Abstract
Ribosomal protein S19 (RPS19) is mutated in patients with Diamond-Blackfan anemia (DBA). We hypothesized that decreased levels of RPS19 lead to a coordinated down-regulation of other ribosomal (r-)proteins at the subunit level. We show that siRNA knock-down of RPS19 results in a relative decrease of small subunit r-proteins (S20, S21 and S24) when compared to large subunit r-proteins (L3, L9, L30 and L38). This correlates with a relative decrease in 18S rRNA with respect to 28S rRNA. The r-protein mRNA levels remain relatively unchanged indicating a post-transcriptional regulation of r-proteins at the level of subunit formation.
Keywords: Diamond-Blackfan anemia (DBA), RPS19, haploinsufficiency, ribosome biogenesis, antibodies
1. Introduction
Ribosomal proteins are fundamental components in cellular metabolism and ribosome synthesis is critical for cell growth and development. The ribosomal (r-)proteins are responsible for the correct folding and cleavage of rRNA as well as for subunit assembly [1]. The first disease to be associated with mutations in a ribosomal protein is Diamond-Blackfan anemia (DBA; OMIM #205900), a rare congenital bone marrow failure characterized by decreased numbers or absence of erythroid precursor cells [2]. Approximately 25% of patients with DBA carry heterozygous mutations in the r-protein S19 (RPS19) gene [3] and other subsets of patients were recently shown to carry mutations in different r-protein genes [4–7]. Until now, a variety of mutations in RPS19 have been described [8] some of which presumably result in RPS19 haploinsufficiency [2,9]. Ribosome profiling of cells with RPS19 insufficiency show a specific reduction of the small subunit [10,11] supporting RPS19 to be critical for formation of the 40S subunit. Furthermore, induced depletion of RPS19 by means of small interfering RNAs (siRNA) [12] leads to a delay in pre-rRNA maturation and a perturbed biosynthesis of the small ribosomal subunit [10,11,13–15]. Thus, it has been suggested that insufficiency of one r-protein is rate limiting for subunit formation as this requires stoichiometric amounts of structural components [16,17].
Here, we report on the quantification of different r-proteins in i) a TF-1-B cell model with inducible siRNAs against RPS19 and ii) lymphoblastoid cell lines (LCLs) derived from RPS19 deficient DBA patients. We used a set of newly obtained antibodies directed against eight specific r-proteins to clarify mechanisms for the coordinated and subunit specific regulation of r-protein levels. We also quantified the corresponding mRNA levels as well as the levels of 18S rRNA to 28S rRNA. We show that siRNAs against RPS19 significantly reduces the levels of other small subunit (SSU) r-proteins but not large subunit (LSU) r-proteins. This correlates to a reduction in 18S rRNA whereas the levels of small r-protein mRNAs are relatively unaltered. The patient derived LCLs showed a skewed ratio of SSU to LSU r-proteins that is similar but less marked to that observed after RPS19 knock-down in TF-1 cells. Our combined data show that reduced levels of a single r-protein leads to a decrease in levels of other r-proteins at the level of subunit formation. This is independent of transcription of r-protein mRNAs and supports that subunit imbalance is a critical pathophysiological mechanism in DBA.
2. Materials and Methods
2.1. Cell Culture, siRNA induction and Western Blotting
The TF-1-B cell line was cultured as described and stably transduced with inducible siRNAs against RPS19 (kindly provided by Prof. Stefan Karlsson, Lund, Sweden) [18]. siRNA expression was induced using doxycycline and cells were harvested after 7 days of induction. EBV transformed lymphoblastoid cells (LCLs) were cultured in RPMI 1640 (GIBCO) supplemented with 10% fetal bovine serum (GIBCO), 2mM L-glutamine (GIBCO) and 20 IU/ml of penicillin and streptomycin solution (GIBCO). TF-1-B cells and lymphoblastoid cells were lysed in RIPA buffer (50mM Tris-HCl pH 7.5, 150mM NaCl, 1% Triton X-100, 1% sodium deoxycholate and 0.1% SDS) supplemented with MG132 proteasome inhibitor (SIGMA), phosphatase inhibitor cocktail 1 (SIGMA), 0.1 mM Sodium vanadate (SIGMA) and protease inhibitor cocktail (SIGMA). Cell debris was removed by centrifugation at 13 000 × g for 10 minutes at 4°C and the supernatant was collected. Cell lysates were separated on a 10% Bis-Tris SDS-PAGE (NuPage gel; Invitrogen), and transferred to PVDF Immobilon-FL membranes (Millipore) according to manufacturer’s protocols. The membranes were hybridized with primary antibodies against eight ribosomal proteins as well as fibrillarin (Abcam) and β-actin (Abcam). Proteins detected by the antibodies were visualized using Alexa Fluor 680 (α-rabbit, α-goat) and IRD 800 labeled (α-mouse) secondary antibodies (Molecular probes and LiCor Bioscience, respectively). Western blots were analyzed using the Odyssey® infrared imaging system, determining integrated intensities for each protein following the instructions manual (Li-Cor Bioscience). The relative amounts of proteins were estimated after normalization to the intensity of β–actin within the same cell line. At least two independent measurements were performed.
2.2. Quantitative RT/PCR (qRT/PCR) and rRNA quantification
Total RNA was isolated from induced and non-induced TF-1-B cells and lymphoblastoid cells using Trizol® reagent (Invitrogen). The 18S and 28S rRNAs were quantified using the Agilent RNA 6000 nano kit and the Agilent 2100 bioanalyser according to manufacturer’s instructions. cDNA was synthesized with M-MULV reverse transcriptase (MBI Fermentas) using random hexamer primers and 2 µg of total RNA following the manufacturer’s recommendations. Quantitative real-time PCR was performed in triplicates using platinum SYBR green qPCR supermix UDG (Invitrogen) according to the protocol supplied by the manufacturer.
2.3. Antibodies
Antibodies against eight human r-proteins used were raised in rabbits by the Swedish Human Protein Resource (HPR) program [19]. The following antibodies with their HPR designations were used: α-RPS19 (HPRK580005); α-RPS20 (HPA003570); α-RPS21 (HPA003371); α-RPS24 (HPA003364); α-RPL3 (HPA003365); α-RPL9 (HPA003372); α-RPL30 (HPA002651) and α-RPL38 (HPRK580013).
2.4. Statistical analysis
The results obtained from Western blots or qRT/PCR are displayed as mean ± standard deviation. The two tailed student’s t-test was employed to compare expression levels of individual r-proteins, mRNAs and ratios.
3. Results and Discussion
3.1. siRNA- mediated knock-down of RPS19 results in reduced levels of small subunit r-proteins and 18S rRNA but not in levels of small subunit r-protein mRNAs
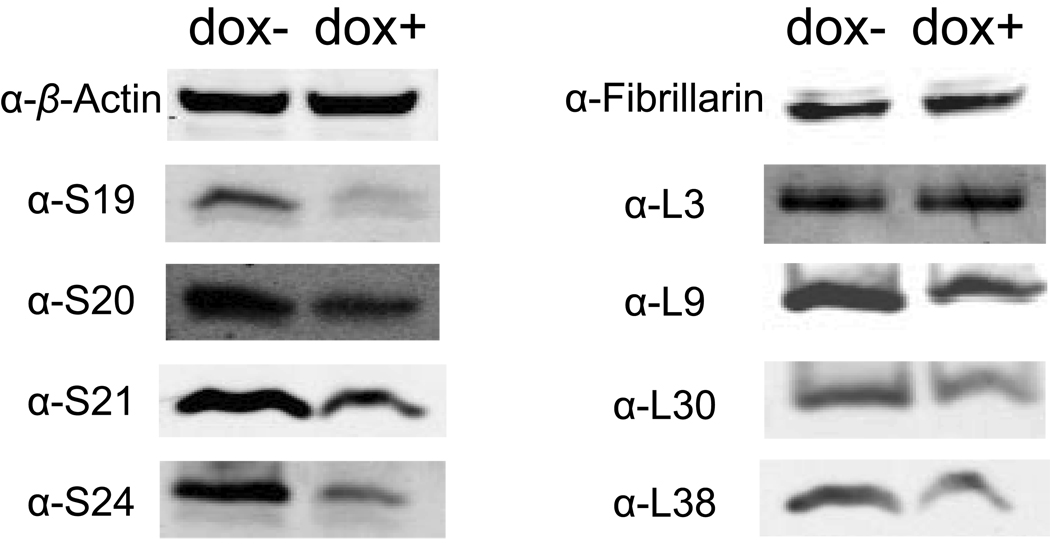
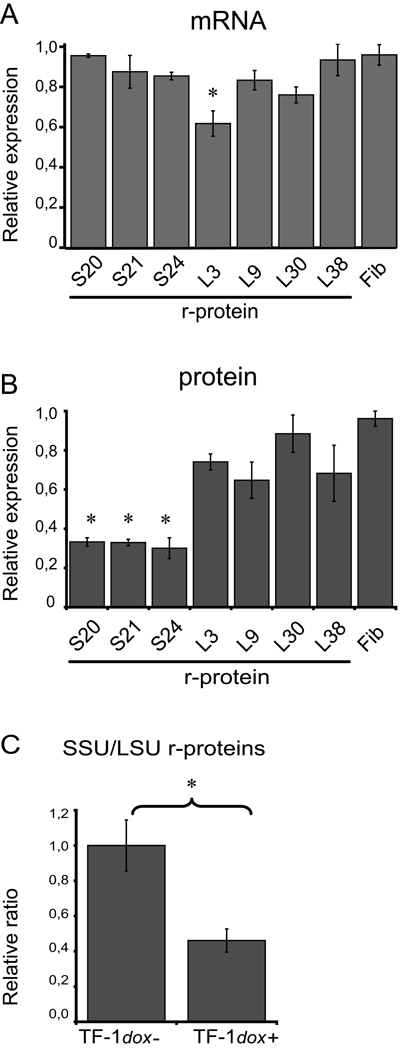
Approximately 25% of patients with Diamond-Blackfan anemia carry heterozygous mutations in RPS19 some of which presumably result in haploinsufficiency [8,9]. The molecular mechanisms mediated by r-protein haploinsufficiency are not fully understood but it has been shown that r-protein deficient cells accumulate rRNA precursors and show a perturbed formation of ribosomal subunits [10,11,20]. We obtained a set of eight novel antibodies directed against human r-proteins to study their expression in cell systems with reduced levels of r-protein S19 [19]. The erythroleukemia derived cell line TF-1-B was induced by doxycycline to express siRNAs targeted against the 3’-end of the RPS19 mRNA [18]. The model allows for a marked down regulation of S19 after siRNA induction and thereby for the detection of subtle effects in S19 deficiency [18]. We analyzed the levels of RPS19 mRNA and r-protein S19 after 2, 4 and 7 days of induction in TF-1-B cells (Figure 1 and Supplementary Figure 1 and Supplementary Figure 2 ). We observed that the siRNA mediated knock-down of RPS19 leads to a gradual decrease of S19 levels with the most pronounced effect after 7 days. Cells induced with doxycycline for 7 days showed RPS19 mRNA at levels approximately 5% of that in non-induced cells (qPCR; data not shown) and RPS19 protein at levels approximately 30% of that in non-induced cells (Figure 1 and Supplementary Figure 1 ). This confirms a marked knock-down of S19 as previously observed in TF-1-B cells [18]. The induced cells were subsequently analyzed for mRNA and protein levels of three additional r-proteins from the small subunit (S20, S21 and S24) and four r-proteins from the large subunit (L3, L9, L30 and L38) by quantitative RT/PCR and western blotting (Figure 1 and Supplementary Figure 1 ). The mRNA levels for the seven r-proteins in induced cells did not show significant changes when compared to non-induced cells with the exception of RPL3 (Figure 2A). However, the protein levels of the three small subunit r-proteins, were significantly reduced when S19 was down-regulated (Figure 2B). The three SSU r-proteins were reduced to approximately 30% of normal levels and similar to the reduction of S19 levels. siRNA induced knock-down of RPS19 was not associated with a significant reduction in levels of large subunit r-proteins although a tendency was observed for three of the four LSU r-proteins (Figure 2B). The mRNA and protein levels of fibrillarin, used as marker for general protein synthesis, were not changed after siRNA induction against RPS19 (Figure 1 and Figure 2). The marked decrease in levels of SSU r-proteins with respect to LSU r-proteins results in a significantly skewed ratio of SSU to LSU r-proteins (Figure 2C).
Figure 1.
Levels of r-proteins after siRNA induced knock-down of RPS19 in TF-1-B cells. Western blot analysis of induced (dox+) and non-induced (dox-) TF-1-B cells, respectively, using the primary antibodies targeting r-proteins, β-actin and fibrillarin. Doxycycline induction results in a reduction of S19 r-protein levels to 30% of levels in non-induced cells (S; small subunit protein: L; large subunit protein. Pictures of the complete Western blots are presented in Supplementary Figure 1).
Figure 2.
Levels of mRNA and protein corresponding to seven r-proteins and fibrillarin (Fib) after siRNA mediated knock-down of RPS19 (7 days of induction). Three SSU r-proteins (S) and four LSU proteins (L) are analyzed. (A) Amounts of mRNA levels in doxycycline induced TF-1-B cells related to the amounts in non-induced cells (set to 1.0). A significant down regulation was observed for RPL3 mRNA only (* denotes p<0.05; three independent experiments). Quantification was performed by qRT/PCR (primer sequences are available upon request) and normalized to β-actin. (B) Amounts of r-protein in doxycycline induced TF-1-B cells related to the amounts in untreated cells (set to 1.0) as determined by western blot analysis using specific antibodies. A significant reduction was observed for the three SSU r-proteins (* denotes p<0.05; three independent experiments) but not for any of the LSU r-proteins using two tailed student’s t-test. (C) Ratio of pooled SSU r-protein levels to pooled LSU r-protein levels in non-induced TF-1-B cells (set to 1,0) and induced TF-1-B cells, respectively (* denotes p<0.05; two tailed student’s t-test). The levels of individual r-proteins are normalized to β-actin and averaged before pooling.
A previous study has used siRNA against RPS19 in HeLa cells for the subsequent analysis of r-protein levels [21]. In this study a single LSU protein (L26) was analyzed suggesting unchanged levels in response to RPS19 knock down. It is known that ribosome biosynthesis, at least in part, is regulated at the transcriptional level [22,23] and the turnover of r-protein mRNAs is assumed to occur quite rapidly [15]. Our observations showing a marked down regulation of SSU r-proteins but not their mRNAs imply a post-transcriptional event.
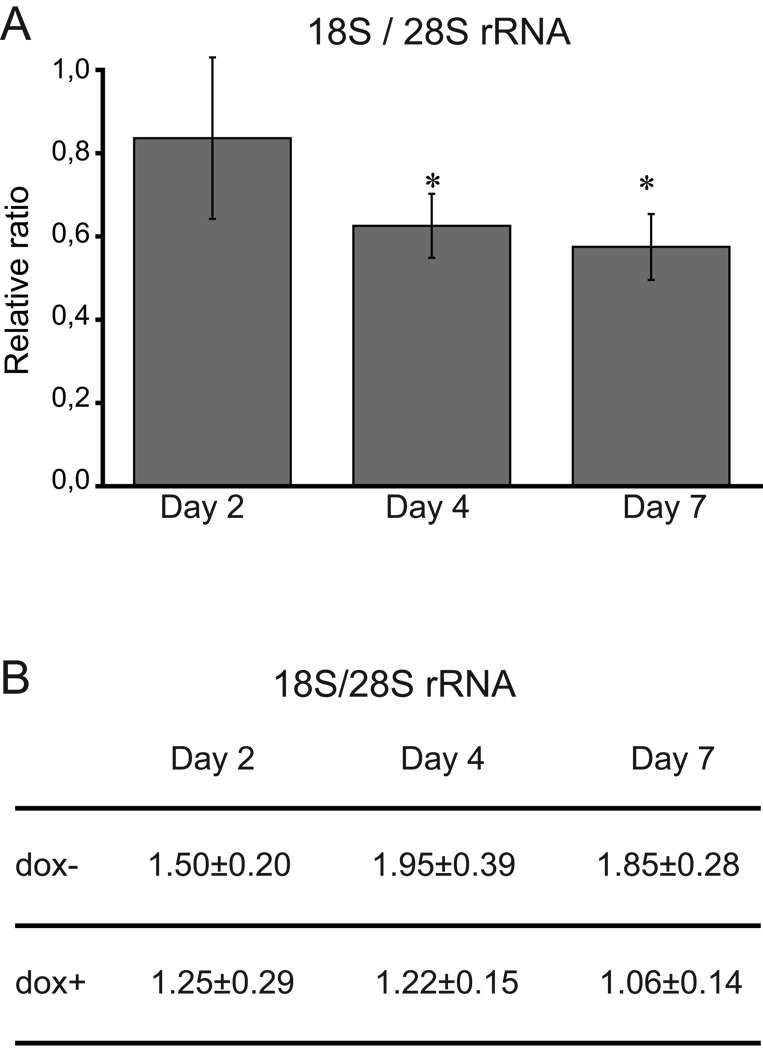
The ribosome comprises four rRNAs and approximately eighty different r-proteins. The r-proteins are structural components of the ribosome that must be coordinated to assure a proper assembly of the ribosomal subunits. It is assumed that the appropriate amounts and stoichiometry of r-proteins is maintained during the assembly of free r-proteins into the 40S and 60S subunits, respectively. Excess proteins are degraded during this process [23]. This mechanism would explain the marked down-regulation of the SSU r-proteins in the siRNA induced TF-1-B cells, resulting in a skewed SSU to LSU r-protein ratio (Figure 2C). To investigate whether the observed SSU r-protein down-regulation can be ascribed to a decrease in the SSU we quantified the amounts of 18S rRNA with respect to 28S rRNA in induced and uninduced TF-1-B cells. The ratio of 18S to 28S rRNA decreased gradually from day 2–7 after induction when compared to uninduced cells (Figure 3). After 7 days of induction the ratio of 18S to 28S rRNA was reduced to 57%. This suggests that the SSU turnover can explain a substantial part of the simultaneous downregulation of SSU r-proteins which was 30% of that observed in non-induced cells (Figure 2B). However, the combined results suggests that additional mechanisms may contribute to the co-regulation of SSU r-protein levels independently of the rRNA levels and subunit assembly.
Figure 3.
Amounts of 18S rRNA to S28 rRNA in TF-1-B cells after siRNA mediated knock-down of RPS19. Total RNA was isolated from doxycycline induced and uninduced TF-1-B cells after 2, 4 and 7 days, respectively. The amounts of rRNAs were quantified using the Agilent 2100 bioanalyser. (A) The 18S to 28S rRNA ratio in induced cells as compared to uninduced cells set to 1 (* denotes p<0.05; two tailed student’s t-test). (B) The mean values of the 18S to 28S ratio in uninduced (dox-) and induced (dox+) cells at day 2, 4 and 7 with standard deviations. The mean values at each day is based on three different experiments.
The observed skewed ratio of SSU to LSU r-proteins would theoretically result in reduced amounts of functional ribosomes and a reduced translational capacity. Such a general effect on protein synthesis has been reported in LCLs derived from DBA patients [24]. However, we did not detect any alteration in non-ribosomal protein levels from the analysis of fibrillarin and β-actin. Surprisingly, RPS19 knock-down seems to result in a slight but non-significant down-regulation of three LSU r-proteins. The biosynthesis of the large subunit proceeds independently of the small subunit and no mechanism is yet known which adjusts levels of LSU r-proteins to SSU r-proteins. One possible explanation is that the marked depletion of RPS19 (30% of normal) after siRNA induction leads to a relative adaptation of LSU protein levels.
3.2. Lymphoblastoid cells from DBA patients with RPS19 mutations show a skewed ratio of SSU to LSU r-proteins
We next asked whether cells with heterozygous RPS19 mutations derived from DBA patients show a skewed SSU/LSU ratio similar to that observed in cells subjected to siRNA knock-down of RPS19. Lymphoblastoid cell lines (LCLs) from four patients carrying different mutations in the RPS19 gene (patient 1 with a 5’splice site mutation (c.411+1G>A); patient 2 with a start codon mutation (c.1A>G); patient 3 with a 3,3 Mb deletion spanning the entire gene [25]; patient 4 with an insertion in exon 3 (c.104_105insA)) and LCLs from three healthy controls were analyzed by both qRT/PCR and western blotting. The four mutations associated with DBA predict a truncated S19 or haploinsufficiency for S19. Analysis using qRT/PCR revealed a significant down regulation in the levels of mRNA for RPS19 and RPS21 in mutant LCLs when compared to control cells. The levels of the other six mRNAs encoding r-proteins were not altered ( Supplementary figure 3A ). We were unable to detect any significant differences in the 18S to 28S ratio when comparing mutant to non-mutant LCLs.
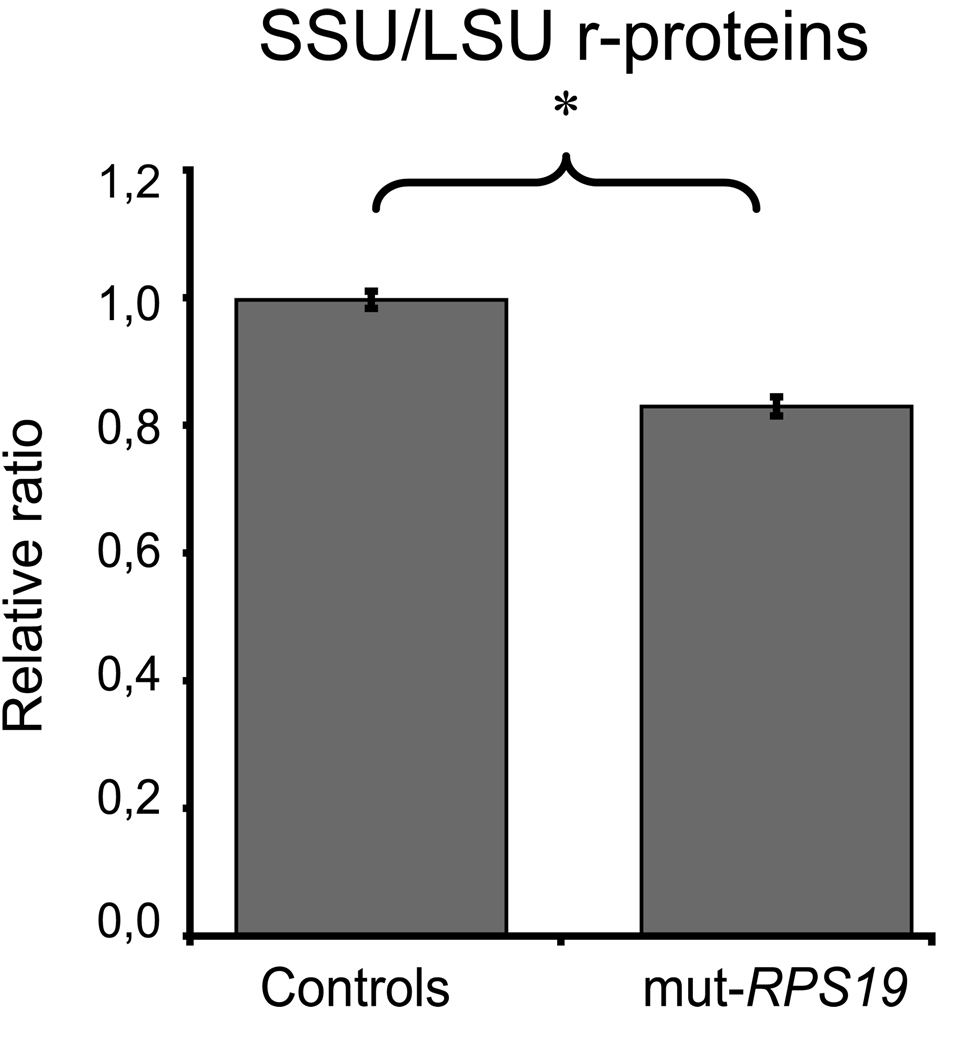
When analyzing the r-proteins we could not detect any significant reduction of individual r-protein levels in RPS19 mutant cell lines when compared to controls ( supplementary figure 3B ) with the eight specific r-protein antibodies. A tendency towards reduced levels was observed for RPS21 and RPS24 but this was non-significant. One possible explanation for this finding is a large variation in expression of individual r-proteins which we observe when comparing the cell lines. It may also be hypothesized that the allelic haploinsufficiency due to RPS19 mutations results in a relative reduction in levels of RPS19 rather than haploinsuffiency because of post-transcriptional compensatory mechanisms. However, we then pooled the r-protein levels from the SSU and from the LSU, respectively, and we compared the ratio of SSU/LSU levels between LCLs from patients and controls. This revealed a significant reduction of the SSU to LSU r-protein ratio in cells carrying RPS19 mutations (p<0.05; Figure 4). Although significant, the effect of heterozygous RPS19 mutations on SSU/LSU ratio is less marked in LCLs when compared to the TF-1-B cell model (Figure 2C).
Figure 4.
Ratio of pooled SSU r-protein levels to pooled LSU r-protein levels in lymphoblastoid cell lines (LCLs) from normal controls and RPS19 deficient DBA patients. The ratio in LCLs from healthy controls (left) is significantly higher than that in DBA patients with RPS19 mutations (right; * denote p<0.05). The levels of individual r-proteins are normalized to β-actin and averaged before pooling. The ratio in controls is set to 1.0.
In summary, we obtained eight antibodies which provide a new and sensitive tool for the analysis of r-protein expression in model systems for DBA. Our combined results imply that RPS19 deficiency results in a down regulation of other r-proteins from the same subunit. This down regulation does not correlate with mRNA levels but partly with a reduction in 18S rRNA. The findings predicts subunit assembly to be one major mechanism in the co-regulation of r-protein levels. However, our results also imply other contributing posttranscriptional mechanisms in the coordination of r-protein levels.
Supplementary Material
Detection of r-proteins using the new set of antibodies obtained from the Swedish Human Proteome Resource (HPR) Programme in TF-1-B cell lysates after induction with doxycycline for 7 days. Primary antibodies raised in rabbit detecting r-proteins S19, S20, S21, S24, L3, L9, L30, L38 or fibrillarin were used and visualized by a secondary IRD800 labeled antibody (green). Membranes were co-hybridised with a mouse anti-β-actin primary antibody and an IRD700 labeled secondary antibody (red). M denotes a protein marker and bands corresponding to the marker size are mentioned in kDa. The expected sizes of the r-proteins are indicated.
Levels of r-proteins after siRNA induced knock-down of RPS19 in TF-1-B cells for 2 and 4 days, respectively. Western blot analysis of induced (dox+) and non-induced (dox-) TF-1-B cells, respectively, using the indicated primary antibodies targeting r-proteins, β-actin or fibrillarin. Expression levels in uninduced TF-1-B cells are set to 1.
Relative levels of mRNA and protein for r-proteins in LCLs from RPS19 deficient DBA patients. The levels in controls are set to 1.0. (A) Relative mRNA levels in RPS19-mutant LCLs with respect to control LCLs . A significant down regulation was only observed for the mRNAs encoding RPS19 and RPS21, respectively. Quantification was performed by qRT/PCR (primer sequences are available upon request) and normalized to β-actin. The black staple denotes relative levels of RPS19 mRNA (B) Relative r-protein levels in RPS19-mutant LCLs with respect to control LCLs as determined by western blot analysis using specific antisera. No significant down regulation was observed for any of the individual proteins analyzed when comparing RPS19 mutant and control LCLs. The black staple denotes relative levels of RPS19 not included in the calculation of the SSU to LSU ratio.
Acknowledgements
We thank all patients and their families for providing samples. Oskar Eriksson is acknowledged for technical support. We thank Professor Stefan Karlsson, Lund University, for sharing the TF-1-B cell line. We also thank two anonymous reviewers for valuable comments and the Swedish Human Proteome Resource (HPR) program (http://www.proteinatlas.org/object.php) for providing α-r-protein antibodies and. This work was supported by the National Institutes of Health (5R01-HL079567-02), Swedish Research Council, Swedish Cancer foundation, Swedish Children’s Cancer foundation, R and T Söderbergs Fund, Marcus Borgström Foundation (to J.S.), Uppsala University and University Hospital. The study was approved by the regional ethical committee of Uppsala.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steitz TA, Moore PB. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem Sci. 2003;28:411–418. doi: 10.1016/S0968-0004(03)00169-5. [DOI] [PubMed] [Google Scholar]
- 2.Ellis SR, Lipton JM. Diamond Blackfan anemia: a disorder of red blood cell development. Curr Top Dev Biol. 2008;82:217–241. doi: 10.1016/S0070-2153(07)00008-7. [DOI] [PubMed] [Google Scholar]
- 3.Draptchinskaia N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 4.Gazda HT. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28:1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 6.Farrar JE. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112:1582–1592. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gazda HT. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campagnoli MF. RPS19 mutations in patients with Diamond-Blackfan anemia. Hum Mutat. 2008;29:911–920. doi: 10.1002/humu.20752. [DOI] [PubMed] [Google Scholar]
- 9.Gazda HT. RNA and protein evidence for haplo-insufficiency in Diamond-Blackfan anaemia patients with RPS19 mutations. Br J Haematol. 2004;127:105–113. doi: 10.1111/j.1365-2141.2004.05152.x. [DOI] [PubMed] [Google Scholar]
- 10.Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis SR. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109:980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choesmel V. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flygare J. Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia. Blood. 2005;105:4627–4634. doi: 10.1182/blood-2004-08-3115. [DOI] [PubMed] [Google Scholar]
- 13.Ebert BL, Lee MM, Pretz JL, Subramanian A, Mak R, Golub TR, Sieff CA. An RNA interference model of RPS19 deficiency in Diamond-Blackfan anemia recapitulates defective hematopoiesis and rescue by dexamethasone: identification of dexamethasone-responsive genes by microarray. Blood. 2005;105:4620–4626. doi: 10.1182/blood-2004-08-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Idol RA, Robledo S, Du HY, Crimmins DL, Wilson DB, Ladenson JH, Bessler M, Mason PJ. Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol Dis. 2007;39:35–43. doi: 10.1016/j.bcmd.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Miyake K, Utsugisawa T, Flygare J, Kiefer T, Hamaguchi I, Richter J, Karlsson S. Ribosomal protein S19 deficiency leads to reduced proliferation and increased apoptosis but does not affect terminal erythroid differentiation in a cell line model of Diamond-Blackfan anemia. Stem Cells. 2008;26:323–329. doi: 10.1634/stemcells.2007-0569. [DOI] [PubMed] [Google Scholar]
- 16.Granneman S, Tollervey D. Building ribosomes: even more expensive than expected? Curr Biol. 2007;17:R415–R417. doi: 10.1016/j.cub.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 18.Miyake K. Development of cellular models for ribosomal protein S19 (RPS19)-deficient diamond-blackfan anemia using inducible expression of siRNA against RPS19. Mol Ther. 2005;11:627–637. doi: 10.1016/j.ymthe.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Uhlen M. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Choesmel V, Fribourg S, Aguissa-Toure AH, Pinaud N, Legrand P, Gazda HT, Gleizes PE. Mutation of ribosomal protein RPS24 in Diamond- Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet. 2008;17:1253–1263. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- 21.Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14:1918–1929. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Nierras CR, Warner JR. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol Cell Biol. 1999;19:5393–5404. doi: 10.1128/mcb.19.8.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry RP. Balanced production of ribosomal proteins. Gene. 2007;401:1–3. doi: 10.1016/j.gene.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cmejlova J, Dolezalova L, Pospisilova D, Petrtylova K, Petrak J, Cmejla R. Translational efficiency in patients with Diamond-Blackfan anemia. Haematologica. 2006;91:1456–1464. [PubMed] [Google Scholar]
- 25.Gustavsson P. Diamond-Blackfan anaemia: genetic homogeneity for a gene on chromosome 19q13 restricted to 1.8 Mb. Nat Genet. 1997;16:368–371. doi: 10.1038/ng0897-368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection of r-proteins using the new set of antibodies obtained from the Swedish Human Proteome Resource (HPR) Programme in TF-1-B cell lysates after induction with doxycycline for 7 days. Primary antibodies raised in rabbit detecting r-proteins S19, S20, S21, S24, L3, L9, L30, L38 or fibrillarin were used and visualized by a secondary IRD800 labeled antibody (green). Membranes were co-hybridised with a mouse anti-β-actin primary antibody and an IRD700 labeled secondary antibody (red). M denotes a protein marker and bands corresponding to the marker size are mentioned in kDa. The expected sizes of the r-proteins are indicated.
Levels of r-proteins after siRNA induced knock-down of RPS19 in TF-1-B cells for 2 and 4 days, respectively. Western blot analysis of induced (dox+) and non-induced (dox-) TF-1-B cells, respectively, using the indicated primary antibodies targeting r-proteins, β-actin or fibrillarin. Expression levels in uninduced TF-1-B cells are set to 1.
Relative levels of mRNA and protein for r-proteins in LCLs from RPS19 deficient DBA patients. The levels in controls are set to 1.0. (A) Relative mRNA levels in RPS19-mutant LCLs with respect to control LCLs . A significant down regulation was only observed for the mRNAs encoding RPS19 and RPS21, respectively. Quantification was performed by qRT/PCR (primer sequences are available upon request) and normalized to β-actin. The black staple denotes relative levels of RPS19 mRNA (B) Relative r-protein levels in RPS19-mutant LCLs with respect to control LCLs as determined by western blot analysis using specific antisera. No significant down regulation was observed for any of the individual proteins analyzed when comparing RPS19 mutant and control LCLs. The black staple denotes relative levels of RPS19 not included in the calculation of the SSU to LSU ratio.