post irradiation compared to HeLa and HeLa/vector cells (Figure 3B). To avoid artifact from our engineered HeLa/HMGA2 cells, profiles for γ-H2AX clearance in various cancer cell lines that are endogenously overexpressing HMGA2 were then examined to realistically reflect the physiological relevance of HMGA2 in tumor cells. Higher levels of γ-H2AX were detected in extracts both prior to and post irradiation in Hep3B and HepG2 (HMGA2-expressing) compared to CL48 cells (Figure 3C), presumably due to the slower rate and/or reduced extent of clearance in Hep3B and HepG2 cells. The higher basal level of γ-H2AX and its delayed clearance after irradiation were also reproduced in HMGA2-expressing Hs578T, but not in HMGA2-underexpressing HCC1419, breast cancer cells (Figure 3D). These findings demonstrated that endogenous expression of HMGA2 in tumor cells induced higher basal levels of γ-H2AX and delayed γ-H2AX elimination post irradiation, a phenotype recapitulating DNA-PK deficiency (Figure 3A).
Figure 3. HMGA2 delayed clearance of γ-H2AX post X-ray exposure.
Western blot analysis of γ-H2AX elimination in (A) DNA-PK(+/+) M059K and DNA-PK(-/-) M059J cells; (B) HeLa, HeLa/vector, HeLa/HMGA2, and HeLa/HMGA2(4P/A) cells; (C) HMGA2-non-expressing CL48, HMGA2-expressing Hep3B and HepG2 hepatoma cells; and (D) HMGA2-non-expressing HCC1419 and HMGA2-expressing Hs578T breast cancer cells. Cells were either nonirradiated or irradiated with 3-Gy X-rays; protein lysates were prepared at different time points post X-ray exposure as indicated.
HMGA2 causes spontaneous chromosome aberrations in WI-38 cells
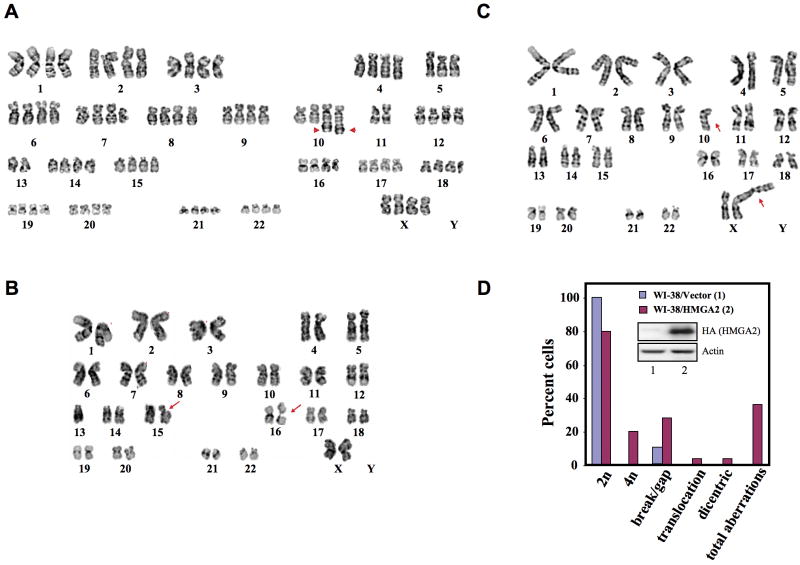
For proof-of-concept that HMGA2 renders chromosome instability, we ectopically expressed HMGA2 or a vector in normal human lung fibroblast WI-38 cells via lentiviral transduction to generate WI-38/HMGA2 and WI-38/vector cells, respectively. HMGA2 expression in transduced cells was confirmed by Western analysis (Figure 4D, inset), and both vector- and HMGA2-transduced cells were pooled and passaged twice, at 1:3 dilution, prior to cytogenetic studies to avoid cloning bias. Twenty-five randomly selected metaphases from each pool were analyzed by cytogenetical characterizations to screen for chromosome aberrations.
Figure 4. Ectopic expression of HMGA2 in WI-38 cells induces chromosomal aberrations.
(A) A near 4n WI38/HMGA2 cell with part of chromosome 13 translocated to chromosome 10 (arrows), der(10)t(10;13)(q26;q12)×2. (B) Chromosomal breaks at chromosomes 15 and 16 (arrows), observed in WI-38/HMGA2 cells. (C) A dicentric chromosome with chromosomes 10 and X fused together (arrows). (D) Quantification of chromosome aberrations detected in ectopically expressed HMGA2 in WI-38 cells. Inset, expression of HMGA2.
Of the 25 WI-38/HMGA2 cells, five showed a karyotype with 92 chromosomes, 4n (Supplementary Figure S5B). In addition, nine chromosome aberrations, including one translocation (Figure 4A), seven nonclonal chromosome breaks and gaps (Figure 4B), and one dicentric chromosome (Figure 4C) were documented. Some metaphases had more than one event of a particular type of chromosome aberration (Figure 4A). Parallel studies of 25 WI-38/vector cells all showed a normal female karyotype within the limits of banding resolution (Supplementary Figure S5A) with three cells showing minimal instability of a single break in the centromeric region. The percentage of metaphases with normal 2n or with any structural abnormality in WI-38/HMGA2 and WI-38/vector cells is summarized in Figure 4D. These results unequivocally demonstrated that overexpression of HMGA2 promotes the accumulation of chromosomal aberrations.
Discussion
Herein, we present evidence in support of the fact that a distinguishing feature of HMGA2 oncoprotein is to dysregulate DNA damage response by impeding NHEJ, leading to genomic instability, a hallmark of tumorigenesis (15). The majority of DSB joining is completed in a rapid manner to restore the structural integrity of the chromosome (21, 29), thus preventing illegitimate joining of chromosomes containing unrepaired DSBs. Cui et al. demonstrated that the presence of DNA-PKcs at DNA ends effectively blocked the access of either processing nucleases or ligases (23). Importantly, DNA-PKcs mutants, incapable of repairing DSBs, displayed rapid initial accumulation, identical to wt DNA-PKcs, but delayed disappearance from the DSB sites (21). In agreement, the failure to release DNA-PKcs from DSB sites in HMGA2-expressing cells (Figure 2) likely reflected the HMGA2-conferred inability of NHEJ to timely repair DNA lesions.
There are several other possibilities that could account for the observations that HMGA2 facilitates DNA lesion accumulation. For example, HMGA2 could affect HR repair or alter chromatin structures. Although HR involvement was not directly examined, it was effectively ruled out as all experiments were performed using cells enriched in G1-phase. While there are several lines of evidence to suggest that HMGA2 is associated with senescence associated heterochromatic foci (30) and heterochromatin serves as a barrier for NHEJ to repair DSB (31), we demonstrated that HMGA2 impaired DSB repair in several cancer cell lines from different origins (Figures 1 and 3). As cellular senescence is proposed to prevent tumorigenesis in vivo (32), we postulated that the accumulation of HMGA2-mediated DNA lesions in HMGA2-expressing cancer cells was not likely result from an HMGA2-associated increase in heterochromatin. Moreover, our biochemical evidence demonstrated hyper-phosphorylation of DNA-PKcs (Figure 1A-C) and a decrease in the steady-state complex of Ku80 with DNA ends (Figure 1D). Recently, Chen et al. reported that treatment with histone deacetylase inhibitor SAHA increased γ-H2AX accumulation and further demonstrated that Ku70 was a target of SAHA-treatment (33). Acetylation of Ku70 following SAHA treatment resulted in the decrease of Ku70 binding to DNA ends. Furthermore, our preliminary in vitro results confirmed that pre-treatment of SAHA only affected DNA-binding of Ku in HeLa, but not HeLa/HMGA2 cells (data not shown). Therefore, it is also possible that acetylated Ku70 (due to HMGA2-repressed HDAC activities (data not shown)) affected the dislodging of DNA-PKcs from DSB. Although the exact sequence of events associated with HMGA2 is not well-established, our data clearly demonstrated that HMGA2 impaired DNA-PK function and therefore diminished cellular ability to repair DSBs.
Chromosomal aberrations, a karyotypic hallmark of oncogenesis, come in many forms and contribute to neoplasia. Ferguson et al. reported that Ku70-/- and ligase 4-/- MEFs exhibited dramatic increases in spontaneous chromosomal fragmentation and nonreciprocal translocation compared with wt controls (34). The role of NHEJ in maintaining genomic stability was further confirmed by recent observations that oncogenic c-myc/IgH translocations were observed in the absence of Ku or Xrcc4 (35) and that formation of NHEJ-derived chromosomal translocation occurred in the absence of Ku70 (36). The high rate of chromosomal breaks and/or gaps in WI-38/HMGA2 cells (Figure 4B) implicates HMGA2 expression in defects in DNA repair and eventual genome instability. Furthermore, one of the most obvious differences between normal and cancer cells is aneuploidy. A wealth of data suggests that tetraploidy (4n) is a genetically unstable intermediate that can precede the development of gross chromosomal aneuploidy (37). In agreement with Figure 4D, DNA-PKcs-impaired cells exhibited DNA content beyond 4n (38). The frequent appearance of cells with 4n DNA in WI-38/HMGA2 cells (Figure 4D) supports a role of HMGA2 in providing a route to aneuploidy. Aside from repair, DNA-PK and Ku are also involved in masking chromosome ends to prevent them from being recognized as DSBs (39). HMGA2 could potentially affect the role of DNA-PK and Ku at the telomere and eventually lead to chromosome end-to-end fusion (Figure 4C). These observations suggest that HMGA2 promotes neoplastic transformation or malignancy due to its effect on the DNA damage response, hence underscoring the oncogenic role of HMGA2.
Current research suggests DNK-PK is not only involved in DNA repair, but is also a molecular sensor for DNA damage, enhancing the signal via phosphorylation of many downstream targets (40). NHEJ has been reported to be involved in surviving topoisomerase II (topo-II)-mediated DNA damage (41), and Dox is known to intercalate with DNA and interact with topo-II (42). Our observations that HMGA2-expressing cells were more sensitive to Dox treatment (Supplementary Figure S2) supported the contention that HMGA2 interfered with DNA-PK and NHEJ. Consistent with our observations, Friesen et al. demonstrated that inhibition of DNA-PK by Wortmannin overcame Dox-resistance in Nalm6 cells (43). Conceivably, HMGA2 impairs NHEJ repair, which fails to efficiently repair DSBs, leading to sensitivity to DNA-damaging agents. This is in agreement with the decrease in cell viability in response to Dox and cisplatin observed in HMGA2-expressing Hep3B and HepG2, but not HMGA2-underexpressing CL48 cells (Supplementary Figure S2A). Moreover, HMGA2 failed to convey enhanced sensitivity towards the anti-microtubulin agent taxol (Supplementary Figure S2D), further confirming that HMGA2 specifically affects cellular responses to DSBs. In light of that DSB-induced genome instability is believed to be important for tumor progression (15, 34), it is conceivable that the activation of apoptosis pathway serves the purpose to eliminate cells with chromosomal damage that risk errant repair and oncogenic transformation. Apoptosis induction was demonstrated in HMGA2-expressing cells (data not shown); therefore, it is easy to appreciate the dual role of HMGA2 in both facilitating oncogenic transformation and favoring DNA-damage-induced cell death. This raises an intriguing therapeutic issue while HMGA2 sensitizes tumor cells to radiation or chemotherapy; HMGA2 also increases the risk of further transformation. This contention is supported by the observation that the enriched breast cancer self-renewing tumor-initiating cells express higher level of HMGA2 (44). Furthermore, the accumulation of basal DSBs as evidenced by an increase in Olive Tail Moment in Pa-4/HMGA2 cells (Supplementary Figure S1A) provides mechanistic support for the observation that HMGA2 increases spontaneous chromosomal aberrations in WI-38/HMGA2 cells (Figure 4), supporting our contention that HMGA2-expressing tumors progress faster by accumulating additional mutations caused by DSBs.
In summary, the present study provides new findings concerning HMGA2 oncogenic function. First and foremost, HMGA2 was shown to be a regulator of NHEJ that impairs DNA-PK dynamics by altering Ku binding to DNA ends. Second, the inhibition of NHEJ by HMGA2 also facilitated the accumulation of chromosomal aberrations, highlighting the importance of HMGA2 as a molecular switch that promotes tumorigenesis. Further delineating the molecular signaling network connecting HMGA2 with the DNA damage response is an exciting area for future study that could impact the development of therapeutics against HMGA2-positive malignancies.
Supplementary Material
Acknowledgments
We thank Drs. Michael Lieber and Hsiu-Ming Shih for critical reading and valuable suggestions; members of Ann's laboratory, Drs. Lucio Comai, Michael A. O'Reilly, Sheau-Yann Shieh, and Kinya Hotta for sharing reagents and helpful discussion, Dr. Shikha Gaur for assistance in the X-irradiation experiments, Dr. Silvia R. da Costa for editing and NIH Research Grants R01 DE 10742 and DE 14183 (to DK Ann), CA 72767 (to Y Yen), and CA 50519 (to DJ Chen). LM Boo is a recipient of Post-Doctoral Training Fellowship (T32 CA 09659).
References
- 1.Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review) Int J Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 2.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–4. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 3.Monzen K, Ito Y, Naito AT, et al. A crucial role of a high mobility group protein HMGA2 in cardiogenesis. Nat Cell Biol. 2008;10:567–74. doi: 10.1038/ncb1719. [DOI] [PubMed] [Google Scholar]
- 4.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–39. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fedele M, Visone R, De Martino I, et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–71. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 6.Zaidi MR, Okada Y, Chada KK. Misexpression of full-length HMGA2 induces benign mesenchymal tumors in mice. Cancer Res. 2006;66:7453–9. doi: 10.1158/0008-5472.CAN-06-0931. [DOI] [PubMed] [Google Scholar]
- 7.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 8.Young AR, Narita M. Oncogenic HMGA2: short or small? Genes Dev. 2007;21:1005–9. doi: 10.1101/gad.1554707. [DOI] [PubMed] [Google Scholar]
- 9.Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008;283:33437–46. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–9. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 13.Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell. 2001;8:1163–74. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 14.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 15.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–54. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 16.Boo LM, Lin HH, Chung V, et al. High mobility group A2 potentiates genotoxic stress in part through the modulation of basal and DNA damage-dependent phosphatidylinositol 3-kinase-related protein kinase activation. Cancer Res. 2005;65:6622–30. doi: 10.1158/0008-5472.CAN-05-0086. [DOI] [PubMed] [Google Scholar]
- 17.Zentner MD, Lin HH, Deng HT, Kim KJ, Shih HM, Ann DK. Requirement for high mobility group protein HMGI-C interaction with STAT3 inhibitor PIAS3 in repression of alpha-subunit of epithelial Na+ channel (alpha-ENaC) transcription by Ras activation in salivary epithelial cells. J Biol Chem. 2001;276:29805–14. doi: 10.1074/jbc.M103153200. [DOI] [PubMed] [Google Scholar]
- 18.Chan DW, Chen BP, Prithivirajsingh S, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–8. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen JL, Lin HH, Kim KJ, Lin A, Forman HJ, Ann DK. Novel roles for protein kinase Cdelta-dependent signaling pathways in acute hypoxic stress-induced autophagy. J Biol Chem. 2008;283:34432–44. doi: 10.1074/jbc.M804239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore SR, Ritter LE, Gibbons CF, Grosovsky AJ. Spontaneous and radiation-induced genomic instability in human cell lines differing in cellular TP53 status. Radiat Res. 2005;164:357–68. doi: 10.1667/rr3422.1. [DOI] [PubMed] [Google Scholar]
- 21.Uematsu N, Weterings E, Yano K, et al. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–29. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–15. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol. 2005;25:10842–52. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turchi JJ, Henkels KM, Zhou Y. Cisplatin-DNA adducts inhibit translocation of the Ku subunits of DNA-PK. Nucleic Acids Res. 2000;28:4634–41. doi: 10.1093/nar/28.23.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004;3:959–67. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Cowell IG, Durkacz BW, Tilby MJ. Sensitization of breast carcinoma cells to ionizing radiation by small molecule inhibitors of DNA-dependent protein kinase and ataxia telangiectsia mutated. Biochem Pharmacol. 2005;71:13–20. doi: 10.1016/j.bcp.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 27.Koike M, Sugasawa J, Yasuda M, Koike A. Tissue-specific DNA-PK-dependent H2AX phosphorylation and gamma-H2AX elimination after X-irradiation in vivo. Biochem Biophys Res Commun. 2008;376:52–5. doi: 10.1016/j.bbrc.2008.08.095. [DOI] [PubMed] [Google Scholar]
- 28.Sturgeon CM, Knight ZA, Shokat KM, Roberge M. Effect of combined DNA repair inhibition and G2 checkpoint inhibition on cell cycle progression after DNA damage. Mol Cancer Ther. 2006;5:885–92. doi: 10.1158/1535-7163.MCT-05-0358. [DOI] [PubMed] [Google Scholar]
- 29.Riballo E, Kuhne M, Rief N, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–24. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 30.Funayama R, Saito M, Tanobe H, Ishikawa F. Loss of linker histone H1 in cellular senescence. J Cell Biol. 2006;175:869–80. doi: 10.1083/jcb.200604005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodarzi AA, Noon AT, Deckbar D, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–77. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 32.Braig M, Schmitt CA. Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res. 2006;66:2881–4. doi: 10.1158/0008-5472.CAN-05-4006. [DOI] [PubMed] [Google Scholar]
- 33.Chen CS, Wang YC, Yang HC, et al. Histone deacetylase inhibitors sensitize prostate cancer cells to agents that produce DNA double-strand breaks by targeting Ku70 acetylation. Cancer Res. 2007;67:5318–27. doi: 10.1158/0008-5472.CAN-06-3996. [DOI] [PubMed] [Google Scholar]
- 34.Ferguson DO, Sekiguchi JM, Chang S, et al. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci U S A. 2000;97:6630–3. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jankovic M, Nussenzweig A, Nussenzweig MC. Antigen receptor diversification and chromosome translocations. Nat Immunol. 2007;8:801–8. doi: 10.1038/ni1498. [DOI] [PubMed] [Google Scholar]
- 36.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–81. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–62. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 38.Kuhne C, Tjornhammar ML, Pongor S, Banks L, Simoncsits A. Repair of a minimal DNA double-strand break by NHEJ requires DNA-PKcs and is controlled by the ATM/ATR checkpoint. Nucleic Acids Res. 2003;31:7227–37. doi: 10.1093/nar/gkg937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–10. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 40.Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24:949–61. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- 41.Malik M, Nitiss KC, Enriquez-Rios V, Nitiss JL. Roles of nonhomologous end-joining pathways in surviving topoisomerase II-mediated DNA damage. Mol Cancer Ther. 2006;5:1405–14. doi: 10.1158/1535-7163.MCT-05-0263. [DOI] [PubMed] [Google Scholar]
- 42.Bodley A, Liu LF, Israel M, et al. DNA topoisomerase II-mediated interaction of doxorubicin and daunorubicin congeners with DNA. Cancer Res. 1989;49:5969–78. [PubMed] [Google Scholar]
- 43.Friesen C, Uhl M, Pannicke U, Schwarz K, Miltner E, Debatin KM. DNA-ligase IV and DNA-protein kinase play a critical role in deficient caspases activation in apoptosis-resistant cancer cells by using doxorubicin. Mol Biol Cell. 2008;19:3283–9. doi: 10.1091/mbc.E08-03-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.