Abstract
Background and purpose:
Monoacylglycerol lipase (MGL) is a presynaptic serine hydrolase that inactivates the endocannabinoid neurotransmitter, 2-arachidonoyl-sn-glycerol. Recent studies suggest that cysteine residues proximal to the enzyme active site are important for MGL function. In the present study, we characterize the role of cysteines in MGL function and identify a series of cysteine-reactive agents that inhibit MGL activity with nanomolar potencies by interacting with cysteine residue 208.
Experimental approach:
A series of cysteine traps were screened for the ability to inhibit MGL in vitro. Rapid dilution assays were performed to determine reversibility of inhibition. Molecular modelling and site-directed mutagenesis were utilized to identify cysteine residues targeted by the inhibitors.
Key results:
The screening revealed that 2-octyl-4-isothiazolin-3-one (octhilinone) inhibited purified rat recombinant MGL (IC50= 88 ± 12 nM) through a partially reversible mechanism. Initial structure–activity relationship studies showed that substitution of the n-octyl group of octhilinone with a more lipophilic oleoyl group increased inhibitor potency (IC50= 43 ± 8 nM), while substitution with a methyl group produced the opposite effect (IC50= 239 ± 68 nM). The inhibitory potency of octhilinone was selectively decreased by mutating cysteine 208 in MGL to glycine (IC50; wild-type, 151 ± 17 nM; C208G, 722 ± 74 nM), but not by mutation of other cysteine residues (C32, C55, C201, C208 and C242).
Conclusions and implications:
The results indicated that cysteine 208 plays an important role in MGL function and identified a novel class of isothiazolinone-based MGL inhibitors with nanomolar potency in vitro.
Keywords: monoacylglycerol lipase, 2-arachidonoylglycerol, endocannabinoids
Introduction
Monoacylglycerol lipase (MGL) is a cytosolic serine hydrolase that cleaves monoacylglycerols into fatty acid and glycerol through a catalytic mechanism that involves a classical serine-aspartate-histidine triad (Karlsson et al., 1997). MGL is a member of the α/β-hydrolase family of enzymes (Ollis et al., 1992) and is distantly related to microbial lysophospholipases and haloperoxidases (Karlsson et al., 1997). Because of its abundant expression in mammals in lipid-metabolizing tissues, such as white fat and liver, MGL is thought to catalyse the final step of the lipolytic cascade that releases fatty acids from triacylglycerol stores (Chon et al., 2007). In the brain, however, a primary role of MGL may be to carry out the hydrolysis and inactivation of the endocannabinoid neurotransmitter, 2-arachidonoyl-sn-glycerol (2-AG) (Dinh et al., 2002).
Several findings implicate MGL in the termination of neuronal 2-AG signalling. First, virally induced MGL overexpression attenuates receptor-dependent accumulation of 2-AG in primary cultures of rat cortical neurons (Dinh et al., 2002) and, conversely, RNAi-mediated silencing of MGL increases 2-AG levels in HeLa cells (Dinh et al., 2004). Second, electron microscopy studies have shown that MGL is localized to presynaptic nerve terminals of glutamatergic projection neurons and γ-aminobutyric acid (GABA)-ergic interneurons, where cannabinoid type-1 (CB1) receptors targeted by 2-AG are also found (Gulyas et al., 2004). Third, blockade of MGL activity by pharmacological inhibitors, such as biphenyl-3-ylcarbamic acid cyclohexyl ester (URB602), elevates 2-AG levels in rat brain (Hohmann et al., 2005; Makara et al., 2005; King et al., 2007) and enhances 2-AG-mediated signalling in both acutely dissected hippocampal slices (Makara et al., 2005; Hashimotodani et al., 2007) and midbrain periaqueductal grey substance in vivo (Hohmann et al., 2005).
Potent and selective MGL inhibitors are not currently available. Nevertheless, such inhibitors would be useful to uncover physiological functions of 2-AG (see Freund et al., 2003; Piomelli, 2003) and validate MGL as a target for therapeutic drugs (for review, see Piomelli, 2005). Three classes of chemical compounds that interact with MGL have been described thus far. The compound URB602, mentioned above, inhibits MGL activity through a non-competitive, partially reversible mechanism (Hohmann et al., 2005; Makara et al., 2005; King et al., 2007). By contrast, lipophilic serine-reactive agents, such as methyl arachidonylfluorophosphonate (MAFP), irreversibly block MGL, presumably by forming a covalent bond with the catalytic nucleophile, serine 122 (Goparaju et al., 1999). An irreversible mechanism has also been postulated to explain the ability of cysteine-reactive agents, such as N-arachidonylmaleimide (NAM) (Saario et al., 2005) and tetraethylthiuram disulphide (disulfiram) (Labar et al., 2007), to inhibit MGL with nanomolar potencies. In particular, computational and mass spectrometry studies have shown that NAM forms a Michael addition product with cysteine 242 in MGL, which is located in close proximity of the catalytic serine 122 (Saario et al., 2005; Zvonok et al., 2008).
In the present study, we combined structure–activity relationship (SAR) and computational strategies to further characterize the role of cysteine residues in MGL function and identify a new family of isothiazolinone-based MGL inhibitors. Kinetic and site-directed mutagenesis studies showed that the prototype in this class of compounds, octhilinone, inhibits MGL through a partially reversible mechanism that involves a specific interaction with cysteine 208.
Methods
MGL expression and purification
Purified MGL was prepared as previously described (King et al., 2007). Briefly, a full-length rat MGL cDNA was subcloned into the pEF15b vector (Novagen, La Jolly, CA, USA) containing an N-terminal histidine tag and transformed in DH5αEscherichia coli cells. Positive clones were expressed in Rosetta 2(De3)pLysS E. coli cells (Novagen) using 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). Cells were lysed by using a French press, and membrane fractions were collected and resuspended in 50 mM HEPES pH 7.4, 300 mM NaCl, 3 mM β-mercaptoethanol, 1% Triton X-100. Solubilized supernatant was loaded onto a TALON column (Clontech, Mountain View, CA, USA), and MGL was eluted from the column by using a stepwise gradient of imidazole (10 to 200 mM) at a concentration around 75 mM.
MGL assay
Monoacylglycerol lipase activity was measured as previously described (King et al., 2007). Briefly, either 10 ng of purified MGL or 2.5–50 µg of protein from MGL-transfected HeLa cell lysates were preincubated with inhibitors for 10 min at 37°C in assay buffer (50 mM Tris-HCL, pH 8.0, 0.5 mg·mL−1 bovine serum albumin, fatty acid-free). Following preincubation, 2-oleoylglycerol (2-OG) substrate (10 µM final) was added, and samples were incubated for an additional 10 min at 37°C. Reactions were stopped with chloroform : methanol (2:1, vol : vol), containing heptadecanoic acid (5 nmol) as an internal standard. In some experiments with cell lysates, 1(3)heptadecanoylglycerol (1-HG) and heptadecenoic acid were used as substrate and internal standard respectively. Samples were subjected to centrifugation at 2000×g at 4°C for 10 min, and the organic layers were collected and dried under a stream of N2. The residues were suspended in chloroform : methanol (1:3, vol : vol) and analysed by liquid chromatography/mass spectrometry (LC/MS).
LC/MS analysis
We used a reversed-phase Eclipse C18 column (30 × 2.1 mm i.d., 1.8 µM, Agilent Technologies, Wilmington, DE, USA) eluted with 95% of solvent A and 5% solvent B for 0.6 min at a flow rate of 0.6 mL·min−1 with column temperature set at 50°C. Solvent A consisted of methanol containing 0.25% acetic acid and 5 mM ammonium acetate. Solvent B consisted of water containing 0.25% acetic acid and 5 mM ammonium acetate. Under these conditions, analytes eluted from the column at the following retention times: oleic acid, 0.34 min; heptadecanoic acid, 0.37 min; heptadecenoic acid, 0.29 min. Electrospray ionization was in the negative mode, capillary voltage was set at 4 Kv, and fragmentor voltage was 100 V. N2 was used as drying gas at a flow rate of 13 L·min−1 and a temperature of 350°C. Nebulizer pressure was set at 60 psi. For quantification purposes, we monitored the [M-H]− ions of m/z= 281.3 for oleic acid, m/z= 269 for heptadecanoic acid, and m/z= 267 for heptadecenoic acid.
Rapid dilution assay
Rapid dilution assays were performed as previously described (Copeland, 2005). Briefly, samples containing of purified MGL (100-fold concentrated compared with standard assays) were preincubated with 10-fold the IC50-equivalent concentration of octhilinone, MAFP or vehicle (dimethylsulphoxide, DMSO, final concentration 2%) for 20 min at 37°C. Samples were then diluted 100-fold with assay buffer containing substrate to initiate reactions, and the time course of product formation was measured by LC/MS.
Molecular modelling
The crystal structure of chloroperoxidase L (EC 1.11.1.10) from Streptomyces lividans (PDB code 1A88) was used as a template (Hofmann et al., 1998; Saario et al., 2005) to produce a first-generation model of the P20-W289 MGL segment, based on the 1A88-rMGL alignment previously reported (Saario et al., 2005). MODELLER 7.0 software was used for 3D model building, and standard loop modelling settings were applied (Sali and Blundell, 1993). A second-generation alignment was created, moving a small gap from one loop (between S185 and R186, in the loop connecting helices α5 and α6) to another (between A203 and G204, in the loop connecting helices α6 and α7) within the lid domain. This modification resulted in C201 being buried within the active site rather than being exposed to the solvent, as in the first-generation alignment, while retaining the same identity, similarity and gap scores, and maintaining comparable matches in the secondary structure between chloroperoxidase L and rMGL as suggested by Saario (Saario et al., 2005) (Figure S1). Five second-generation 3D models were produced. PROCHECK (Laskowski et al., 1993) was used to assess the overall geometric quality of the structures, and the best model (G-factor of −0.23, 86% of allowed residues and 0.8% of disallowed residues) was selected and utilized for modelling purposes. Hydrogen atoms were added by the Biopolymer module of Sybyl (Version 7.2, Tripos Inc., St. Louis, MO, USA), choosing the tautomeric states of histidines that maximized the number of hydrogen bonds within the protein; then the geometry was relaxed by energy minimization. The 2-AG substrate was interactively docked into the rMGL-binding site by using the Dock_minimize module of Sybyl. The docking procedure was followed by an energy minimization of the complex and by 20 cycles of simulated annealing (heating phases of 1000 fs at 700 K followed by cooling phases of 1000 fs at 200 K). The annealed rMGL/2-AG complex was finally submitted to 500 ps of molecular dynamics at 310 K followed by energy minimization. Octhilinone was docked in the active site by using the Dock_minimize procedure. Starting from the resulting complex, a covalent adduct was built, modifying the topology of the inhibitor and C208. The resulting adduct was submitted to a final energy minimization. In all the energy minimizations, simulated annealing and molecular dynamics calculations, the backbone atoms of the protein were kept frozen to preserve the rMGL tertiary structure. MMFF94s (Halgren, 1999) and MMFF94 (Halgren, 1996) force fields were employed, with the dielectric constant set to 1. Energy minimizations were performed to a gradient of 0.1 kcal·(mol·Å)−1.
Site-directed mutagenesis
A full-length rat MGL cDNA was subcloned into the pEF6/V5-His vector (Invitrogen, San Diego, CA, USA). The MGL gene was modified by using the QwikChange II XL Site-Directed Mutagenesis Kit (Strategene, La Jolla, CA, USA) following the manufacturer's instructions. Primers were commercially synthesized (Invitrogen). Following DNA sequencing, positive clones were identified and transfected into HeLa cells by using the Superfect transfectant reagent (Qiagen, Valencia, CA, USA), following standard protocol. Cells were resuspended in ice-cold Tris-HCL (50 mM, pH 8.0) containing 0.32 M sucrose and harvested by using a cell scraper. Lysates were sonicated on ice for 1 min followed by three rounds of freeze thawing. Samples were subjected to centrifugation at 100 000×g at 4°C for 30 min to separate membrane (pellet) and cytosolic (supernatant) fractions.
Protein analysis
Protein concentrations were measured by using the BCA Protein Assay kit (Pierce, Rockford, IL, USA). Samples (5 µg) were electrophoresed on a 4–20% SDS-polyacrylamide gel (Invitrogen) and transferred to a polyvinylidene difluoride membrane (Amersham, Piscataway, NJ, USA). Membranes were blocked in 10% milk and incubated in the presence of anti-V5 monoclonal antibody (1:3000, 12 h; Invitrogen). Immunoreactive bands were visualized by using the ECL-Plus kit (Amersham), exposed to high performance chemiluminescence film (Amersham) and developed with a Mini-Medical film processor (AFP Imaging, Elmsford, NY, USA). As a loading control, membranes were stripped for 15 min at room temperature and probed again by using an anti-actin monoclonal antibody (1:10 000, 12 h; Calbiochem, La Jolla, CA, USA)
Statistical analyses
All results are expressed as mean ± SEM. Non-linear regression analyses were performed by using Prism (GraphPad Software Inc., La Jolla, CA, USA). Statistical significance was assessed by the Student's t-test.
Materials
Methyl arachidonylfluorophosphonate, NAM and NEM were purchased from Cayman Chemical (Ann Arbor, MI, USA). Compounds 1, 2, 6–8 and dithiothreitol (DTT) were from Sigma-Aldrich (St. Louis, MO, USA). Compound 5 was from Acros Organics (Morris Plains, NJ, USA). 2-OG and 1-HG were from Indofine Chemical (Hillsborough, NJ, USA) and Nu-Chek Prep (Elysian, MN, USA) respectively. All drug and molecular target nomenclature follows the Guide to Receptors and Channels (Alexander et al., 2008).
Compound 3 (2-octadec-9-enyl-isothiazol-3-one) was prepared by cyclization of N-oleyl-propiolamide as described below. (i) Preparation of N-octadec-9-enyl-propiolamide: N-oleylamine (16 mmol) was added under nitrogen atmosphere to a cooled (−40°C) mixture of propiolic acid (16 mmol), triethylamine (16 mmol) and ethylchloroformate (16 mmol), and the resulting mixture was stirred for 90 min at −40°C and then for 16 h at −10°C. Solvent evaporation and purification by flash chromatography (SiO2, DCM : MeOH 98:2) gave N-octadec-9-enyl-propiolamide (yield: 68%) as yellow oil. 1H NMR (300 Mz, CDCl3): δ 0.88 (t, 3H, J= 6.39 Hz), 1.28 (m, 22H), 1.52 (m, 2H), 2.01 (m, 4H), 2.77 (s, 1H), 3.29 (q, 2H, J= 6.93), 5.36 (m, 2H), 6.12 (bs, 1H). APCI-MS m/z+ 320.5 [MH]+. (ii) Preparation of 2-octadec-9-enyl-isothiazol-3-one: a mixture of N-octadec-9-enyl-propiolamide (1.5 mmol) and ammonium thiosulphate (1.5 mmol) in water-dimethylformamide (DMF) was stirred at 0°C for 21 h under nitrogen atmosphere; HCl (37%) was added and the mixture refluxed for 10 min. Neutralization by sodium bicarbonate solution, solvent evaporation and flash chromatography (SiO2, dicloromethane : methanol 98:2) afforded a yellow oil, which was further purified by semi-preparative HPLC (Supelco Discover Bio-Wide Pore, 25 cm × 10 mm, 10 microm, 3 mL·min−1 flow rate, acetonitrile : water 85:15, l = 254 nm), giving 2-octadec-9-enyl-isothiazol-3-one (yield 7%) as a colourless oil. 1H NMR (300 Mz, CDCl3): d; 0.88 (t, 3H, J= 6.36 Hz), 1.28 (m, 22H), 1.72 (m, 2H), 2.01 (m, 4H), 3.78 (t, J= 7.26, 2H), 5.34 (m, 2H), 6.25 (d, 1H, J= 6.24), 8.03 (d, 1H, J= 6.21). APCI-MS m/z+ 352.4 [MH]+. Anal. (C21H37NOS) C, H, N.
2-octyl-benzo[d]isothiazol-3-one (4) was prepared by adding benzo[d]isothiazol-3-one (Sigma-Aldrich, 10 mmol) and anhydrous potassium carbonate (16 mmol) to a mixture of 1-chlorooctane (Sigma-Aldrich, 10 mmol) and sodium iodide (16 mmol), which had been refluxed for 30 min in anhydrous DMF, and the reaction vessel was heated by microwaves in two steps of 10 min, setting the temperature to 100°C and 120°C respectively (CEM Discover apparatus). After cooling and filtration the mixture was diluted with water and ice and extracted with dichloromethane. Solvent evaporation afforded a crude oil, which was eluted by flash chromatography (SiO2, dichloromethane : methanol 99.5:0.5) and distilled under reduced pressure (b.p. 240°C at 1 mmHg). 1H NMR (CDCl3, 300 MHz): δ 0.82–0.87 (m, 3H), 1.24–1.40 (m, 10H), 1.74 (q, 2H, J= 7.4 Hz), 3.87 (t, 2H, J= 7.3 Hz), 7.38 (ddd, 1H, J= 7.9, 6.9, 1.4 Hz), 7.53 (ddd, 1H, J= 8.1, 1.3, 0.8 Hz), 7.58 (ddd, 1H, J= 8.1, 6.8, 1.2 Hz), 8.03 (d, 1H, J= 7.9 Hz). Anal. (C15H21NOS) C, H, N.
Results
MGL inhibition by cysteine trapping compounds
Previous studies have shown that rat cerebellar membranes contain an MGL-like activity that is weakly inhibited by the cysteine-reactive agent N-ethylmaleimide, but is highly sensitive to inhibition by the lipophilic maleimide NAM (Saario et al., 2005). We replicated these results using recombinant purified rat MGL overexpressed in E. coli (NEM, IC50= 2.7 ± 0.5 µM; NAM, IC50= 46 ± 7 nM; n= 3) (Figure 1A). Based on computational (Saario et al., 2005) and structural data (Zvonok et al., 2008), the inhibitory effect of NAM has been attributed to the ability of this compound to react with C242 and form a Michael addition product. To further examine the functional role of cysteine residues, we screened a series of structurally different cysteine traps for their ability to inhibit MGL activity (Table 1). All compounds contained an electrophilic centre that could covalently bind sulphydryl groups. Only one, however, 2-octyl-4-isothiazolin-3-one (octhilinone, 1) was potent in inhibiting MGL, that is, IC50 in nM range (n= 5; Figure 1B). By contrast, compounds 5 (an epoxysuccinate) and 6 (an alkylidene-thioxothiazolidinone) displayed weak inhibitory activities (IC50 in µM range; n= 3), while compounds 7 (a thiadiazole) and 8 (a propoxyphthalide) were inactive at all concentrations tested (up to 1 mM, Table 1).
Figure 1.
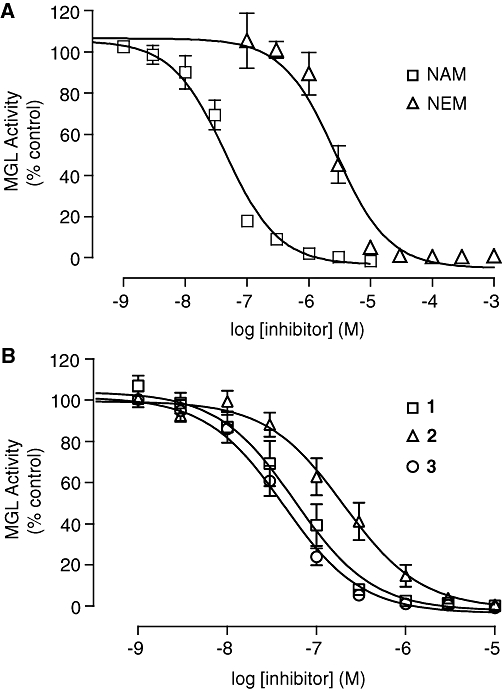
Inhibition of monoacylglycerol lipase (MGL) activity by cysteine-reactive agents. Concentration–response curves for the inhibition of purified MGL overexpressed in Escherichia coli by (A) the maleimides N-arachidonylmaleimide (NAM) and NEM and by (B) the isothiazolinones octhilinone (1), 2-methyl-4-isothiazolin-3-one (2) and 2-oleoyl-4-isothiazolin-3-one (3). Results are expressed in percentage of activity in the presence of vehicle (dimethylsulphoxide, final concentration 1%) (mean ± SEM, n= 3–5).
Table 1.
Inhibition of purified recombinant MGL by cysteine trap compounds
| Compound | Structure | IC50 |
|---|---|---|
| N-arachidonylmaleimide | 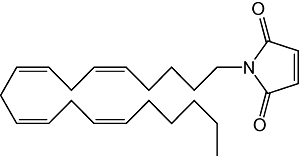 |
46 ± 7 nM |
| NEM | 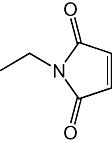 |
3 ± 0.3 µM |
| 1(Octhilinone) | 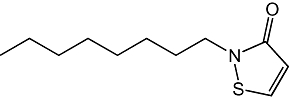 |
88 ± 12 nM |
| 2 | 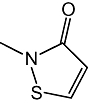 |
239 ± 68 nM |
| 3 | 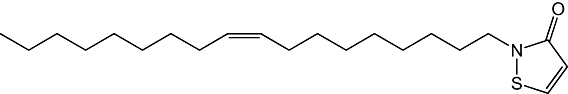 |
43 ± 8 nM |
| 4 | 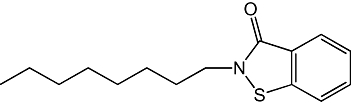 |
59 ± 7 nM |
| 5 | 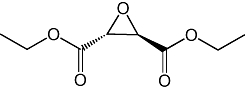 |
20 ± 7 µM |
| 6 | 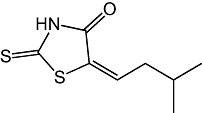 |
28 ± 0.5 µM |
| 7 | 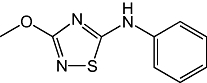 |
>100 µM |
| 8 | 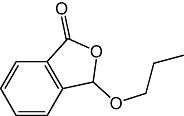 |
>1 mM |
Results are expressed as mean ± SEM (n= 3–4).
Figure 1B shows the results of a preliminary SAR study, which investigated the effects of select chemical modifications of octhilinone on MGL inhibition. Substitution of the n-octyl group with a methyl group (2) resulted in a threefold decrease in potency compared with the parent compound (Table 1; P < 0.05, n= 4). By contrast, introduction of a larger and more lipophilic oleoyl group at the same site (3) resulted in a twofold increase in potency (Table 1; P < 0.05, n= 4). Replacement of the isothiazolinone moiety with a benzisothiazolinone group (4) also yielded a potent inhibitor (Table 1; n= 3).
Mechanism of MGL inhibition by octhilinone
We utilized a rapid dilution assay (Copeland, 2005) to assess whether octhilinone inhibits MGL through a reversible or irreversible mechanism. Purified MGL was first preincubated with a concentration of octhilinone that was 10-fold higher than its IC50 value, and then diluted 100-fold (to 10% of its IC50 value). As shown in Figure 2A, the dilution produced a partial recovery of enzyme activity. By contrast, when MGL was incubated with the irreversible serine-reacting probe MAFP, MGL activity did not recover after dilution (Figure 2A). The partial reversibility of the effect of octhilinone is not consistent with the chemical reactivity of a Michael addition product, suggesting that octhilinone may form instead a reducible disulphide bond with a cysteine residue in MGL (Figure 2B). Two findings support this possibility. First, the benzisothiazolinone 4, which cannot undergo Michael addition, was as potent as octhilinone at inhibiting MGL activity (Table 1). Second, adding a low concentration of the reducing agent DTT (10 µM) to the incubation mixture produced a 47-fold decrease in the inhibitory potency of octhilinone (Figure 2C) without affecting basal MGL activity (data not shown). As expected, incubation with DTT produced no change in the potency of MAFP (Figure 2C). We interpret these results to indicate that octhilinone inhibits MGL activity by interacting with a cysteine residue in this enzyme.
Figure 2.
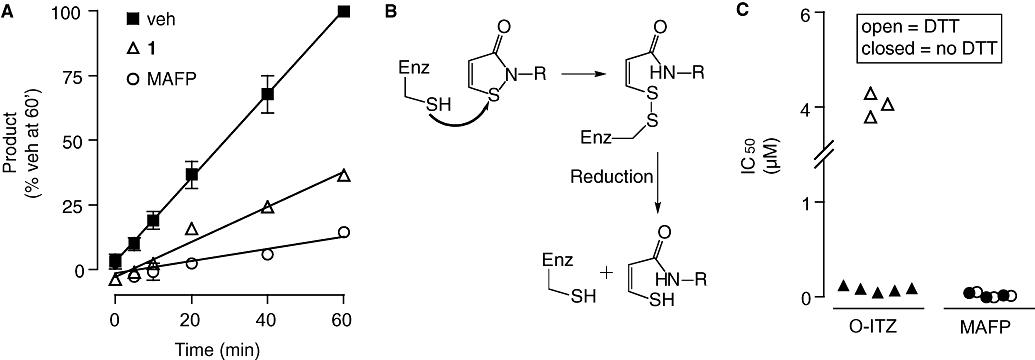
Reversibility of monoacylglycerol lipase (MGL) inhibition by octhilinone. (A) Rapid dilution assays of purified MGL in the presence of vehicle (dimethylsulphoxide, final concentration 2%), octhilinone or methyl arachidonylfluorophosphonate (MAFP). See text for details. Results are expressed in percentage of product generated after a 60 min incubation with vehicle (mean ± SEM, n= 3). (B) Scheme illustrating the formation of a disulphide adduct between octhilinone and cysteine. (C) Effects of dithiothreitol (DTT, 10 µM) on inhibitory potency (IC50, µM) of octhilinone and MAFP towards purified MGL. Results are from individual experiments, each performed in duplicate.
Localization of cysteine residues in MGL
There are six cysteine residues in rat and mouse MGL, four of which are conserved in their human orthologue (Figure 3A). A homology model of MGL constructed using as a template the crystal structure of chloroperoxidase L from S. lividans (Figure 3B) (Saario et al., 2005) positions C242 on a conserved loop of the enzyme, in close proximity of a histidine residue (H269) that is part of the catalytic triad (S122, D239, H269) (Karlsson et al., 1997). Residues C201 and C208 are located within the lid domain, wherefrom they might be able to extend their sulphydryl groups towards the substrate-binding site, while residues C32 and C55 are positioned in two structurally conserved regions: C32 in the second β-strand and C55 in a loop before the beginning of the first α-helix. C301 is not included in the model because it occupies a region of MGL that is not present in the chloroperoxidase template. Consistent with previous studies (Saario et al., 2005), our computational analyses identify three cysteines (C201, C208 and C242) which, based on their location on the tertiary structure of the enzyme, may exert regulatory roles in MGL function and might, therefore, be targeted by isothiazolinone-based compounds.
Figure 3.
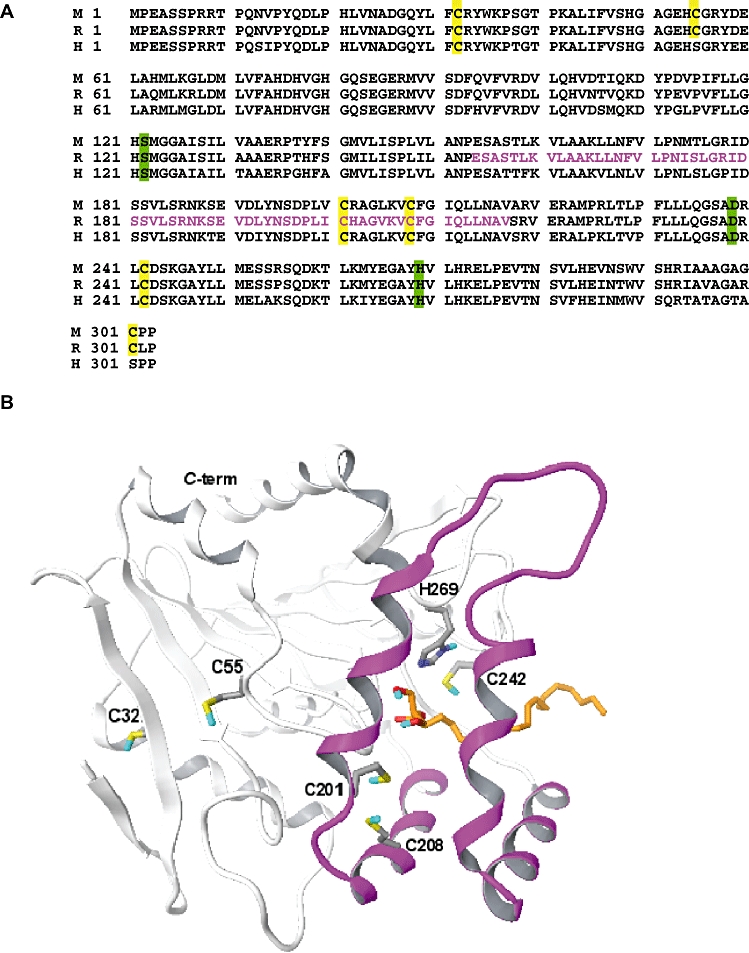
(A) Primary amino-acid sequence alignment of mouse (M), rat (R) and human (H) monoacylglycerol lipase (MGL). Cysteines (yellow) and catalytic triad residues (green) are highlighted. The putative lid domain, based on the crystal structure of chloroperoxidase L from Streptomyces lividans, is shown in magenta. (B) Computational three-dimensional model of rat MGL. The α/β core structure of the enzyme, shown in white, contains the catalytic triad. 2-arachidonoyl-sn-glycerol is shown in orange. The lid domain is in magenta. Carbon atoms are coloured grey; oxygen, red; sulphur, yellow; hydrogen, blue.
Site-directed mutagenesis
To test these possibilities, we generated a series of MGL mutants in which individual cysteines were replaced with glycines. We expressed the mutants in HeLa cells and evaluated the impact of each mutation on MGL activity. Wild-type (WT) and mutant proteins displayed comparable levels of heterologous expression (Figure 4A). Nevertheless, the C242G mutation produced a striking reduction in MGL activity (Figure 4B), which was due to a decrease in maximal reaction velocity (VMAX in pmol·min−1·µg−1; WT, 58 ± 7; C242G, 4 ± 1; n= 3–4) rather than a change in Michaelis constant (KM in µM; WT, 115 ± 32; C242G, 98 ± 28; n= 3–4). Mutations affecting C201 and C208 also caused significant decreases in enzyme activity, albeit smaller than that produced by C242G (Figure 4C). By contrast, mutations to cysteines located distal to the active site of MGL (C32G, C55G and C301G) had no effect on MGL activity (Figure 4B). The results highlight the important functional role of cysteine residues proximal to the MGL active site, as previously suggested by computational (Saario et al., 2005) and structural studies (Zvonok et al., 2008).
Figure 4.
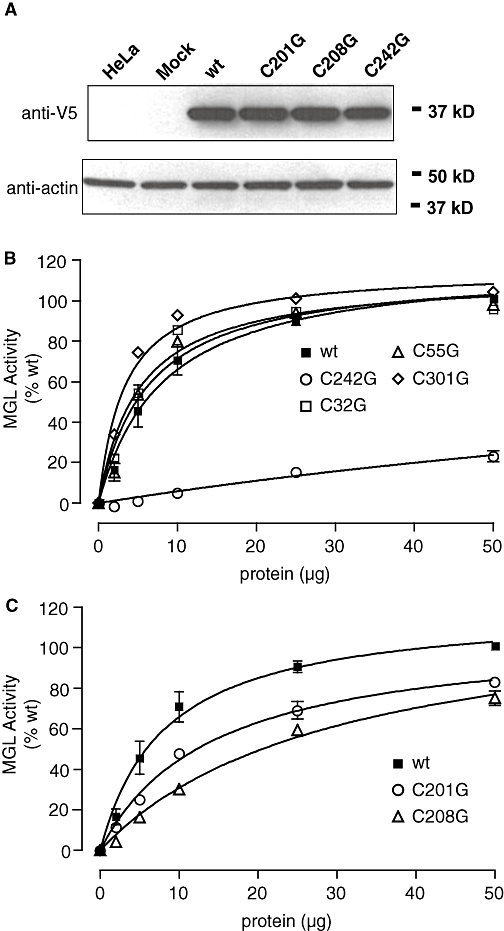
Effects of cysteine mutations (to glycine) on MGL activity. (A) Heterologous expression of monoacylglycerol lipase (MGL) in HeLa cells. Transiently transfected cells were lysed and subjected to SDS-polyacrylamide gel electrophoresis. Recombinant MGL was visualized by Western blot analysis by using an anti-V5 antibody. Loading efficiency was controlled for using an anti-actin antibody. (B) MGL activity in MGL mutants C242G, C32G, C55G and C301G. (C) MGL activity in MGL mutants C201G and C208G. Results are expressed in percentage of wild-type (wt) activity (50 µg protein) (mean ± SEM, n= 3).
Interaction of octhilinone with cysteine 208
We next examined whether cysteine mutations influence the ability of octhilinone to inhibit MGL. As illustrated in Figure 5, mutating C208 to glycine reduced the inhibitory potency of octhilinone, compared with WT MGL (WT, IC50= 151 ± 17 nM; C208G, IC50= 722 ± 74 nM, P < 0.0001; n= 4) (Figure 5A), whereas mutation of all other cysteines produced no significant effect (Figure 5B). Interestingly, the C208G mutation had no effect on the inhibitory potency of NAM, which was selectively reduced instead by mutation of C242 or C201 (WT, IC50= 30 ± 3 nM; C242G, IC50= 107 ± 17 nM, P < 0.05; C201G, IC50= 442 ± 79 nM, P < 0.01; n= 3) (Figure 5C).
Figure 5.
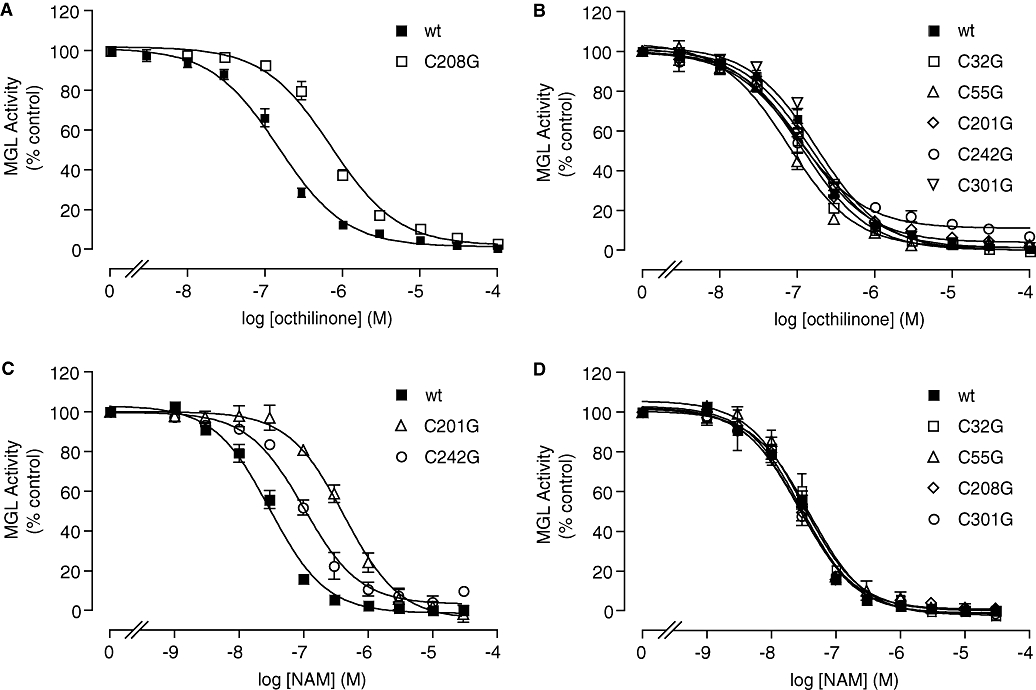
Effect of cysteine mutations on monoacylglycerol lipase (MGL) inhibition by octhilinone and N-arachidonylmaleimide (NAM). Concentration-response curves for the inhibition of MGL activity by octhilinone (A,B) or NAM (C,D) in wild-type MGL (wt) and individual MGL mutants. Results are expressed in percentage of control activity (dimethylsulphoxide, final concentration 1%) (mean ± SEM, n= 3–5).
Discussion
In the present study, we have further characterized the role of cysteine residues in MGL function and identified a novel family of highly potent isothiazolinone-based inhibitors of MGL activity. Preliminary SAR studies on one of these compounds, octhilinone, revealed that increasing lipophilicity of the carbon side chain enhanced MGL inhibitory potency. Indeed, substitution of the n-octyl group with a more lipophilic oleoyl chain increased potency, while substitution with a methyl group decreased it. A role for lipophilicity in modulating the inhibitory potency of cysteine traps towards MGL has been previously noted by Saario et al. (2005), who explored the SAR of maleimide-based compounds such as NAM.
N-arachidonylmaleimide reacts with C242, which gives a Michael addition to the carbon–carbon double bond (Zvonok et al., 2008). The isothiazolinone group of octhilinone presents a sulphenamide fragment and a conjugated double bond, both potentially able to make an electrophilic attack on a cysteine sulphydryl group. It has been shown that 2-methyl-isothiazolin-3-one (2) reacts with N-acetylcysteine in aqueous solutions, yielding a disulphanyl derivative resulting from attack at the sulphenamide fragment (Alvarez-Sanchez et al., 2002). Yet the close resemblance between the structures of isothiazolinone and maleimide suggests that derivatives of these compounds could inhibit MGL through a Michael addition. To explore the mechanism by which octhilinone inhibits MGL, we first performed rapid dilution assays, which test the reversibility of enzyme–inhibitor interactions (Copeland, 2005). The results showed that octhilinone blocked MGL activity through a partially reversible mechanism. We hypothesized that this partial reversibility might arise from the formation of a reducible disulphide bond with a cysteine sulphydryl group in MGL, rather than from the generation of a Michael addition product. Accordingly, increasing the reducing potential of the assay buffer by addition of a low concentration of DTT (10 µM) produced a marked decrease in the inhibitory potency of octhilinone. In further support of our hypothesis, the benzisothiazolinone 4, which cannot undergo Michael addition because its conjugated double bond is involved in benzene aromaticity, was as potent as octhilinone at inhibiting MGL activity. Together, the results suggest that the sulphenamide fragment in octhilinone forms a disulphide bond with one or more cysteine residues in MGL.
To identify such residues, we systematically mutated all cysteines in MGL and transiently expressed the mutants in HeLa cells. We found that mutations affecting cysteines that are located in close proximity of the active site caused a loss in basal MGL activity. This result is in apparent contrast with a recent study, which reported that mutating either C208 or C242 does not affect the basal activity of human MGL (Zvonok et al., 2008). The discrepancy might be attributed, however, to the considerable differences in enzyme assay protocols utilized in the two studies. Specifically, we employed an LC/MS-based assay, which quantifies both substrates and products in the MGL reaction, while Zvonok et al. (2008) used a spectrophotometric assay, which measures hydrolysis of the fluorogenic reporter substrate arachidonoyl,7-hydroxy-6-methoxy-4-methylcoumarin ester.
In addition to its effect on baseline MGL activity, mutation of C208 caused a marked decrease in the inhibitory potency of octhilinone. The remaining inhibitory effect of octhilinone on the C208G mutant was likely due to non-selective interactions with other amino acid residues in MGL, because mutations targeting other cysteines did not affect the compound's inhibitory potency. Thus, the high-potency component of the inhibitory effect of octhilinone on MGL appears to be due to a preferential interaction of this compound with C208. A docking position for a disulphide adduct of octhilinone with C208 is illustrated in Figure 6.
Figure 6.
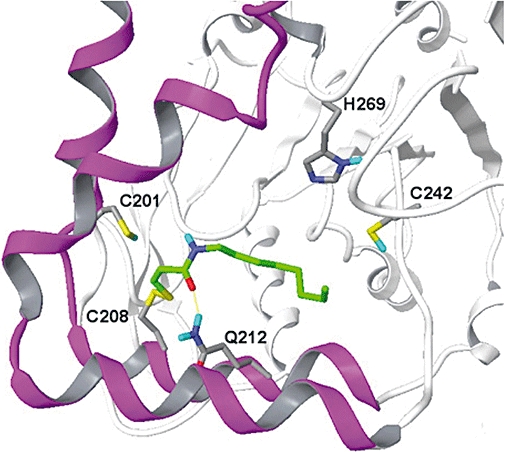
Computational model showing the orientation of a possible disulphide adduct between octhilinone and C208 of monoacylglycerol lipase. The protein is rotated by 90° around its vertical axis with respect to Figure 3B. Protein carbon atoms are shown in grey, octhilinone carbon atoms in green. Lid domain, magenta; α/β core structure, white; nitrogen, dark blue; oxygen, red; sulphur, yellow; hydrogen, blue.
Previous studies have demonstrated that NAM inhibits MGL by interacting with C242 (Saario et al., 2005; Zvonok et al., 2008). Our result with the C242G mutant, which shows a ∼3.5-fold decrease in NAM potency, supports the conclusion that inhibition of MGL by NAM occurs via C242. However, we also see a significant rightward shift in potency resulting from mutation of C201. This unexpected observation suggests a previously unrecognized interaction between NAM and C201, which deserves further investigation.
In conclusion, the serine lipase MGL may be a crucial component of the presynaptic mechanism by which neurons inactivate the endocannabinoid 2-AG. Thus, targeting this enzyme might have a variety of therapeutic applications, including analgesia, anti-inflammation and neuroprotection (Hohmann et al., 2005; Comelli et al., 2007; Mechoulam and Shohami, 2007; Desroches et al., 2008). The present results underscore the functional importance of select cysteine residues in MGL activity and introduce a novel isothiazolinone-based scaffold, which may be used to design novel MGL inhibitors.
Acknowledgments
This work was supported by a grant from the National Institute on Drug Abuse (R01 DA-012447). The authors would like to thank the S.I.T.I. and C.I.M. (University of Parma) for supplying molecular modelling software licences.
Glossary
Abbreviations:
- 1-HG
1(3)heptadecanoylglycerol
- 2-AG
2-arachidonoyl-sn-glycerol
- 2-OG
2-oleoylglycerol
- DMF
dimethylformamide
- DTT
dithiothreitol
- MGL
monoacylglycerol lipase
- MAFP
methyl arachidonylfluorophosphonate
- NAM
N-arachidonylmaleimide
Conflicts of interest
None.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1 Sequence alignments of rat monoacylglycerol lipase (MGL) (P20-W289 segment) and Chloroperoxidase L from Streptomyces lividans. rMGL1: first-generation alignment [Saario 2005]: identity score: 17%; similarity score: 32%; gap score: 7% (calculated by GeneDoc: http://www.nrbsc.org/gfx/genedoc). rMGL2: second-generation alignment: identity score: 17%; similarity score: 31%; gap score: 7%. Conserved residues are highlighted in bold, cysteine residues of rMGL are coloured in red. S185-R186 and A203-G204 residues are underlined and coloured in blue. Secondary structure elements of Chloroperoxidase L, derived from the crystallographic coordinates (PDB code 1A88), are denoted by the letters E (b sheet) or H with (a helix). Numbers refers to rMGL1 sequence.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alexander SPH, Mathie A, Peters JA. Guide to receptors and channels (GRAC), 3rd edn. Br J Pharmacol. 2008;153(Suppl. 2):S1–S209. doi: 10.1038/sj.bjp.0707746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Sanchez R, Basketter D, Pease C, Lepoittevin JP. Studies of chemical electivity of hapten, reactivity, and skin sensitization potency. 3. Synthesis and studies on the reactivity toward model nucleophiles of the 13C-labeled skin sensitizers, 5-chloro-2-methylisothiazol-3-one (MCI) and 2-methylisothiazol-3-one (MI) Chem Res Toxicol. 2002;16:627–636. doi: 10.1021/tx0256634. [DOI] [PubMed] [Google Scholar]
- Chon SH, Zhou YX, Dixon JL, Storch J. Intestinal monoacylglycerol metabolism: developmental and nutritional regulation of monoacylglycerol lipase and monoacylglycerol acyltransferase. J Biol Chem. 2007;282:33346–33357. doi: 10.1074/jbc.M706994200. [DOI] [PubMed] [Google Scholar]
- Comelli F, Giagnoni G, Bettoni I, Colleoni M, Costa B. The inhibition of monoacylglycerol lipase by URB602 showed an anti-inflammatory and anti-nociceptive effect in a murine model of acute inflammation. Br J Pharmacol. 2007;152:787–794. doi: 10.1038/sj.bjp.0707425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland RA. Lead optimization and SAR for reversible inhibitors. In: Copeland RA, editor. Evaluation of Enzyme Inhibitors in Drug Discovery. Hoboken, NJ: John Wiley and Sons Inc.; 2005. pp. 125–128. [Google Scholar]
- Desroches J, Guindon J, Lambert C, Beaulieu P. Modulation of the anti-nociceptive effects of 2-arachidonoyl glycerol by peripherally administered FAAH and MGL inhibitors in a neuropathic pain model. Br J Pharmacol. 2008;155:913–924. doi: 10.1038/bjp.2008.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, et al. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci USA. 2002;99:10819–10824. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TP, Kathuria S, Piomelli D. RNA interference suggests a primary role for monoacylglycerol lipase in the degradation of the endocannabinoid 2-arachidonoylglycerol. Mol Pharmacol. 2004;66:1260–1264. doi: 10.1124/mol.104.002071. [DOI] [PubMed] [Google Scholar]
- Freund TF, Katona I, Piomelli D. Role of endogenous cannabinoids in synaptic signaling. Physiol Rev. 2003;83:1017–1066. doi: 10.1152/physrev.00004.2003. [DOI] [PubMed] [Google Scholar]
- Goparaju SK, Ueda N, Taniguchi K, Yamamoto S. Enzymes of porcine brain hydrolyzing 2-arachidonoylglycerol, an endogenous ligand of cannabinoid receptors. Biochem Pharmacol. 1999;57:417–423. doi: 10.1016/s0006-2952(98)00314-1. [DOI] [PubMed] [Google Scholar]
- Gulyas AI, Cravatt BF, Bracey MH, Dinh TP, Piomelli D, Boscia F, et al. Segregation of two endocannabinoid-hydrolyzing enzymes into pre- and postsynaptic compartments in the rat hippocampus, cerebellum and amygdala. Eur J Neurosci. 2004;20:441–458. doi: 10.1111/j.1460-9568.2004.03428.x. [DOI] [PubMed] [Google Scholar]
- Halgren TA. Merck molecular force field. I. Basis, form, scope, parametrization, and performance of MMFF94. J Comput Chem. 1996;17:490–519. [Google Scholar]
- Halgren TA. MMFF VI. MMFF94s option for energy minimization studies. J Comput Chem. 1999;20:720–729. doi: 10.1002/(SICI)1096-987X(199905)20:7<720::AID-JCC7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Hashimotodani Y, Ohno-Shosaku T, Kano M. Presynaptic monoacylglycerol lipase activity determines basal endocannabinoid tone and terminates retrograde endocannabinoid signaling in the hippocampus. J Neurosci. 2007;27:1211–1219. doi: 10.1523/JNEUROSCI.4159-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann B, Tolzer S, Pelletier I, Altenbuchner J, van Pee KH, Hecht HJ. Structural investigation of the cofactor-free chloroperoxidases. J Mol Biol. 1998;279:889–900. doi: 10.1006/jmbi.1998.1802. [DOI] [PubMed] [Google Scholar]
- Hohmann AG, Suplita RL, Bolton NM, Neely MH, Fegley D, Mangieri R, et al. An endocannabinoid mechanism for stress-induced analgesia. Nature. 2005;435:1108–1112. doi: 10.1038/nature03658. [DOI] [PubMed] [Google Scholar]
- Karlsson M, Contreras JA, Hellman U, Tornqvist H, Holm C. cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J Biol Chem. 1997;272:27218–27223. doi: 10.1074/jbc.272.43.27218. [DOI] [PubMed] [Google Scholar]
- King AR, Duranti A, Tontini A, Rivara S, Rosengarth A, Clapper JR, et al. URB602 inhibits monoacylglycerol lipase and selectively blocks 2-arachidonoylglycerol degradation in intact brain slices. Chem Biol. 2007;14:1357–1365. doi: 10.1016/j.chembiol.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labar G, Bauvois C, Muccioli GG, Wouters J, Lambert DM. Disulfiram is an inhibitor of human purified monoacylglycerol lipase, the enzyme regulating 2-arachidonoylglycerol signaling. Chembiochem. 2007;8:1293–1297. doi: 10.1002/cbic.200700139. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Makara JK, Mor M, Fegley D, Szabo SI, Kathuria S, Astarita G, et al. Selective inhibition of 2-AG hydrolysis enhances endocannabinoid signaling in hippocampus. Nat Neurosci. 2005;8:1139–1141. doi: 10.1038/nn1521. [DOI] [PubMed] [Google Scholar]
- Mechoulam R, Shohami E. Endocannabinoids and traumatic brain injury. Mol Neurobiol. 2007;36:68–74. doi: 10.1007/s12035-007-8008-6. [DOI] [PubMed] [Google Scholar]
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, et al. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- Piomelli D. The molecular logic of endocannabinoid signalling. Nat Rev Neurosci. 2003;4:873–884. doi: 10.1038/nrn1247. [DOI] [PubMed] [Google Scholar]
- Piomelli D. The endocannabinoid system: a drug discovery perspective. Curr Opin Investig Drugs. 2005;6:672–679. [PubMed] [Google Scholar]
- Saario SM, Salo OMH, Nevalainen T, Poso A, Laitinen JT, Järvinen T, et al. Characterization of the sulfhydryl-sensitive site in the enzyme responsible for hydrolysis of 2-arachidonoyl-glycerol in rat cerebellar membranes. Chem Biol. 2005;12:649–656. doi: 10.1016/j.chembiol.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Zvonok N, Pandarinathan L, Williams J, Johnston M, Karageorgos I, Janero DR, et al. Covalent inhibitors of human monoacylglycerol lipase: ligand-assisted characterization of the catalytic site by mass spectrometry and mutational analysis. Chem Biol. 2008;15:854–862. doi: 10.1016/j.chembiol.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


