Abstract
The ability of cells to sense and respond to mechanical force underlies diverse processes such as touch and hearing in animals, gravitropism in plants, and bacterial osmoregulation1, 2. In bacteria, mechanosensation is mediated by the mechanosensitive channels of large (MscL), small (MscS), potassium-dependent (MscK), and mini (MscM) conductances. These channels act as “emergency relief valves” protecting bacteria from lysis upon acute osmotic downshock3. Among them, MscL has been intensively studied since the original identification and characterization 15 years ago by Kung and co-workers4. MscL is reversibly and directly gated by changes in membrane tension. In the open state, MscL forms a nonselective 3 nS-conductance channel which gates at tensions close to the lytic limit of the bacterial membrane. An earlier crystal structure at 3.5 Å resolution of a pentameric MscL from Mycobacterium tuberculosis (TbMscL) represents a closed-state or nonconducting conformation5, 6. MscL has a complex gating behaviour; it exhibits several intermediates between the closed and open states, including one putative nonconductive expanded state and at least three sub-conducting states7. Although our understanding of the closed5, 6 and open8-10 states of MscL has been increasing, little is known about the structures of the intermediate states despite their importance in elucidating the complete gating process of MscL. Here we present the crystal structure of a truncation mutant (Δ95-120) of MscL from Staphylococcus aureus (SaMscL-CΔ26) at 3.8 Å resolution. Strikingly, SaMscL-CΔ26 forms a tetrameric channel with both transmembrane (TM) helices tilted away from the membrane normal at angles close to that inferred for the open state9, likely corresponding to a nonconductive but partially expanded intermediate state.
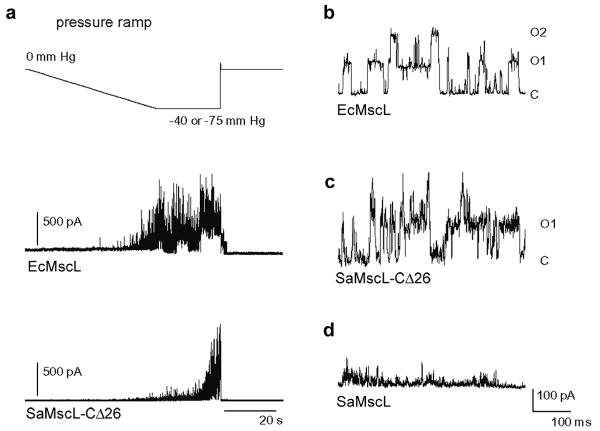
The full-length SaMscL protein is 120 amino acid residues long and shares 40% and 51% homology with TbMscL and the Escherichia coli MscL (EcMscL), respectively. Initial electrophysiological characterization of SaMscL by Moe et al11 established that it exhibited characteristic membrane tension dependent MscL activity with significantly shorter open dwell times compared to EcMscL. Excised patches from azolectin liposomes containing SaMscL-CΔ26 show a stereotypical pressure dependent increase in channel activity, similar to patches containing wild-type EcMscL, with a single channel conductance comparable to EcMscL (Fig. 1). The open state of SaMscL-CΔ26 appears more stable than that of wild-type SaMscL (Fig. 1c and d), suggesting a role for the carboxy-terminal truncation in the gating behaviour change. Nevertheless, as shown in supplementary Figure 1, both full-length and CΔ26 SaMscL are capable of rescuing a mechanosensitive channel knockout strain of E. coli cells from osmotic downshock12, indicating that they are both functional in vivo. Consequently, truncation of 26 residues from the carboxy-terminus of SaMscL does not impair its function, consistent with previous reports for EcMscL13, 14.
Figure 1. SaMscL-CΔ26 is a pressure sensitive channel.
a, Pressure sensitive response of EcMscL and SaMscL-CΔ26 proteins reconstituted into azolectin. Excised patches were held at +60 mV and pressure clamped from 0 to -40 mm Hg (EcMscL) or -75 mm Hg (SaMscL-CΔ26). In both cases, channel activity steadily increases with greater negative pressure and decreases when the pressure is removed. b-d, Single channel recordings of EcMscL (b), SaMscL-CΔ26 (c), and wild-type SaMscL (d). EcMscL and SaMscL-CΔ26 channels open and close with discreet jumps in current. Calculated single channel conductances for EcMscL and SaMscL-CΔ26 are 2.9 and 2.6 nS respectively. In contrast, single channel SaMscL openings flicker rapidly and are too brief to calculate a conductance, as observed previously11. C denotes the closed state, while O1 and O2 indicate the opening of one and two channels, respectively.
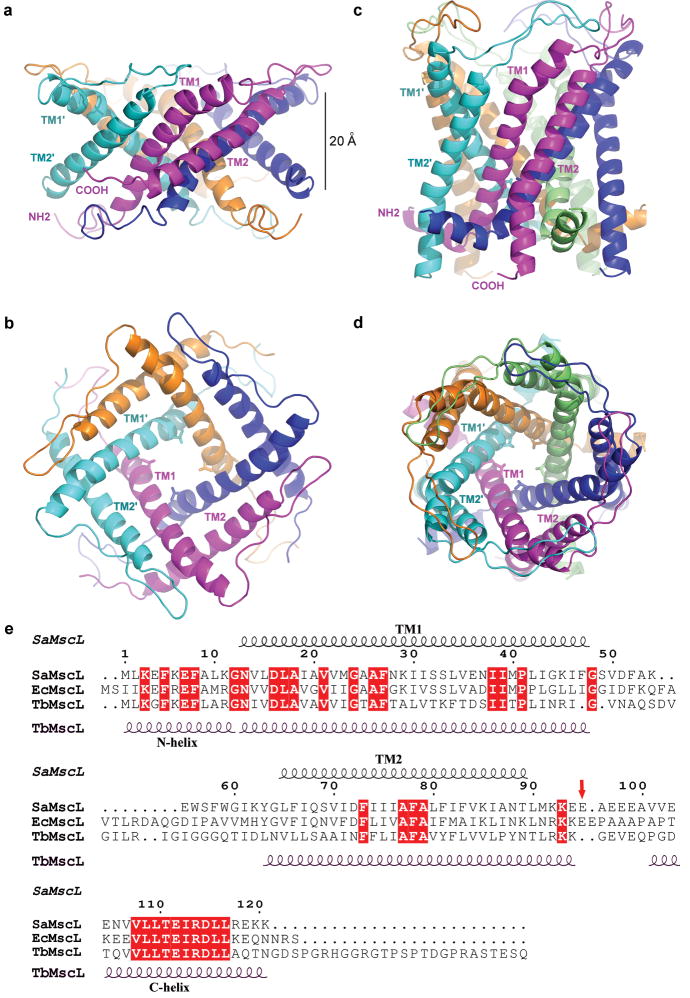
SaMscL-CΔ26 forms a tetramer in the crystal with the molecular symmetry axis coincident with the crystallographic four-fold axis (supplementary Fig. 2), so that the asymmetric unit contains one subunit. The SaMscL-CΔ26 tetramer measures ∼69 Å wide in diameter and ∼37 Å thick (Fig. 2a, b), while the TbMscL pentamer, omitting carboxyl-terminal residues 102-151 that form a cytoplasmic helical bundle, measures ∼52 Å wide and ∼50 Å thick (Fig. 2c, d). Excluding the carboxy-terminal residues which have been deleted in SaMscL-CΔ26, the major differences between the amino acid sequences of SaMscL and TbMscL are localized to the periplasmic ends of TM1 and TM2 and their connecting loop. In vitro crosslinking experiments indicate that both CΔ26 and full-length SaMscL are tetrameric in detergent solution and that removal of the carboxy-terminal domain does not change the oligomeric state of SaMscL (supplementary Fig. 3). Although unexpected, it is not unprecedented that certain multimeric membrane proteins form distinct oligomers in different species. For example, the CorA Mg2+ transporter forms a pentamer in Thermotoga maritima and a tetramer in Bacillus subtilis and E. coli15, while the bacterial light-harvesting complex II may be either octameric or nonameric16. The ring of c subunits of the membrane embedded F0-ATPase also exhibits variable stoichiometry17.
Figure 2. Structures of SaMscL-CΔ26 and TbMscL.
a and b, The crystal structure of tetrameric SaMscL-CΔ26 viewed along the membrane plane and the pore axis (from the periplasmic side), respectively. c and d, The membrane spanning domain of the TbMscL structure (residues 1-101 of PDB set 2OAR) viewed as in a and b. Val 21 is presented as a stick model while the polypeptide chains are shown as ribbons. e, Structure-based sequence alignment of SaMscL, EcMscL and TbMscL. The red arrow indicates the truncation point (Glu 95) of the SaMscL-CΔ26 mutant.
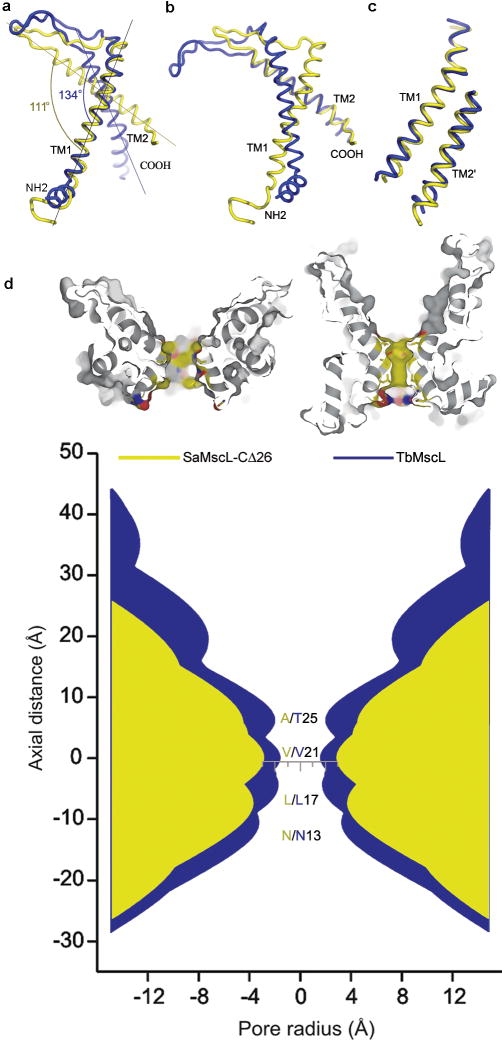
Figure 3. Alignment of SaMscL-CΔ26 (yellow) and TbMscL (blue).
a, b, Intrasubunit conformational change visualized by aligning the two monomeric structures on TM1 and TM2, respectively. c, The superposition of intersubunit TM1-TM2′ pair from the two structures. d, The pore profiles of SaMscL-CΔ26 and TbMscL along the central axes. The top two surface presentations are the sectional views of SaMscL-CΔ26 (left) and TbMscL (right) with the atoms around the constriction colored in yellow, red and blue for carbon, oxygen and nitrogen respectively. In the bottom profiles, the regions corresponding to SaMscL-CΔ26 and TbMscL are colored yellow and blue, respectively. The residues lining the constriction are labeled in the same color codes.
The secondary structure of the TM region of SaMscL-CΔ26 resembles that of TbMscL (Fig. 2e) with a few exceptions. TM1 of SaMscL-CΔ26 begins at the cytoplasmic surface and extends from Asn 13 to Phe 47, kinking near Pro 41 on the periplasmic side. The polypeptide chain continues through the periplasmic loop from Gly 48 to Gly 64 and crosses the membrane again through TM2 from Leu 65 to Leu 90. TM1 has the same length as in TbMscL, while the periplasmic loop and TM2 are 4 residues and 7 residues shorter respectively, than the corresponding parts of TbMscL. The largest changes in the two structures are the first 12 residues (which adopt an irregular structure in SaMscL-CΔ26 but a short α-helix in TbMscL) and the detailed conformations of the periplasmic loop. In addition, the intrasubunit crossing angle between TM1 and TM2 changes from 134° in TbMscL to 111° in SaMscL-CΔ26 (Fig. 3a, b). An overlay of TbMscL and SaMscL-CΔ26 monomers on TM1s suggests a counterclockwise pivoting of TM2 away from the membrane normal and a translation towards the periplasmic surface (Fig. 3a). In both structures, TM1 and TM2 from the adjacent subunit (TM2′) form an antiparallel pair of helices with crossing angle = 169/185° in SaMscL-CΔ26/TbMscL. The TM1-TM2′ pair of SaMscL-CΔ26 aligns well with that of TbMscL (root mean square deviation of α-carbon atoms = 1.5 Å) indicating this pair might move as a rigid body during the conformational change (Fig. 3c and ref.18, 19). The extensive network of largely conserved hydrophobic residues at the TM1-TM2′ interface presumably reflects the stability of this antiparallel helical pair (supplementary Fig. 4).
The SaMscL-CΔ26 tetramer is ∼13 Å thinner along the membrane normal than the membrane spanning domain of TbMscL pentamer but up to 17 Å wider on the periplasmic surface. The periplasmic surface area of tetrameric SaMscL-CΔ26 is ∼2200 Å2, while that of pentameric TbMscL is ∼2000 Å2. As such, the membrane spanning region of SaMscL-CΔ26 tetramer is expanded within the membrane plane and compressed along the pore axis as compared to the TbMscL pentamer (Fig. 2 a-d). This expansion is also evident in the pore profiles of both channels (Fig. 3d). The constriction of SaMscL-CΔ26 at Val 21 is widened to ∼6 Å diameter as compared to ∼3 Å in TbMscL. Ala 25 and Leu 17, positioned to either side of Val 21 in the permeation pathway, move further away from the central axis, opening the pore to ∼8 Å around those residues. The corresponding residues in TbMscL, Thr 25 and Leu 17, respectively, lie closer to the axis and hence the pore diameter in this structure narrows to ∼4 Å in this region. Although the constriction in SaMscL-CΔ26 is wider than in TbMscL, theoretical studies indicate hydrophobic pores with diameters less than 9 Å and 13 Å are impermeable to water and ions20, respectively, implying that both structures are likely non-conducting since they each have narrow and hydrophobic pore constrictions. The expanded state of SaMscL-CΔ26 channel reflects the higher tilt angles of TM1 and TM2, relative to the membrane normal. In SaMscL-CΔ26, TM1 and TM2 are tilted 49° and 59° with respect to the pore axis, significantly larger than the 36-38° for the TM helices of TbMscL6, and close to the angles in an open-state model of EcMscL (∼51° for TM1/TM2)9. Consequently, the crossing angle between the axes of two adjacent TM1s in SaMscL-CΔ26 (-63°) changes by ∼-21° relative to -42° in TbMscL. The combination of the constricted pore and expanded conformation suggests SaMscL-CΔ26 represents an intermediate state during the transition from the resting closed state to the sub-conducting or open states.
The SaMscL-CΔ26 structure is clearly expanded in the crystal, but why was this conformation trapped? It is possible that removal of 26 residues from the carboxy-terminus lowers the energy barrier for the transition from the closed to expanded state and crystal packing facilitates this specific expanded conformation. There are several lines of evidence consistent with this idea. Examination of the TbMscL structure suggests that the carboxy-terminal bundle acts as a plug or pre-filter to the permeation pathway which may dissociate to permit the passage of large solutes, including small proteins21-23, through the channel. This is consistent with recent electron microscopy work, which showed that a pore is present at the center of a gain-of-function mutant (G22N) of EcMscL, whereas the cytoplasmic protrusion is absent24. We suggest that the carboxy-terminal helix bundle stabilizes the channel in the closed state and restrains the TMs from tilting into the expanded conformation. The dissociation of this bundle triggered by the mechanical force transmitted from the lipid bilayer removes the restraint, allowing the TMs to tilt and transit to the expanded state. Indeed, SaMscL-CΔ26 reconstituted in liposomes is more stable in the open state than the wild-type channel, which displays fast flickering from the closed to open state11 (Fig. 1c and d), suggesting that the truncation mutant may sample the expanded state more readily under crystallization conditions. The relatively small detergent micelle of n-dodecyl-N,N-dimethylamine-N-oxide (LDAO)25 might also favor the expanded state by exerting curvature stress on the channel.
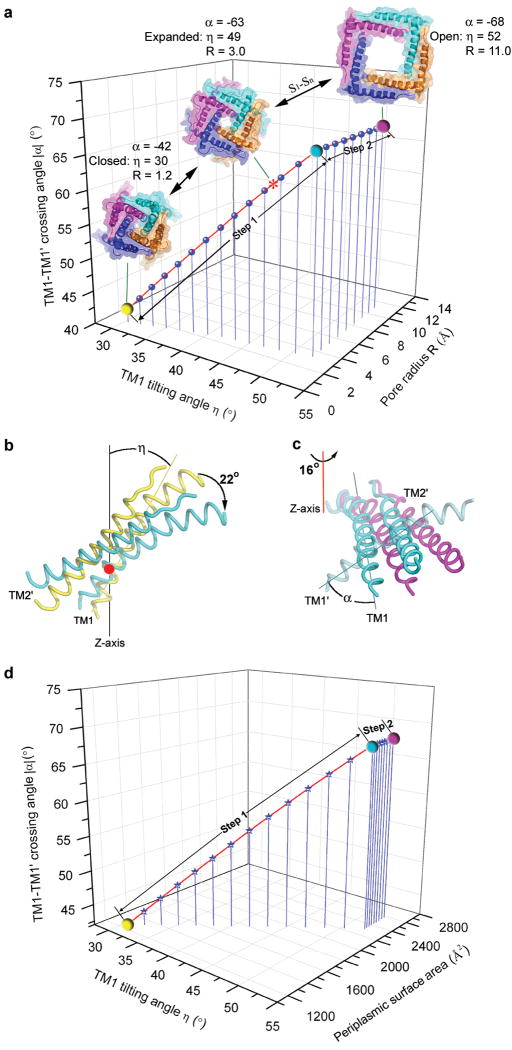
To model the gating process of MscL, a series of structural models of tetrameric SaMscL from closed to open state were constructed using relationships between the helix tilting angles (η), interhelical-crossing angles (α) and the minimum pore radius (R) of the oligomeric proteins26. We use equations cosα = cos2η + sin2ηcosθ and R = d(tanη cot(α/2) − 1)/2, where θ = 360°/N with N = 4 or 5 for a tetramer or pentamer and d is the helix diameter26, as the basis for modelling a series of tetrameric structures with the α angles determined from the pentameric structures at different states. Here we propose a two step helix-pivoting model of SaMscL gating. In this model, it is assumed that the TM1-TM2′ pair rotates and shifts as a rigid body18, 19. The first pivoting step occurs during the transition from the closed state to a state near the expanded intermediate (Fig. 4a, b). TM1 pivots clockwise about Val 21 by ∼22° and slides along the surface of TM2 (of the same subunit) from Gln 68 down to Ile 75. This induces TM2 to rotate counterclockwise about the pore axis and incline toward the membrane plane along with TM1. Increasing the TM1 tilt angle requires that the helices move farther apart to maintain a symmetric oligomer. As TM2 is in close interaction with TM1 from the next subunit, these movements are synchronized throughout the tetramer. The crossing angle between two adjacent TM1s increases from -42° to -68° at this stage. This angle reaches -67° in a previous open-state model8, 18 and -55° in an alternative model9, bracketing the value (-63°) observed in the expanded state structure, suggesting that it is likely near the edge of the open state. At the end of this first step, the periplasmic loop becomes fully stretched between intrasubunit TM1 and TM2, and if tilting continues, the channel would become thinner than the membrane.
Figure 4. The two-step helix-pivoting model of SaMscL gating.
a, The relationship among TM1 tilting angle η, TM1-TM1′ crossing angle α, and the pore radius R at Val 21 derived from the gating models. The * symbol indicates the position of the SaMscL-CΔ26 structure on the curve. The yellow, cyan and magenta spheres indicate the positions of the resting closed, turning-point intermediate and open states on the curve. S1-Sn denotes the multiple subconducting states between the expanded and open states. At step 1, η and α are greatly changed due to the helix tilting motion, while R is only slightly increased. As the helices swing away from the pore axis at the step 2, R increases rapidly, while η and α remain constant. b, The first pivoting step of TM1-TM2′ occurs about Val 21 (red point). The pair pivots as a rigid body rotating 22° from the resting closed (yellow, based on TbMscL structure5, 6) to the turning-point intermediate state (cyan, near the SaMscL-CΔ26 structure). c, The second pivoting step occurs about the red-line axis through Gly 48, from the intermediate to open state (magenta, based on the Sukharev and Guy model8). d, The relationship among η, α and the periplasmic surface area of the models. At step 1, the periplasmic surface area expands more drastically than at step 2.
The constraint of the membrane thickness leads to a proposed second pivoting step, defined as a counterclockwise rotation of TM1-TM2′ pair about an axis running normal to the membrane and through Gly 48 near the carboxy-terminal end of TM1. The helices rotate by 16° (Fig. 4a and c). The highly conserved Gly 48 (Fig. 2e) appears well suited to serve as the pivot for the outward swinging of the TM1-TM2′ pair. This second-step action expands the pore diameter to 22 Å, compatible with the observed large conductance. In MscS, the pore-lining helix also pivots around a glycine residue and moves outwards from the central axis to expand the pore constriction27. The periplasmic loop of MscL provides the flexibility for the substantial relative movements of the TM1 and TM2 observed within the same subunit, but also defines the steric limit of channel expansion in the fully open state. Previous functional studies on the periplasmic loop suggested it may act as a spring, influencing channel kinetics and mechanosensitivity13, 28-30. During the first step of this transition, the periplasmic surface of the SaMscL expands significantly while the channel constriction remains essentially closed until the second step takes place (Fig. 4a, d).
In summary, we have determined the crystal structure of a carboxy-terminal truncation mutant of S. aureus MscL and demonstrated that SaMscL forms a tetrameric pressure gated channel. This structure represents a likely expanded intermediate conformation of MscL between the resting closed state and the open state. A two step helix-pivoting model of the gating mechanism has been proposed.
Methods Summary
The SaMscL gene was inserted into a pET15b vector with a stop codon at Glu 95 introduced by site-directed mutagenesis to generate the SaMscL-CΔ26 mutant. The channel was overexpressed in BL21-Gold cells and purified in the detergent LDAO by immobilized metal affinity and gel filtration chromatographies. Crystals were grown using the hanging-drop vapor diffusion method. The structure was solved by the method of multiple isomorphous replacement with anomalous scattering.
Crosslinking was performed using N,N-dicyclohexylcarbodiimide at room temperature for 30 min-1 h and the products were analyzed by SDS-polyacrylamide gel electrophoresis.
The MJF465 (mscl-, mscs-, msck-) (DE3) strain was used for the osmotic downshock assay. Cells hosting EcMscL and an empty vector were included as positive and negative control sets, respectively. For the osmotic shock, the culture was diluted 1:500 into sterile double-distilled H2O. For the mock shock, LB media with 500 mM NaCl was used instead of H2O.
MscL protein was reconstituted into giant azolectin liposomes via the dehydration/rehydration method and assayed by patch clamp electrophysiology. Patches containing at least 1-10 channels were excised and currents measured at various pressures. Recording and pipette solutions both contained 200 mM KCl, 40 mM MgCl2, 5 mM HEPES, pH 7.2.
Supplementary Material
Acknowledgments
We thank A. Shih for early cloning work, T. Walton for biochemical analysis, A. Lee for initial efforts on the MscL project, J. Choe for expression and purification protocols, O. Lewinson for manuscript reading, Y. Poon and J. Lai for treating MJF465 with λDE3 and the osmotic downshock assay protocol, J. Kaiser for suggestions on structure refinement, R. Phillips and E. Haswell for discussions, P. Blount for the MJF465 strain, and the staff at SSRL, the Advanced Light Source (ALS) and the Advanced Photon Source (APS) for technical support with crystal screening and data collection. We would like to acknowledge the Gordon and Betty Moore Foundation for support of the Molecular Observatory at Caltech. Operations at SSRL, ALS and APS are supported by the U.S. Department of Energy and NIH. This work was supported in part by grants from the Howard Hughes Medical Institute and the National Institutes of Health (GM084211). C.S.G. was supported in part by postdoctoral fellowships from the National Institutes of Health and the Beckman Foundation. D.C.R. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Author Contributions Z. F. L. designed and performed the experiments in molecular biology, biochemistry, crystallography and the structure analysis. Z.F.L. and C.S.G. conducted the downshock assays. C. S. G. was responsible for the protein reconstitution and electrophysiology. D. C. R. coordinated the project and contributed to the structure analysis. The manuscript was written by Z. F. L, C. S. G and D. C. R.
Author Information The atomic coordinates and structure factors have been deposited at the Protein Data Bank with accession numbers 3HZQ. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Kung C. A possible unifying principle for mechanosensation. Nature. 2005;436:647–654. doi: 10.1038/nature03896. [DOI] [PubMed] [Google Scholar]
- 2.Sukharev S, Corey DP. Mechanosensitive channels: multiplicity of families and gating paradigms. Sci STKE. 2004;2004:1–24. doi: 10.1126/stke.2192004re4. [DOI] [PubMed] [Google Scholar]
- 3.Booth IR, Edwards MD, Black S, Schumann U, Miller S. Mechanosensitive channels in bacteria: signs of closure? Nat Rev Micro. 2007;5:431–440. doi: 10.1038/nrmicro1659. [DOI] [PubMed] [Google Scholar]
- 4.Sukharev SI, Blount P, Martinac B, Blattner FR, Kung C. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature. 1994;368:265–268. doi: 10.1038/368265a0. [DOI] [PubMed] [Google Scholar]
- 5.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 6.Steinbacher S, Bass R, Strop P, Rees DC. In: Current Topics in Membranes Mechanosensitive Ion Channels, Part A. Hamill OP, editor. Academic Press; London: 2007. pp. 1–24. [Google Scholar]
- 7.Sukharev SI, Sigurdson WJ, Kung C, Sachs F. Energetic and spatial parameters for gating of the bacterial large conductance mechanosensitive channel, MscL. J Gen Physiol. 1999;113:525–540. doi: 10.1085/jgp.113.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sukharev S, Betanzos M, Chiang CS, Guy HR. The gating mechanism of the large mechanosensitive channel MscL. Nature. 2001;409:720–724. doi: 10.1038/35055559. [DOI] [PubMed] [Google Scholar]
- 9.Perozo E, Cortes DM, Sompornpisut P, Kloda A, Martinac B. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature. 2002;418:942–948. doi: 10.1038/nature00992. [DOI] [PubMed] [Google Scholar]
- 10.Anishkin A, Sukharev S. State-stabilizing interactions in the bacterial mechanosensitive channel gating and adaptation. J Biol Chem. 2009 doi: 10.1074/jbc.R109.009357. doi: R109.009357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moe PC, Blount P, Kung C. Functional and structural conservation in the mechanosensitive channel MscL implicates elements crucial for mechanosensation. Mol Microbiol. 1998;28:583–592. doi: 10.1046/j.1365-2958.1998.00821.x. [DOI] [PubMed] [Google Scholar]
- 12.Levina N, et al. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. Embo J. 1999;18:1730–1737. doi: 10.1093/emboj/18.7.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blount P, Sukharev SI, Schroeder MJ, Nagle SK, Kung C. Single residue substitutions that change the gating properties of a mechanosensitive channel in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:11652–11657. doi: 10.1073/pnas.93.21.11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Häse CC, Le Dain AC, Martinac B. Molecular dissection of the large mechanosensitive ion channel (MscL) of E. coli: mutants with altered channel gating and pressure sensitivity. J Membr Biol. 1997;157:17–25. doi: 10.1007/s002329900212. [DOI] [PubMed] [Google Scholar]
- 15.Niegowski D, Eshaghi S. The CorA family: structure and function revisited. Cell Mol Life Sci. 2007;64:2564–2574. doi: 10.1007/s00018-007-7174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogdell RJ, et al. The structure and function of the LH2 (B800-850) complex from the purple photosynthetic bacterium Rhodopseudomonas acidophila strain 10050. Prog Biophys Mol Biol. 1997;68:1–27. doi: 10.1016/s0079-6107(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 17.Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 18.Sukharev S, Durell SR, Guy HR. Structural models of the MscL gating mechanism. Biophys J. 2001;81:917–936. doi: 10.1016/S0006-3495(01)75751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strop P, Bass R, Rees DC. Prokaryotic mechanosensitive channels. Adv Protein Chem. 2003;63:177–209. doi: 10.1016/s0065-3233(03)63008-1. [DOI] [PubMed] [Google Scholar]
- 20.Beckstein O, Sansom MSP. The influence of geometry, surface character, and flexibility on the permeation of ions and water through biological pores. Phys Biol. 2004;1:42–52. doi: 10.1088/1478-3967/1/1/005. [DOI] [PubMed] [Google Scholar]
- 21.Cruickshank CC, Minchin RF, Le Dain AC, Martinac B. Estimation of the pore size of the large-conductance mechanosensitive ion channel of Escherichia coli. Biophys J. 1997;73:1925–1931. doi: 10.1016/S0006-3495(97)78223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ajouz B, Berrier C, Garrigues A, Besnard M, Ghazi A. Release of thioredoxin via the mechanosensitive channel MscL during osmotic downshock of Escherichia coli cells. J Biol Chem. 1998;273:26670–26674. doi: 10.1074/jbc.273.41.26670. [DOI] [PubMed] [Google Scholar]
- 23.van den Bogaart G, Krasnikov V, Poolman B. Dual-color fluorescence-burst analysis to probe protein efflux through the mechanosensitive channel MscL. Biophys J. 2007;92:1233–1240. doi: 10.1529/biophysj.106.088708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimura K, Usukura J, Sokabe M. Gating-associated conformational changes in the mechanosensitive channel MscL. Proc Natl Acad Sci USA. 2008;105:4033–4038. doi: 10.1073/pnas.0709436105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strop P, Brunger AT. Refractive index-based determination of detergent concentration and its application to the study of membrane proteins. Protein Sci. 2005;14:2207–2211. doi: 10.1110/ps.051543805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spencer RH, Rees DC. The α-helix and the organization and gating of channels. Annu Rev Biophys Biomol Struct. 2002;31:207–233. doi: 10.1146/annurev.biophys.31.082901.134329. [DOI] [PubMed] [Google Scholar]
- 27.Wang W, et al. The structure of an open form of an E. coli mechanosensitive channel at 3.45 Å resolution. Science. 2008;321:1179–1183. doi: 10.1126/science.1159262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurer JA, Elmore DE, Lester HA, Dougherty DA. Comparing and contrasting Escherichia coli and Mycobacterium tuberculosis mechanosensitive channels (MscL) J Biol Chem. 2000;275:22238–22244. doi: 10.1074/jbc.M003056200. [DOI] [PubMed] [Google Scholar]
- 29.Tsai IJ, et al. The role of the periplasmic loop residue glutamine 65 for MscL mechanosensitivity. Eur Biophys J. 2005;34:403–412. doi: 10.1007/s00249-005-0476-x. [DOI] [PubMed] [Google Scholar]
- 30.Ajouz B, Berrier C, Besnard M, Martinac B, Ghazi A. Contributions of the different extramembranous domains of the mechanosensitive ion channel MscL to its response to membrane tension. J Biol Chem. 2000;275:1015–1022. doi: 10.1074/jbc.275.2.1015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.