Abstract
Rock proteins are Rho GTPase-dependent serine/threonine kinases with crucial roles in F-actin dynamics and cell transformation. By analogy with other protein kinase families, it can be assumed that Rock proteins act, at least in part, through the regulation of gene expression events. However, with the exception of some singular transcriptional targets recently identified, the actual impact of these kinases on the overall cell transcriptome remains unknown. To address this issue, we have used a microarray approach to compare the transcriptomes of exponentially growing NIH3T3 cells that had been untreated or treated with Y27632, a well known specific inhibitor for Rock kinase activity. We show here that the Rock pathway promotes a weak impact on the fibroblast transcriptome, since its inhibition only results in changes in the expression of 2.3% of all the genes surveyed in the microarrays. Most Y27632-dependent genes are downregulated at moderate levels, indicating that the Rock pathway predominantly induces the upregulation of transcriptionally active genes. Although functionally diverse, a common functional leitmotiv of Y27632-dependent genes is the implication of their protein products in cytoskeletal-dependent processes. Taken together, these results indicate that Rock proteins can modify cytoskeletal dynamics by acting at post-transcriptional and transcriptional levels. In addition, they suggest that the main target of these serine/threonine kinases is the phosphoproteome and not the transcriptome.
Keywords: Rock, Rho/Rac GTPases, Microarray, Gene expression, Transcription, Cytoskeleton
Introduction
The Rho/Rac family is a large group of GTP-binding proteins specialised in the regulation of a wide spectrum of cellular functions such as cytoskeletal organisation, cell proliferation, vesicle trafficking and cytokinesis [1–6]. Rho/Rac proteins are regulated by extracellular stimulus-dependent changes in their bound guanosine nucleotides. In non-stimulated cells, these proteins are bound to GDP molecules and in an inactive conformation [1, 7]. In addition, they remain sequestered in the cytosol due to their interaction with Rho GDP dissociation inhibitors (RhoGDIs) [1, 8, 9]. In stimulated cells, these proteins are released from RhoGDIs, translocate from the cytosol to cellular membranes and undergo the exchange of GDP by GTP. This exchange of nucleotides promotes a conformational change in the GTPase switch regions that, in turn, allows the binding of downstream effectors [1]. This activation step, as well as its subsequent inactivation by hydrolysis of the bound GTP molecules, is catalysed by guanosine nucleotide exchange factors and GTPase activating proteins, respectively [10, 11]. According to structural criteria, Rho/Rac proteins can be subdivided in Rho (RhoA, RhoB, RhoC), Rac (Rac1, Rac2, Rac3, RhoG) and Cdc42 (Cdc42, TTF) subfamilies [1]. Within the Rho subfamily, RhoA is perhaps the best characterised in terms of three-dimensional structure, effectors and its participation in normal and pathological-related biological responses [1, 12–15].
The elucidation of the regulation and function of RhoA effectors is important to understanding the intracellular pathways that control RhoA-dependent cellular, physiological and pathological responses. Two of the main RhoA effectors are the highly related serine/threonine kinases RockI (also known as Rokβ and p160Rock) and RockII (also referred to as both Rokα and Rho kinase) [16, 17]. These proteins become activated during signal transduction by the binding to Rho subfamily members [16, 17] and/or second messengers such as arachidonic acid [18] and sphingo-sylphosphorylcholine [19]. Their activities are also subjected to negative regulation by specific subsets of GTPases (RhoE, Gem, Rad) [20, 21] and cell cycle inhibitors (p21Cip1) [22].
Rock proteins induce intracellular pathways that mediate the formation of stress fibres and focal adhesions, thereby participating in cell-to-cell and cell-to-substratum adhesion, cell migration and motility, phagocytosis and neurite retraction [16, 17]. They also work in cell division and cytokinesis processes by regulating centrosomal functions and actomyosin ring contraction, respectively [16, 17]. Deregulated signalling outputs from these kinases appear to be important for some pathologies, such as hypertension, Alzheimer’s disease and cancer [12, 14–17]. Demonstrating the role of these proteins in these diseases, it has been shown that the use of chemical inhibitors for Rock proteins alleviates cardiovascular pathologies such as pulmonary hypertension, vasospasms and angina pectoris [14, 16, 17]. Rock inhibitors also block tumorigenesis in vitro [23] and appear to be potentially useful for the treatment of other medical conditions including Alzheimer’s disease, stroke and neuropathic pain [17].
Several Rock downstream targets have been identified, including regulators of the F-actin cytoskeleton (myosin light chain (MLC), the MLC phosphatase, Lim kinases 1 and 2), intermediate filament components (vimentin, glial fibrillary acidic protein and neurofilaments) and microtubule-associated proteins (Tau, microtubule-associated protein 2) [16, 17]. Whereas the phosphorylation of MLC and its phosphatase by Rock proteins promotes the formation of F-actin fibres, the phosphorylation of other protein classes appears to induce neurofilament disassembly and to halt microtubule polymerisation. Thus, the phosphoproteome induced by Rock proteins is fully consistent with the assigned roles of these proteins in cell migration and morphology [16, 17].
Similar to other serine/threonine kinases involved in signal transduction (i.e., Erk, p38MAPK), it is possible that Rock could also promote the long-term regulation of gene expression. Consistent with this view, it has been shown that Rock activity is important for the stimulation of c-Myc by the constitutively active, oncogenic version of RhoA (Q63L mutant) [24, 25] and for the expression of a small subset of the transcriptome of NIH3T3 cells transformed by the chronic expression of the rhoA oncogene [24]. Other studies have also shown that the expression of specific RhoAQ63L-dependent genes is abrogated upon inhibition of the Rock pathway [26, 27]. In the present study, we aimed at expanding these results to non-transformed fibroblasts. To this end, we used microarray technology to assess the effect of Y27632, a chemical inhibitor commonly used to block Rock kinase activity [28], in the transcriptome of exponentially growing NIH3T3 cells. This cell line has been widely utilised before for the characterisation of the biological properties of both Rho and Rock proteins. Previous observations by us and others have shown that Y27632 treatments inhibit several Rock-dependent responses in this cell line, including MLC phosphorylation and stress fibre formation [23, 24]. We report here the results obtained from this research avenue.
Materials and methods
Cell lines
Murine NIH3T3 cells were grown under standard temperature/CO2 conditions in Dulbecco’s modified Eagle’s medium supplemented with 1% L-glutamine, 1% penicillin/streptomycin and 10% calf serum. All tissue culture reagents were obtained from Invitrogen. When appropriate, cells were treated for 24 h with 10 µM Y27632 (Cal-biochem) to inhibit endogenous Rock proteins. RhoA-transformed cells have been described before [24]. To confirm the effectiveness of Rock inactivation in this experimental setting, parallel cultures of NIH3T3 and RhoA-transformed cells were analysed by immunoblot and immunofluorescence techniques to corroborate the expected inhibition of the phosphorylation of the myosin light chain and the disassembly of stress F-actin fibres in Y27632-treated cells, as indicated and shown before [24].
Microarray experiments and data analysis
Microarray analyses were performed using RNAs obtained from seven and five independent experiments of untreated and Y27632-treated NIH3T3 cells, respectively. In each independent experiment, three 10-cm diameter plates containing exponentially growing cultures were used to generate the total RNA used in the microarray studies. To this end, cultured cells were washed with phosphate-buffered saline solution and total cellular RNAs isolated using the RNeasy kit (Qiagen) according to the supplier’s specifications. The quantity and quality of the total RNAs obtained were determined using 6000 Nano Chips (Agilent Technologies). Total RNA samples (4 µg) were then processed for hybridisation on MGU75Av2 microarrays (Affymetrix) using standard Affymetrix protocols at the CIC Genomics and Proteomics Facility. Normalisation, filtering and analysis of the raw data obtained from microarrays were carried out with the Bioconductor software (www.bioconductor.com) using the ReadAffy package and the RMA application. We considered a gene to be differentially expressed when exhibiting a signal ≥100 and its fold change respect to the levels of expression of untreated NIH3T3 cells was ≥±1.5 and with p values ≤0.01. Statistical analyses were performed using F-statistics.
For the graphical presentation of microarray data, we performed hierarchical clustering analysis using the WP-GA average-linkage and the standard correlation similarity metric method with the J-Express application. Functional annotation of gene functions was performed manually using internet-available databases such as those maintained by the NCBI (www.ncbi.nlm.nih.gov/sites/entrez?db=omim) and the Weizman Institute of Science (Rehovot, Israel; www.genecards.org). The identification of interactive networks of proteins and common functions was done using the Ingenuity Pathways Analysis program, a web-delivered application that enables discovery, visualisation and exploration of biological interaction networks and biological processes (www.ingenuity.com) [29]. In this case, we considered a network as significant when it fulfilled the following criteria: (i) a minimal score of 15; (ii) at least 10 proteins participating in direct, functional interactions inside the network. In addition, and to reinforce the strength of the functional relationships, we only took into consideration direct, not indirect, relationships among the network components. Bioinformatically identified networks were edited manually to sieve out proteins that, according to published data, did not have a coherent or well defined functional relationship with the other network components.
The comparison of transcriptomes between Y27632-treated non-transformed and RhoAQ63L-transformed NIH3T3 cells was done using the previously published data on the RhoAQ63L-dependent transcriptome [24].
Real-time quantitative RT-PCR
Exponentially growing cells treated and non-treated with Y27632 were lysed and total RNA extracted using the RNAeasy kit (Qiagen). RNAs were quantified by loading aliquots into 6000 Nano Chips. Quantitative polynucleotide chain reactions were performed using the Quanti-Tect SYBR Green RT-PCR kit (Qiagen). 18S rRNA primers were used as controls for both loading and quantitation of relative expression levels of the genes tested. Amplifications were performed using the iCycler machine (Bio-Rad). Raw data were analysed using the iCycler iQ Optical System software (version 3.0a, Bio-Rad). In other cases, quantitative RT-PCR experiments were conducted using a microfluidic card (Applied Biosystems) service available at the Program for Genomics, Proteomics and Bioinformatics of the Spanish Network of Cancer Groups.
Results and discussion
We made use of Affymetrix microarray technology to identify the transcriptomal changes induced by culturing exponentially growing NIH3T3 cells with the Rock inhibitor Y27632 for 24 h. The reason for selecting this time point was two-fold. On the one hand, it ensured effective Rock inhibition, since the dephosphorylation of MLC is detected already 6 h after addition of Y27632 and remains at low levels thereafter [24]. On the other hand, this early time point allowed us to select for primary Rock transcriptional targets rather than detecting secondary transcriptomal changes derived from the deregulation of the expression of putative Rock-dependent transcriptional factors. We also cultured cells in the presence of serum and under non-confluent conditions (approx. 70% confluency) to avoid the activation of strong genetic programs related to serum withdrawal or contact inhibition that may obscure the detection of the Rock-dependent transcriptome [30]. In addition, we isolated total RNAs from seven (in the case of NIH3T3 cells) and five (in the case of Y27632-treated cells) independent cell cultures in order to make a robust statistical treatment of the data generated possible. Total RNAs samples obtained under those conditions were converted into biotinylated-cRNA probes and hybridised independently to Affymetrix Genechip MG U74Av2 arrays, thus allowing the monitoring of the expression status of ≈12,500 mouse genes.
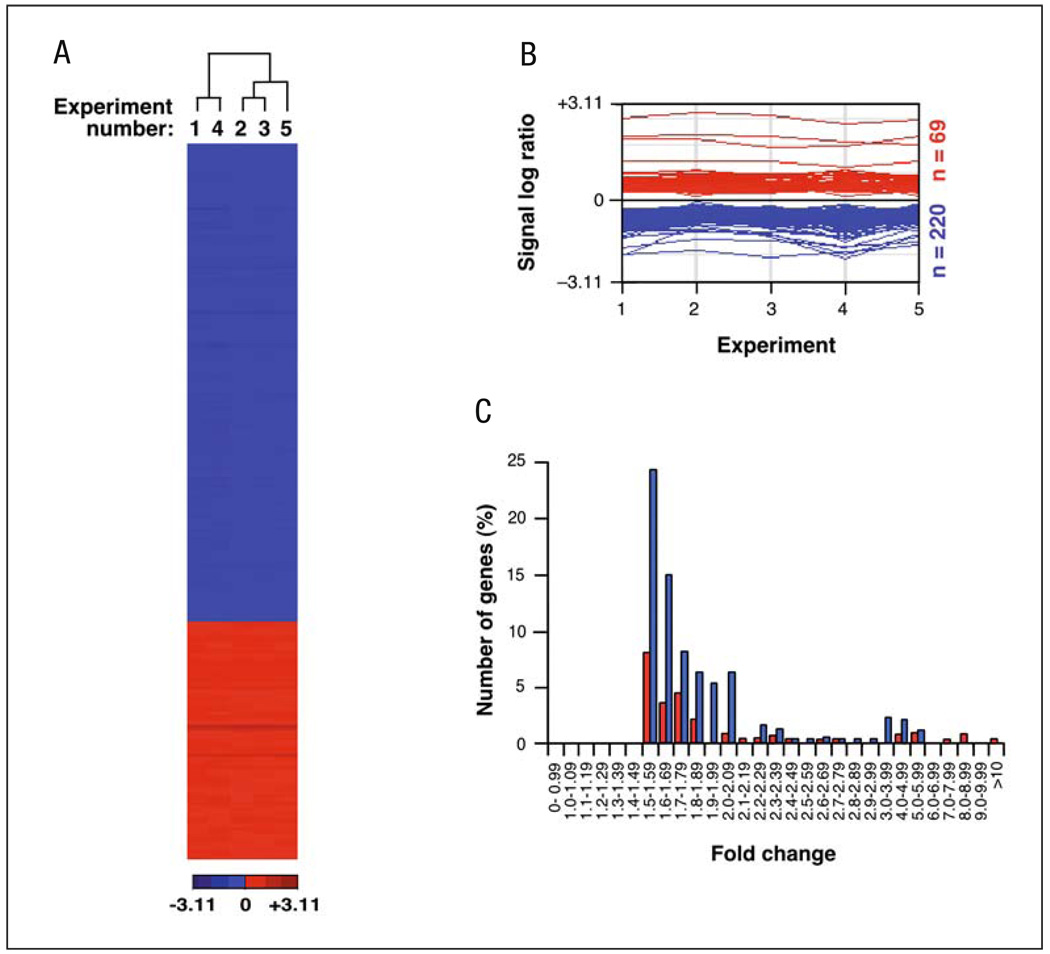
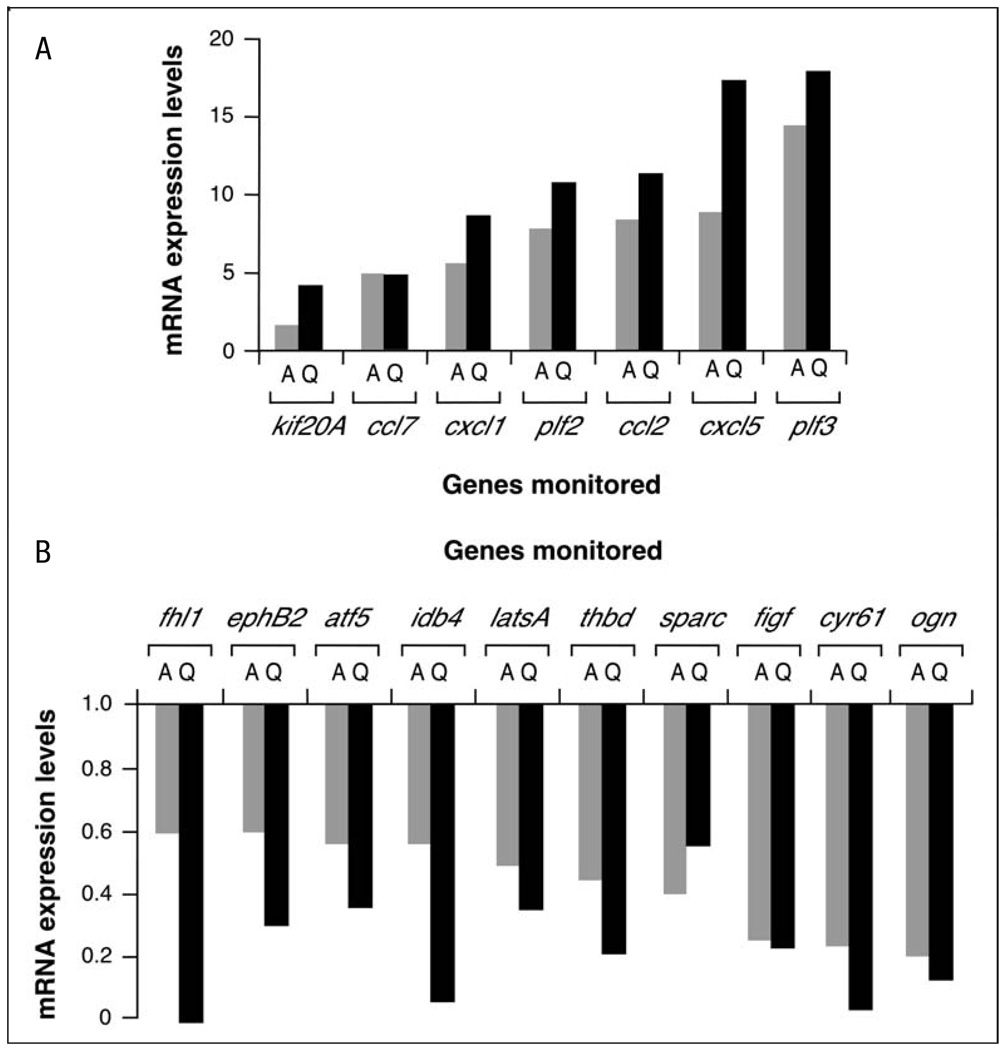
The results from these microarray experiments indicated that the chemical inhibition of Rock led to changes in approximately 2.3% (289 genes) of all genes probed in the arrays (Fig. 1A, B and Table 1). Interestingly, these changes involved mostly the downregulation of transcriptionally active genes (76.1% of all responsive genes), indicating that Rock activity is required primarily to maintain transcription from specific gene subsets in mouse fibroblasts. Instead, the inhibition of Rock determined the activation of a much smaller group of 69 genes. To confirm the microarray results, the expression of seven of the upregulated and ten of the downregulated genes was re-evaluated using real-time quantitative RT-PCR analysis. These analyses confirmed that these 17 genes were indeed Rock-dependent (Fig. 2). Interestingly, we also detected genes identified as Rock targets such as cyr61, c-myc and cyclin D1 [24, 26, 27], further supporting the validity of our microarray data.
Fig. 1.
Transcriptomal changes induced by Y27632 in exponentially growing NIH3T3 cells. A Hierarchical cluster diagram of the 289 genes whose expression levels changed in Y27632-treated cells. Each column represents one experiment and each row a gene. Varying levels of expression are represented on a scale from dark blue (lowest expression) to dark red (highest expression). Note that expression values are represented as signal log ratio numbers (SLR) and that, therefore, the total fold change value is obtained from 2SLR. The experiment number is shown at the top of each column. B Gene graphs showing the induced (red) and repressed (blue) genes in Y27632-treated cells. In each category, the expression values of all deregulated genes are represented as SLR (considering that fold change is 2SLR; y-axis) obtained in each experimental sample (x-axis). The total number of upregulated and downregulated genes in each category is indicated on the right of each panel. C Histogram showing the number of up- (red) and downregulated (blue) genes with a given expression fold change value in Y27632-treated cells
Table 1.
List of genes whose expression is upregulated or downregulated by Y27632 in NIH3T3 cellsa
| Function | Locus ID | Gene | Description | Changeb | p valuec |
|---|---|---|---|---|---|
| Extracellular | |||||
| Cell adhesion | 12825 | Col3a1 | Procollagen, type III, alpha 1 | 0.58 | 0.0017 |
| 12832 | Col5a2 | Procollagen, type V, alpha 2 | 0.55 | 0.0001 | |
| 13003 | Cspg2 | Chondroitin sulfate proteoglycan 2 | 1.52 | <1.00E-04 | |
| 16007 | Cyr61 | Cysteine-rich protein 61 | 0.26 | 0.0001 | |
| 16779 | Lamb2 | Laminin, beta 2 | 0.52 | 0.0014 | |
| 17313 | Mglap | Matrix gamma-carboxyglutamate protein | 0.53 | <1.00E-04 | |
| 17395 | Mmp9 | Matrix metalloproteinase 9 | 1.5 | <1.00E-04 | |
| 114249 | Npnt | Mus musculus transcribed sequences | 0.63 | 0.0001 | |
| 18787 | Serpine1 | Serine (or cysteine) proteinase inhibitor 1 | 0.61 | 0.0074 | |
| 20720 | Serpine2 | Serine (or cysteine) proteinase inhibitor, 2 | 0.38 | 0.0001 | |
| 20692 | Sparc | Secreted acidic cysteine rich glycoprotein | 0.41 | <1.00E-04 | |
| 13602 | Sparcl1 | SPARC-like 1 | 1.54 | 0.0095 | |
| 21810 | Tgfbi | Transforming growth factor beta induced | 1.57 | <1.00E-04 | |
| Immune response | 12266 | C3 | Complement component 3 | 4.55 | <1.00E-04 |
| 19288 | Ptx3 | Pentaxin related gene | 0.21 | <1.00E-04 | |
| 20568 | Slpi | Secretory leukocyte protease inhibitor | 2.39 | <1.00E-04 | |
| 20210 | Saa3 | Serum amyloid A3 | 1.89 | <1.00E-04 | |
| Ion transport | 12870 | Cp | Ceruloplasmin | 4.77 | <1.00E-04 |
| Ligand | 14824 | Grn | Granulin | 0.66 | 0.0039 |
| 23893 | Grem2 | Gremlin 2 homolog | 1.76 | 0.001 | |
| 20750 | Spp1 | Secreted phosphoprotein 1 | 1.82 | 0.0017 | |
| 18812 | Plf2 | Proliferin 2 | 7.7 | <1.00E-04 | |
| 20311 | Cxcl5 | Chemokine (C-X-C motif) ligand 5 | 8.83 | <1.00E-04 | |
| 11535 | Adm | Adrenomedullin | 0.47 | <1.00E-04 | |
| 13642q | Efnb2 | Ephrin B2 | 0.61 | 0.0068 | |
| 20306 | Ccl7 | Chemokine (C-C motif) ligand 7 | 5 | <1.00E-04 | |
| 14825 | Cxcl1 | Chemokine (C-X-C motif) ligand 1 | 5.56 | <1.00E-04 | |
| 20296 | Ccl2 | Chemokine (C-C motif) ligand 2 | 8.34 | <1.00E-04 | |
| 26421 | Mrpplf3 | Proliferin 3 | 14.29 | <1.00E-04 | |
| 21825 | Thbs1 | Thrombospondin 1 | 0.21 | <1.00E-04 | |
| Other | 14118 | Fbn1 | Fibrillin 1 | 0.65 | 0.0064 |
| 50530 | Mfap5 | Microfibrillar associated protein 5 | 0.5 | 0.0023 | |
| 18451 | P4ha1 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase a 1 | 0.64 | <1.00E-04 | |
| 14827 | Pdia3 | Protein disulfide isomerase associated 3 | 0.48 | <1.00E-04 | |
| Receptor activity | 12931 | Crlf1 | Cytokine receptor-like factor 1 | 1.76 | 0.0004 |
| Growth factor | 12977 | Csf1 | Colony stimulating factor 1 | 0.61 | <1.00E-04 |
| 14205 | Figf | C-Fos induced growth factor | 0.27 | 0.0002 | |
| 18295 | Ogn | Osteoglycin | 0.22 | <1.00E-04 | |
| 19242 | Ptn | Pleiotrophin | 0.31 | 0.0015 | |
| Membrane | |||||
| Cell adhesion | 12558 | Cdh2 | Cadherin 2 | 0.47 | <1.00E-04 |
| 16403 | Itga6 | Integrin alpha 6 | 0.63 | 0.0003 | |
| Membrane/cell adhesion | 20320 | Sdfr1 | Stromal cell derived factor receptor 1 | 0.63 | 0.0004 |
| Channel-transporter | 11826 | Aqp1 | Aquaporin 1 | 2.77 | <1.00E-04 |
| 14726 | Gp38 | Glycoprotein 38 | 0.57 | 0.0001 | |
| 16497 | Kcnab1 | Potassium voltage-gated channel beta member 1 | 0.66 | 0.0011 | |
| 20514 | Slc1a7 | Solute carrier family 1, member 7 | 0.45 | 0.0001 | |
| 67760 | Slc38a2 | Solute carrier family 38, member 2 | 0.52 | 0.0004 | |
| 22334 | Vdac2 | Voltage-dependent anion channel 2 | 0.57 | <1.00E-04 | |
| Immune response | 15040 | H2-T23 | Histocompatibility 2, T region locus 23 | 1.86 | 0.0008 |
| 17069 | Ly6e | Lymphocyte antigen 6 complex, locus E | 1.57 | <1.00E-04 | |
| Receptor activity | 11609 | Agtr2 | Angiotensin II receptor, type 2 | 0.43 | 0.0026 |
| 14062 | F2r | Coagulation factor II (thrombin) receptor | 0.64 | <1.00E-04 | |
| 20321 | Frrs1 | Ferric-chelate reductase 1 | 0.6 | 0.0003 | |
| 83924 | Gpr137b | G protein-coupled receptor 137B | 2.39 | <1.00E-04 | |
| 16412 | Itgb1 | Integrin beta 1 | 0.65 | 0.0003 | |
| 16948 | Lox | Lysyl oxidase | 0.66 | 0.0003 | |
| 16974 | Lrp6 | Low density lipoprotein receptor-related protein 6 | 0.64 | 0.0012 | |
| 319900 | Npr3 | Natriuretic peptide receptor 3 | 0.33 | 0.0011 | |
| 18186 | Nrp | Neuropilin | 0.4 | <1.00E-04 | |
| 19220 | Ptgfr | Prostaglandin F receptor | 2.44 | <1.00E-04 | |
| 21824 | Thbd | Thrombomodulin | 0.47 | 0.0001 | |
| 18383 | Tnfrsf11b | Tumour necrosis factor receptor superfamily 1b | 1.57 | 0.0007 | |
| 21937 | Tnfrsf1a | Tumour necrosis factor receptor superfamily 1a | 1.52 | 0.0015 | |
| 14102 | Tnfrsf6 | Tumour necrosis factor receptor superfamily 6 | 1.57 | <1.00E-04 | |
| Signal transducer activity | 11487 | Adam10 | A disintegrin and metalloprotease domain 10 | 0.62 | 0.0004 |
| 17118 | Marcks | Myristoylated alanine rich protein kinase C substrate | 0.42 | 0.0001 | |
| 18858 | Pmp22 | Peripheral myelin protein | 0.64 | 0.0007 | |
| 20338 | Sel1h | Sel1 1 homolog | 0.52 | <1.00E-04 | |
| Other | 11502 | Adam9 | A disintegrin and metalloproteinase domain 9 | 0.45 | <1.00E-04 |
| 319434 | Amot | Angiomotin | 0.6 | ||
| 13244 | Degs | Degenerative spermatocyte homologue | 0.57 | <1.00E-04 | |
| 53872 | Gpiap1 | GPI-anchored membrane protein 1 | 0.58 | 0.0005 | |
| 20014 | Rpn2 | Ribophorin II | 0.64 | 0.0033 | |
| 20324 | Sdpr | Serum deprivation response | 0.37 | <1.00E-04 | |
| 20908 | Stx3 | Syntaxin 3 | 0.53 | 0.0014 | |
| 21838 | Thy1 | Thymus cell antigen 1 theta | 0.5 | <1.00E-04 | |
| 232086 | TM6P1 | Fasting-inducible integral membrane protein TM6P1 | 0.65 | 0.0094 | |
| Nuclear membrane | 67154 | Mtdh | Metadherin | 0.65 | 0.0063 |
| Cytoskeleton | 226251 | Ablim1 | Actin-binding LIM protein 1 | 0.56 | 0.0023 |
| 26357 | Abcg2 | ATP-binding cassette, sub-family G2 | 0.65 | 0.0023 | |
| 11475 | Acta2 | Actin, alpha 2, smooth muscle | 0.18 | <1.00E-04 | |
| 11464 | Actc1 | Actin, alpha, cardiac | 0.57 | <1.00E-04 | |
| 109711 | Actn1 | Actinin alpha 1 | 0.54 | 0.0008 | |
| 12798 | Cnn2 | Calponin 2 | 0.49 | <1.00E-04 | |
| 13007 | Csrp1 | Cysteine and glycine-rich protein 1 | 0.58 | 0.0001 | |
| 13860 | Eps8 | Epidermal growth factor receptor pathway substrate 8 | 0.56 | 0.0001 | |
| 14086 | Fscn1 | Fascin homologue 1 | 0.55 | <1.00E-04 | |
| 56486 | Gabarap | Gamma-aminobutyric acid receptor associated protein | 0.66 | 0.0004 | |
| 19348 | Kif20a | Kinesin family member 20A | 1.64 | 0.0049 | |
| 16571 | Kif4 | Kinesin family member 4 | 1.89 | 0.007 | |
| 16573 | Kif5b | Kinesin family member 5B | 0.57 | 0.0014 | |
| 65970 | Lima1 | LIM domain and actin binding 1 | 0.61 | 0.0009 | |
| 11426 | Macf1 | Microtubule-actin crosslinking factor 1 | 0.64 | < 1.00E-04 | |
| 17886 | Myh9 | Myosin heavy chain IX | 0.49 | <1.00E-04 | |
| 17909 | Myo10 | Myosin X | 0.6 | 0.0002 | |
| 83431 | Ndel1 | Nuclear distribution gene E-like homolog 1 | 0.63 | 0.0001 | |
| 218952 | Plekhc1 | Pleckstrin homology domain containing, family C1 | 0.56 | 0.0001 | |
| 20740 | Spna2 | Spectrin alpha 2 | 0.49 | 0.0004 | |
| 20742 | Spnb2 | Spectrin beta 2 | 0.55 | 0.0005 | |
| 104027 | Synpo | Synaptopodin | 0.59 | 0.0028 | |
| 21345 | Tagln | Transgelin | 0.26 | <1.00E-04 | |
| 21894 | Tln | Talin | 0.63 | <1.00E-04 | |
| 19241 | Tmsb4x | Thymosin, beta 4, X chromosome | 0.51 | <1.00E-04 | |
| 22003 | Tpm1 | Tropomyosin 1, alpha | 0.59 | <1.00E-04 | |
| 22330 | Vcl | Vinculin | 0.44 | <1.00E-04 | |
| Intracellular | |||||
| Signal transducer activity | 12388 | Catns | Catenin src | 0.6 | 0.0035 |
| 83397 | Akap12 | A kinase anchor protein 12 | 0.38 | 0.0001 | |
| 54519 | Apbb1ip | Amyloid beta precursor-binding 1 interacting protein | 0.43 | <1.00E-04 | |
| 80837 | Arhj | Ras homologue gene family J | 0.64 | 0.0046 | |
| 19252 | Dusp1 | Dual specificity phosphatase 1 | 0.6 | 0.0002 | |
| 67603 | Dusp6 | Dual specificity phosphatase 6 | 0.24 | <1.00E-04 | |
| 74155 | Errfi1 | ERBB receptor feedback inhibitor 1 | 0.51 | 0.0027 | |
| 212398 | Frat2 | Frequently rearranged in T-cell lymphomas 2 | 1.57 | <1.00E-04 | |
| 16413 | Itgb1bp1 | Integrin beta 1 binding protein 1 | 0.6 | <1.00E-04 | |
| 50523 | Lats2 | Large tumour suppressor 2 | 0.52 | 0.0001 | |
| 26410 | Map3k8 | Mitogen activated protein kinase kinase kinase 8 | 1.7 | <1.00E-04 | |
| 18647 | Pftk1 | PFTAIRE protein kinase 1 | 0.63 | <1.00E-04 | |
| 18708 | Pik3r1 | P85 alpha | 2.28 | <1.00E-04 | |
| 19046 | Ppp1cb | Protein phosphatase 1, catalytic subunit, beta isoform | 0.47 | 0.0013 | |
| 56044 | Rala | V-ral simian leukaemia viral oncogene homologue A | 0.63 | 0.001 | |
| 218397 | Rasa1 | RAS p21 protein activator 1 | 0.63 | 0.0009 | |
| 56437 | Rrad | Ras-related associated with diabetes | 1.52 | <1.00E-04 | |
| 16765 | Stmn1 | Stathmin 1 | 1.82 | 0.0077 | |
| 21353 | Tank | TRAF NF-kappa B activator | 0.48 | 0.0001 | |
| 68842 | Tulp4 | Tubby like protein 4 | 0.65 | 0.0012 | |
| 59043 | Wsb2 | WD-40-repeat-containing protein with a SOCS box | 0.52 | <1.00E-04 | |
| Protein biosynthesis | 16341 | Eif3s6 | Eukaryotic translation initiation factor 3, subunit 6 | 0.58 | 0.0026 |
| 107508 | Eprs | Glutamyl-prolyl-trna synthetase | 0.66 | 0.0052 | |
| 23874 | Farsl | Phenylalanine-tRNA synthetase-like | 0.65 | 0.003 | |
| Protein folding and modification | 12469 | Cct8 | Chaperonin subunit 8 | 0.64 | 0.0021 |
| 14976 | H2-Ke2 | H2-K region expressed gene 2 | 1.52 | 0.0015 | |
| 14828 | Hspa5 | Heat shock 70 kDa protein 5 | 0.63 | 0.0011 | |
| 15481 | Hspa8 | Heat shock protein 8 | 0.59 | <1.00E-04 | |
| 217664 | Mgat2 | Mannoside acetylglucosaminyltransferase 2 | 0.61 | 0.0005 | |
| 12406 | Serpinh1 | Serine (or cysteine) proteinase inhibitor, clade H 1 | 0.66 | 0.0001 | |
| 54609 | Ubqln2 | Ubiquilin 2 | 0.52 | 0.0004 | |
| Proteolysis and peptidolysis | 67526 | Apg12l | Autophagy 12-like | 1.73 | 0.0004 |
| 17035 | Lxn | Latexin | 0.58 | 0.0069 | |
| 22194 | Ube2e1 | Ubiquitin-conjugating enzyme E2E 1 | 0.52 | 0.0022 | |
| 70620 | Ube2v2 | Ubiquitin-conjugating enzyme E2 variant 2 | 0.66 | 0.004 | |
| 59025 | Usp14 | Ubiquitin specific protease 14 | 0.62 | 0.0025 | |
| Heat shock | 15505 | Hsp105 | Heat shock protein 105 | 0.55 | <1.00E-04 |
| 15519 | Hspca | Heat shock protein 1 alpha | 0.65 | 0.0009 | |
| Apoptosis | 54673 | Bif-1 | SH3-domain GRB2-like B1 | 0.58 | <1.00E-04 |
| 12363 | Casp4 | Caspase 4, apoptosis-related cysteine protease | 1.64 | <1.00E-04 | |
| 114774 | Pawr | PRKC apoptosis WT1 regulator | 0.59 | 0.0011 | |
| Cell cycle | 215387 | Brrn1 | Barren homologue | 1.79 | 0.0086 |
| 12442 | Ccnb2 | Cyclin B2 | 1.82 | 0.0003 | |
| 12443 | Ccnd1 | Cyclin D1 | 0.55 | <1.00E-04 | |
| 12453 | Ccni | Cyclin I | 0.54 | 0.002 | |
| 12532 | Cdc25c | Cell division cycle 25 homologue C | 1.79 | 0.0054 | |
| 12545 | Cdc7 | Cell division cycle 7 | 1.5 | 0.0038 | |
| 59125 | Nek7 | NIMA-related expressed kinase 7 | 0.65 | 0.0001 | |
| 30939 | Pttg1 | Pituitary tumour-transforming 1 | 2 | 0.0002 | |
| DNA replication and repair | 12615 | Cenpa | Centromere autoantigen A | 2 | 0.0007 |
| 69072 | Ebna1bp2 | EBNA1 binding protein 2 | 0.6 | 0.0016 | |
| 15569 | Elavl2 | ELAV-like 2 | 0.62 | 0.0001 | |
| 15078 | H3f3a | H3 histone, family 3A | 0.58 | 0.0011 | |
| 15364 | Hmga2 | High mobility group AT-hook 2 | 0.66 | 0.0005 | |
| 15354 | Hmgb3 | High mobility group box 3 | 2.18 | 0.0024 | |
| 50887 | Nsbp1 | Nucleosome binding protein 1 | 1.54 | 0.0006 | |
| 21974 | Top2b | Topoisomerase (DNA) II beta | 0.63 | 0.0003 | |
| Regulation of transcription | 107503 | Atf5 | Activating transcription factor 5 | 0.58 | <1.00E-04 |
| 12034 | Bcap37 | Dentatorubral pallidoluysian atrophy | 1.58 | 0.0002 | |
| 12051 | Bcl3 | B-cell leukaemia/lymphoma 3 | 1.59 | <1.00E-04 | |
| 12053 | Bcl6 | B-cell leukaemia/lymphoma 6 | 0.61 | <1.00E-04 | |
| 57316 | C1d | C1d nuclear DNA binding protein | 0.64 | 0.0083 | |
| 17684 | Cited2 | Cbp/p300-interacting transactivator 2 | 0.59 | <1.00E-04 | |
| 107765 | Crap | Cardiac responsive adriamycin protein | 0.35 | <1.00E-04 | |
| 13017 | Ctbp2 | C-terminal binding protein 2 | 0.65 | 0.001 | |
| 23871 | Ets1 | E26 avian leukemia oncogene 1 | 0.61 | 0.0054 | |
| 14200 | Fhl2 | Four and a half LIM domains 2 | 0.66 | <1.00E-04 | |
| 15904 | Idb4 | Inhibitor of DNA binding 4 | 0.57 | 0.0003 | |
| 26388 | Ifi202b | Interferon activated gene 202B | 0.52 | 0.0002 | |
| 16978 | Lrrfip1 | Leucine rich repeat interacting protein 1 | 0.49 | <1.00E-04 | |
| 17869 | Myc | Myelocytomatosis oncogene | 0.54 | 0.0055 | |
| 27057 | Ncoa4 | Nuclear receptor coactivator 4 | 0.63 | 0.0001 | |
| 18028 | Nfib | Nuclear factor I/B | 0.57 | 0.0031 | |
| 18035 | Nfkbia | Nuclear factor of kappa B inhibitor alpha | 1.7 | <1.00E-04 | |
| 80859 | Nfkbiz | Nuclear factor of kappa B inhibitor zeta | 2.64 | <1.00E-04 | |
| 353187 | Nr1d2 | Nuclear receptor subfamily 1, group D, member 2 | 1.5 | 0.0001 | |
| 11819 | Nr2f2 | Nuclear receptor subfamily 2, group F, member 2 | 0.5 | <1.00E-04 | |
| 22634 | Plagl1 | Pleiomorphic adenoma gene-like 1 | 0.61 | 0.0093 | |
| 19664 | Rbpsuh | Recombining binding protein suppressor of hairless | 1.67 | <1.00E-04 | |
| 19698q | Relb | Avian reticuloendotheliosis viral (v-rel) oncogene B | 1.62 | <1.00E-04 | |
| 67155 | Smarca2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subf.a, mem2 | 0.43 | 6.00E-04 | |
| 20677 | Sox4 | SRY-box containing gene 4 | 1.79 | <1.00E-04 | |
| 21807 | Tgfb1i4 | Transforming growth factor beta 1 induced transcript 4 | 1.57 | <1.00E-04 | |
| 228775 | Trib3 | Tribbles homologue 3 | 1.67 | <1.00E-04 | |
| 22160 | Twist1 | Ubiquitin-conjugating enzyme E2D 3 | 0.5 | <1.00E-04 | |
| 22433 | Xbp1 | X-box binding protein 1 | 0.62 | <1.00E-04 | |
| RNA metabolism | 14007 | Cugbp2 | CUG triplet repeat,RNA binding protein 2 | 0.52 | 0.0004 |
| 18569 | Pdcd4 | Programmed cell death 4 | 0.63 | 0.0017 | |
| 81898 | Sf3b1 | Splicing factor 3b, subunit 1 | 0.52 | 0.0001 | |
| 20823 | Ssb | Sjogren syndrome antigen B | 0.54 | 0.0016 | |
| 12192 | Zfp36l1 | Zinc finger protein 36, C3H type-like 1 | 1.73 | 0.0001 | |
| Metabolism | |||||
| Lipid metabolism | 208715 | Hmgcs1 | 3-Hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | 0.62 | <1.00E-04 |
| 16956 | Lpl | Lipoprotein lipase | 0.65 | 0.0017 | |
| 67041 | Oxct | 3-Oxoacid coa transferase | 0.66 | 0.0001 | |
| 27273 | Pdk4 | Pyruvate dehydrogenase kinase, isoenzyme 4 | 0.63 | 0.0013 | |
| 18783 | Pla2g4a | Phospholipase A2, group IVA | 0.66 | 0.0008 | |
| 66234 | Sc4mol | Sterol-C4-methyl oxidase-like | 0.63 | <1.00E-04 | |
| 235293 | Sc5d | Sterol-C5-desaturase | 0.47 | 0.0005 | |
| 20397 | Sgpl1 | Sphingosine phosphate lyase 1 | 0.66 | 0.0004 | |
| 56442 | Tde1l | Tumour differentially expressed 2 | 0.63 | 0.0001 | |
| Carbohydrate metabolism | 15926 | Idh1 | Isocitrate dehydrogenase 1 (NADP+) | 0.64 | 0.0005 |
| Nucleic acid-related | 104444 | Rexo2 | RNA exonuclease 2 homologue | 0.57 | < 1.00E-04 |
| Electron transporter | 70495 | Atp6ip2 | Atpase, H+ transporting accessory protein 2 | 0.58 | 0.0003 |
| 11972 | Atp6v0d1 | Atpase, H+ transporting, V0 subunit D isoform 1 | 0.59 | 0.0003 | |
| 11964 | Atp6v1a1 | Atpase, H+ transporting, V1 subunit A, isoform 1 | 0.53 | 0.0004 | |
| 11966 | Atp6v1b2 | Atpase, H+ transporting, V1 subunit B, isoform 2 | 0.62 | 0.0013 | |
| 66335 | Atp6v1c1 | Atpase, H+ transporting, V1 subunit C, isoform 1 | 0.63 | 0.0002 | |
| 12864 | Cox6c | Cytochrome c oxidase, subunit vic | 0.64 | 0.0003 | |
| 13078 | Cyp1b1 | Cytochrome P450, family 1, subfamily b, polypep. 1 | 0.53 | 0.0008 | |
| Glutathione metabolism | 14873 | Gsto1 | Glutathione S-transferase omega 1 | 0.66 | <1.00E-04 |
| Other | 11668 | Aldh1a1 | Aldehyde dehydrogenase family 1, subfamily A1 | 0.23 | 0.001 |
| 56332 | Amotl2 | Angiomotin like 2 | 0.45 | <1.00E-04 | |
| 14199 | Fhl1 | Four and a half LIM domains 1 | 0.62 | 0.0015 | |
| 15939 | Ier5 | Immediate early response 5 | 0.33 | <1.00E-04 | |
| 17161 | Maoa | Monoamine oxidase A | 0.44 | <1.00E-04 | |
| 19317 | Qk | Quaking | 0.58 | 0.001 | |
| 52357 | Wwc2 | WW, C2 and coiled-coil domain containing 2 | 0.61 | <1.00E-04 | |
| Other | 11630 | Aim1 | Absent in melanoma 1 | 1.52 | 0.0001 |
| 11737 | Anp32a | Acidic nuclear phosphoprotein 32 family, member A | 1.5 | <1.00E-04 | |
| 20239 | Atxn2 | Ataxin 2 | 0.66 | <1.00E-04 | |
| 110147 | Bat8 | HLA-B associated transcript 8 | 0.65 | 0.0002 | |
| 223770 | Brd1 | Bromodomain containing 1 | 0.64 | 0.0005 | |
| 12512 | Cd63 | Cd63 antigen | 0.66 | <1.00E-04 | |
| 321022 | Cdv3 | Carnitine deficiency-associated of ventricle 3 | 0.63 | 0.0001 | |
| 13008 | Csrp2 | Cysteine and glycine-rich protein 2 | 1.67 | 0.0001 | |
| 13025 | Ctla2b | Cytotoxic T lymphocyte-associated protein 2 beta | 0.56 | 0.0077 | |
| 14209 | Fin14 | Fibroblast growth factor inducible 14 | 0.63 | 0.0073 | |
| 17263 | Gtl2 | GTL2, imprinted maternally expressed mRNA | 0.65 | 0.0028 | |
| 72568 | Lin9 | Lin-9 homologue | 1.54 | 0.0025 | |
| 17184 | Matr3 | Matrin 3 | 0.65 | 0.006 | |
| 17966 | Nbr1 | Neighbour of Brca1 gene 1 | 0.6 | <1.00E-04 | |
| 30877 | Ns | Nucleostemin | 0.61 | 0.0003 | |
| 18203 | Ntan1 | N-terminal Asn amidase | 0.59 | 0.0001 | |
| 269424 | Phf17 | PHD finger protein 17 | 0.6 | 0.0001 | |
| 56705 | Ranbp9 | RAN binding protein 9 | 0.6 | 0.0003 | |
| 26611 | Rcn2 | Reticulocalbin 2 | 0.59 | 0.0012 | |
| 319714 | Rnase4 | Ribonuclease, rnase A family 4 | 0.51 | 0.0043 | |
| 20715 | Serpina3g | Serine (or cysteine) proteinase inhibitor, clade A-3G | 1.64 | <1.00E-04 | |
| 94186 | Strn3 | Striatin, calmodulin binding protein 3 | 0.52 | 0.0001 | |
| 22134 | Tgoln1 | Trans-Golgi network protein | 0.63 | 0.0005 | |
| 53612 | Vti1b | Vesicle transport t-SNARE interactor, 1B homologue | 0.61 | 0.0001 | |
| 211652 | Wwc1 | WW, C2 and coiled-coil domain containing 1 | 0.47 | <1.00E-04 | |
| 53861 | Zfp265 | Zinc finger protein 265 | 0.61 | 0.0055 | |
| Transport | |||||
| Vesicle transport | 16952 | Anxa1 | Annexin A1 | 0.34 | <1.00E-04 |
| 11745 | Anxa3 | Annexin A3 | 0.27 | <1.00E-04 | |
| 12389 | Cav | Caveolin, caveolae protein | 0.59 | 0.0001 | |
| 13429 | Dnm | Dynamin | 1.62 | <1.00E-04 | |
| 16784 | Lamp2 | Lysosomal membrane glycoprotein 2 | 0.62 | 0.0002 | |
| 53869 | Rab11a | RAB11a, member RAS oncogene family | 0.63 | 0.0004 | |
| 22319 | Vamp3 | Vesicle-associated membrane protein 3 | 0.6 | 0.0036 | |
| Protein-nucleus import | 16649 | Kpna4 | Karyopherin alpha 4 | 0.53 | 0.0002 |
| Protein transport | 216363 | Rab3ip | RAB3A interacting protein | 0.65 | <1.00E-04 |
Genes have been classified into 19 different functional groups. The locus identification number (Locus ID), the gene symbol (gene) and spelled out designation of each gene are shown. For the sake of simplicity, EST clones with unknown functions have not been included in the list.
Fold change in gene expression levels upon a 24-h-long treatment with Y27632.
p-values of genes affected in NIH3T3 cells determined with the F-statistic. p-values lower than 1.00E-04 are shown as <1.00E-04
Fig. 2.
Corroboration of Affymetrix data by quantitative RT-PCR. A, B Expression levels of the indicated up-regulated (A) and downregulated (B) mRNAs by the Y27632 treatment were determined by either microarray (A, grey bars) or quantitative RT-PCR (Q, black bars). Values are expressed as fold change of the appropriate gene with respect to the transcript levels found in untreated NIH3T3 cells
We observed that the Rock-dependent genes were distributed following a Poisson-like distribution when classified in terms of overall fold-change variations (Fig. 1C). Thus, the majority of up- and downregulated genes displayed modest changes (1.5–2.1-fold) when their expression levels were compared between Y27632-treated and untreated NIH3T3 cells. Instead, only a small minority of genes showed fold-change variations outside that interval (Fig. 1C). Upregulated and downregulated genes followed similar trends, although we only observed upregulated loci in the subset of genes displaying variations larger than 6-fold (Fig. 1C). We observed that the genes undergoing the largest upregulation encoded either chemokines (Ccl13, Cxcl6, Ccl7) or secreted factors (Cp and the component 3 of the complement) (Table 1). Instead, the genes with largest repressions encoded for cytoskeletal-related proteins such as actin α2, Ptx3, Thbs1 and Ogn (Table 1).
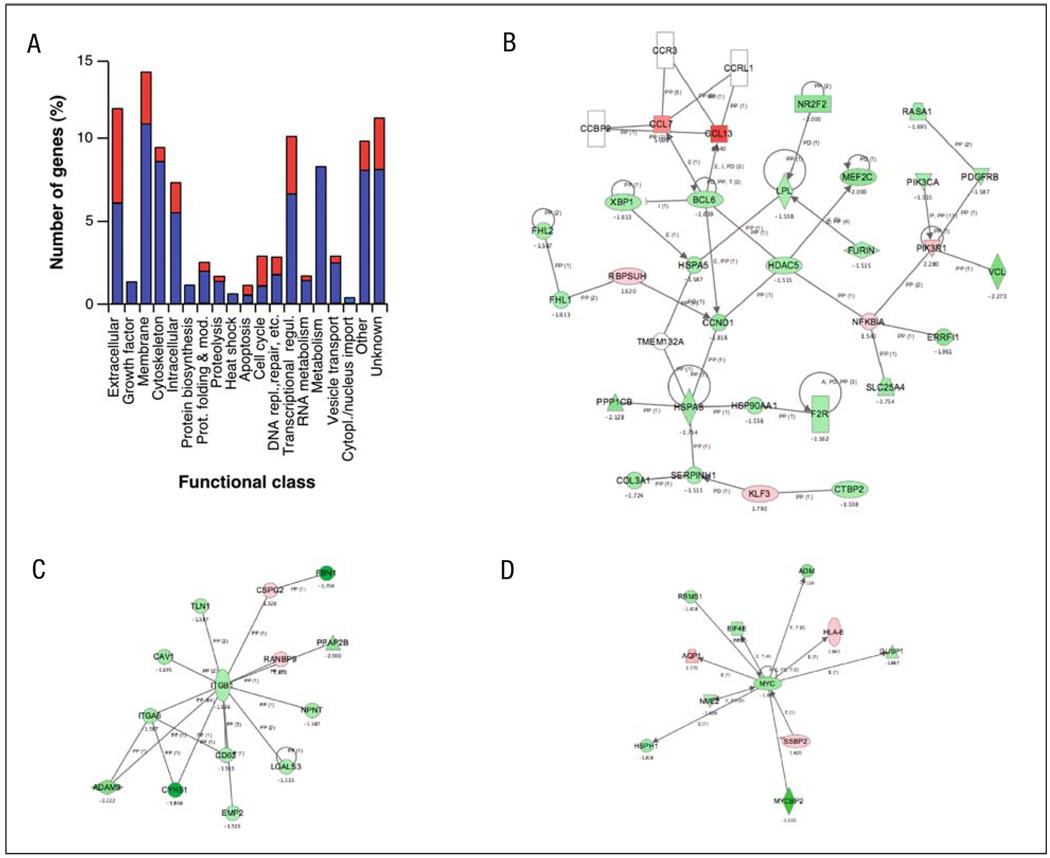
To establish an overview of the transcriptional changes induced by Rock inhibition, we assigned the 289 identified genes regulated by Y27632 to 19 different functional groups using manual annotation procedures (Fig. 3A, Table 1). This analysis revealed that the main functional groups targeted by the Y27632 include those corresponding to extra-cellular matrix proteins, membrane-localised proteins, cytoskeleton, transcriptional regulation and metabolism. With the exception of the apoptosis and cell cycle-related class, all the other functional groups contain a larger number of downregulated than of upregulated genes. In fact, five of those classes (growth factors, protein biosynthesis, heat shock, metabolism, cytoplasmic/nuclear transport) contain only repressed genes (Fig. 3A, Table 1). Interestingly, the largest percentage of upregulated genes is seen in the immune-related (83%), cell cycle (63%), ligand (58%) and membrane receptor (83%) subclasses (Table 1).
Fig. 3.
Functional annotation and characterisation of Y27632-dependent genes. A Classification of up- (red) and downregulated (blue) genes by the Y27632 treatment according to general biological functions. B–D The molecular networks identified using the Ingenuity database in the Y27632-affected transcriptome. Nodes are colour-coded in red (upregulated) or green (downregulated) according to their fold change values
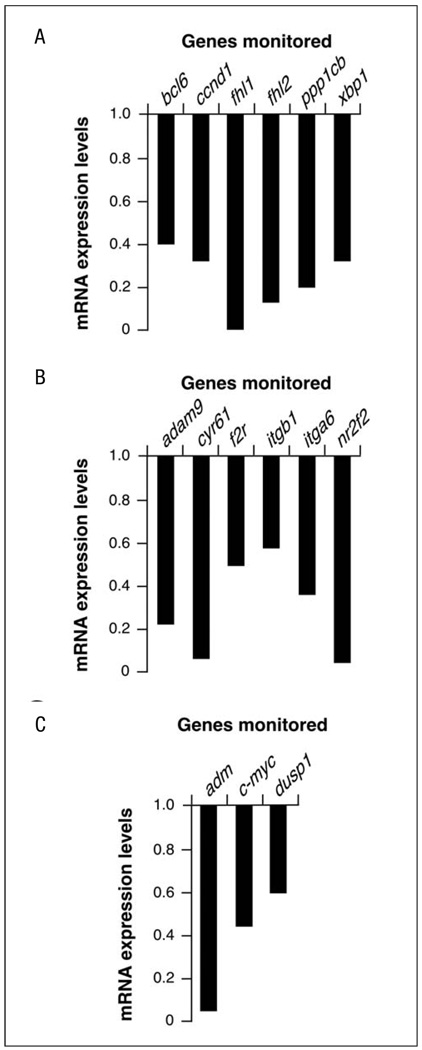
The main problem with functional annotation is that it groups genes according to functional similarity or relatedness. Due to this, this type of analysis usually oversees other important biological information, such as the interconnectivities established among genes of different functional classes, homogeneous alterations of signal transduction pathways, etc. To surmount this problem, we subjected our microarray data to further bioinformatic characterisation using the Ingenuity program. This web-based software allows the identification of common biological processes and molecular networks because it relates each gene entry with a comprehensive database of known protein–protein, transcriptional or enzymatic relationships available for = 10,000 mammalian proteins [29]. At the level of molecular networks, the analysis of the Y27632-targeted transcriptome using this bioinformatics package indicated changes in the expression of genes whose protein products are involved in integrin and c-Myc function (Fig. 3C,D). The composition of the first network is, however, limited to specific integrin subunits and proximal cytoskeletal components (see below), indicating that the signalling cascades located down-stream of integrins are not touched by the inhibition of the Rock route. The detection of the c-Myc network is of interest, because we have shown before that the overexpression of RhoAQ63L promotes this network whereas the inhibition of Rock downmodulates it in rhoA-transformed cells [24]. A third molecular network contained a larger number of protein constituents (30 in total) (Fig. 3B). This network can be further subdivided into two main branches, one that is loosely related with the Ccl7 and Ccl13 chemokines and one of its repressors (the transcriptional factor Bcl6) and another branch connecting the downmodulation of specific nuclear proteins (Xbp1, Nr2f2) with the repression of the expression of extracellular proteins such as collagen, serpin and a lipoprotein lipase (Lpl). Unlike the other two networks, this third molecular conglomerate does include nuclear, cytosolic, membrane and extracellular-located molecules, suggesting that its targeting by the Rock pathway may have some signalling purpose. To our knowledge, however, this network has not been linked to any established biological process so far. Quantitative RT-PCR experiments demonstrated that the selected elements of these three networks are indeed deregulated upon treatment of NIH3T3 cells with the Rock inhibitor (Fig. 4). At the level of biological processes affected, the bioinformatic analysis indicated that the inactivation of Rock by Y27632 alters, in a statistically significant manner (p≤0.001), genes whose protein products are primarily in charge of cellular functions usually regulated by these serine/threonine kinases such as cellular movement (migration, chemotaxis, transmigration, haptotaxis, invasion, scattering), cell morphology and the extracellular matrix. The products of these genes were also either directly (i.e., c-Myc) or indirectly (rest of genes) linked to cell growth and survival processes. However, this latter functional category probably has poor significance from a biological point of view, because we have not detected a large pool of genes directly activated or repressed in proliferating cells (i.e., proteins directly involved in the cell cycle machinery, replication origins, DNA synthesis, etc.) (Table 1). This is consistent with previous observations by us and others indicating that Y27632 treatments do not significantly alter the proliferation rates of fibroblasts [23, 24]. Based on these results, the only obvious common feature observed among these transcriptomal changes is their relationship, direct or indirect, with integral and regulatory components of cellular components usually regulated by Rock proteins such as F-actin cytoskeleton, microtubules and cell movement-related processes. Consistent with this view, it was observed that cytoskeleton-related proteins (actinin, vinculin, calponin, talin, spectrins, actin itself), cytoskeletal regulators (thy-mosin β4, myosin subunits, integrins, transgelin, catenin, Nap125, dynamin, caveolin) and microtubule-related molecules (Macf1, kinesins) showed up in most of the networks and pathways picked up by the Ingenuity software.
Fig. 4.
Corroboration of the molecular networks identified in Fig. 3 by quantitative RT-PCR. A–C RT-PCR-determined expression levels of selected genes belonging to the molecular networks shown in Fig. 3B (A), 3C (B) and 3D (C). Values are expressed as fold change of the appropriate gene with respect to the transcript levels found in untreated NIH3T3 cells
We have previously shown that the inhibition of Rock in RhoAQ63L-transformed NIH3T3 cells with Y27632 also provokes minor changes in the cell transcriptome of these oncogenically transformed cells [24]. The similarity in the experimental conditions used in that work and in the current study made it possible to compare side by side the effects of Y27632 in transformed and non-transformed cells. This analysis indicated that Y27632 induced larger expression changes in the transcriptome of non-transformed (298 genes) than in RhoAQ63L-transformed (179 genes) cells. Moreover, we have observed that the gene subsets targeted by the Rock inhibitor in the parental and the RhoAQ63L-transformed NIH3T3 cells were significantly different, since these two transcriptomes only show 70 coincident target genes. These shared genes showed the same change pattern and belonged to both the upregulated (17 genes) and downregulated (53 genes) classes. This subset of genes included the c-Myc network detected in the Ingenuity analysis, although this interactive molecular network has a larger number of components in RhoA-transformed cells than in the non-transformed parental cells [this work, 24]. Four additional genes, although targeted by Y27632 in both cell types, showed opposite change patterns in normal and RhoAQ63L-transformed cells. The functional classes deregulated by the Y27632 treatment that displayed more disparity between non-transformed and transformed cells were those related with membrane-located, cell adhesion-related proteins, cell cycle, DNA replication and electron transporter. The classes showing more coincident expression changes encompassed those related to extracellular, cell-adhesion-related functions, extracellular ligands, cytoskeleton, regulation of transcription and proteins with unassigned functions. These data indicate that the impact of the Rock pathway on the transcriptome is always small regardless of whether fibroblasts have normal or exacerbated RhoA activity levels. However, the type of genes targeted by this signalling route is significantly different depending on the transformed status of these cells.
In summary, these results indicate that the Rock pathway has a rather weak impact in the transcriptome of normal fibroblasts. Therefore, they are consistent with the idea that the main signalling purpose of this route is to induce phosphoproteomal rather than transcriptomal changes in the cell. Furthermore, the relative small fold change variations found in the majority of Y27632-targeted genes indicated that the transcriptional action of the Rock pathway relies mainly on modulating the levels of activity of already active genes rather than on turning on previously silent loci or turning off active genes. Interestingly, most Rock-dependent transcriptional targets are downmodulated upon inhibition of Rock activity, indicating that the Rock pathway is oriented fundamentally to the upregulation of genes under exponentially growing conditions. Finally, we have observed that the main transcriptional targets affected by the blockage of Rock activity in fibroblasts are related with processes of cell movement, cell shape and F-actin dynamics. Thus, the post-transcriptional action of Rock proteins on the F-actin cytoskeleton appears to be coordinated, in the long-term, with the modulation at the transcriptional level of genes involved in the regulation of those components of the cell architecture. It will be important in the future to complement these studies with others focused on the cell proteome and phosphoproteome to get a comprehensive view of the effects and impact of Rock function in the biology of the mammalian cells.
Acknowledgements
We wish to thank E. Fermiñán (CIC Genomics and Proteomics Unit) and M. Blázquez for array hybridisations and general technical assistance, respectively. This work was supported by aids to XRB from the Ramón Areces Foundation, the Special Action on Genomics and Proteomics of the Spanish Ministry of Education and Science (GEN2003-20239-C06-01), the US National Cancer Institute/NIH (5R01-CA73735-11), the Spanish Ministry of Education and Science (SAF2006-01789), the Castilla-León Autonomous Government (SA053A05) and the Red Temática de Investigación Co-operativa en Cáncer (RTICC) (RD06/0020/0001, Fondo de Investigaciones Sanitarias (FIS), Carlos III Institute, Spanish Ministry of Health). IMB was supported by a FPU fellowship (FP2000-6489) of the Spanish Ministry of Education and Science and by the US National Cancer Institute. All Spanish funding is co-sponsored by the European Union FEDER programme.
Footnotes
The genomic data of this work are deposited in the NCBI Gene Expression Omnibus database (Accession number: GSE5913).
References
- 1.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007 Apr;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 3.Symons M, Rusk N. Control of vesicular trafficking by Rho GTPases. Curr Biol. 2003;13:R409–R418. doi: 10.1016/s0960-9822(03)00324-5. [DOI] [PubMed] [Google Scholar]
- 4.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 5.Coleman ML, Marshall CJ, Olson MF. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat Rev Mol Cell Biol. 2004;5:355–366. doi: 10.1038/nrm1365. [DOI] [PubMed] [Google Scholar]
- 6.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 7.Colicelli J. Human RAS superfamily proteins and related GTPases (250)RE13. 2004 doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Dransart E, Olofsson B, Cherfils J. RhoGDIs revisited: novel roles in Rho regulation. Traffic. 2005;6:957–966. doi: 10.1111/j.1600-0854.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- 10.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 11.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 12.Benitah SA, Valeron PF, van Aelst L, Marshall CJ, Lacal JC. Rho GTPases in human cancer: an unresolved link to upstream and downstream transcriptional regulation. Biochim Biophys Acta. 2004;1705:121–132. doi: 10.1016/j.bbcan.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Boettner B, Van Aelst L. The role of Rho GTPases in disease development. Gene. 2002;286:155–174. doi: 10.1016/s0378-1119(02)00426-2. [DOI] [PubMed] [Google Scholar]
- 14.Budzyn K, Marley PD, Sobey CG. Targeting Rho and Rho-kinase in the treatment of cardiovascular disease. Trends Pharmacol Sci. 2002;27:97–104. doi: 10.1016/j.tips.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 16.Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 17.Mueller BK, Mack H, Teusch N. Rho kinase, a promising drug target for neurological disorders. Nat Rev Drug Discov. 2005;4:387–398. doi: 10.1038/nrd1719. [DOI] [PubMed] [Google Scholar]
- 18.Fu X, Gong MC, Jia T, Somlyo AV, Somlyo AP. The effects of the Rho-kinase inhibitor Y-27632 on arachidonic acid-, GTPgammaS-, and phorbol ester- induced Ca2+-sensitization of smooth muscle. FEBS Lett. 1998;440:183–187. doi: 10.1016/s0014-5793(98)01455-0. [DOI] [PubMed] [Google Scholar]
- 19.Shirao S, Kashiwagi S, Sato M, et al. Sphingosylphosphorylcholine is a novel messenger for Rho-kinase-mediated Ca2+ sensitization in the bovine cerebral artery: unimportant role for protein kinase C. Circulation research. 2002;91:112–119. doi: 10.1161/01.res.0000026057.13161.42. [DOI] [PubMed] [Google Scholar]
- 20.Riento K, Guasch RM, Garg R, Jin B, Ridley AJ. RhoE binds to ROCK I and inhibits downstream signaling. Mol Cell Biol. 2003;23:4219–4229. doi: 10.1128/MCB.23.12.4219-4229.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward Y, Yap SF, Ravichandran V, Matsumura F, Ito M, Spinelli B, Kelly K. The GTP binding proteins Gem and Rad are negative regulators of the Rho-Rho kinase pathway. J Cell Biol. 2002;157:291–302. doi: 10.1083/jcb.200111026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, Helfman DM. Cytoplasmic p21Cip1 is involved in Ras-induced inhibition of the ROCK/LIMK/cofilin pathway. J Biol Chem. 2004;279:1885–1891. doi: 10.1074/jbc.M306968200. [DOI] [PubMed] [Google Scholar]
- 23.Sahai E, Ishizaki T, Narumiya S, Treisman R. Transformation mediated by RhoA requires activity of ROCK kinases. Curr Biol. 1999;9:136–145. doi: 10.1016/s0960-9822(99)80067-0. [DOI] [PubMed] [Google Scholar]
- 24.Berenjeno IM, Nunez F, Bustelo XR. Transcriptomal profiling of the cellular transformation induced by Rho subfamily GTPases. Oncogene. 2007;26:4295–4305. doi: 10.1038/sj.onc.1210194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiariello M, Marinissen MJ, Gutkind JS. Regulation of c-myc expression by PDGF through Rho GTPases. Nat Cell Biol. 2001;3:580–586. doi: 10.1038/35078555. [DOI] [PubMed] [Google Scholar]
- 26.Han JS, Macarak E, Rosenbloom J, Chung KC, Chaqour B. Regulation of Cyr61/CCN1 gene expression through RhoA GTPase and p38MAPK signaling pathways. Eur J Biochem. 2003;270:3408–3421. doi: 10.1046/j.1432-1033.2003.03723.x. [DOI] [PubMed] [Google Scholar]
- 27.Croft DR, Olson MF. The Rho GTPase effector ROCK regulates cyclin A, cyclin D1, and p27Kip1 levels by distinct mechanisms. Mol Cell Biol. 2006;26:4612–4627. doi: 10.1128/MCB.02061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uehata M, Ishizaki T, Satoh H, et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 29.Calvano SE, Xiao W, Richards DR, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005 Oct 13;437(7061):1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 30.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4:e83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]