Abstract
Background and purpose:
We conducted a genome wide gene expression analysis to explore the biological aspects of 15-methoxypinusolidic acid (15-MPA) isolated from Biota orientalis and tried to confirm the suitability of 15-MPA as a therapeutic candidate for CNS injuries focusing on microglia.
Experimental approach:
Murine microglial BV2 cells were treated with 15-MPA, and their transcriptome was analysed by using oligonucleotide microarrays. Genes differentially expressed upon 15-MPA treatment were selected for RT-PCR (reverse transcription-polymerase chain reaction) analysis to confirm the gene expression. Inhibition of cell proliferation and induction of apoptosis by 15-MPA were examined by bromodeoxyuridine assay, Western blot analysis of poly-ADP-ribose polymerase and flow cytometry.
Key results:
A total of 514 genes were differentially expressed by 15-MPA treatment. Biological pathway analysis revealed that 15-MPA induced significant changes in expression of genes in the cell cycle pathway. Genes involved in growth arrest and DNA damage [gadd45α, gadd45γ and ddit3 (DNA damage-inducible transcript 3)] and cyclin-dependent kinase inhibitor (cdkn2b) were up-regulated, whereas genes involved in cell cycle progression (ccnd1, ccnd3 and ccne1), DNA replication (mcm4, orc1l and cdc6) and cell proliferation (fos and jun) were down-regulated. RT-PCR analysis for representative genes confirmed the expression levels. 15-MPA significantly reduced bromodeoxyuridine incorporation, increased poly-ADP-ribose polymerase cleavage and the number of apoptotic cells, indicating that 15-MPA induces apoptosis in BV2 cells.
Conclusion and implications:
15-MPA induced apoptosis in murine microglial cells, presumably via inhibition of the cell cycle progression. As microglial activation is detrimental in CNS injuries, these data suggest a strong therapeutic potential of 15-MPA.
Keywords: 15-methoxypinusolidic acid, microglia, oligonucleotide microarray analysis, cell cycle, growth arrest and DNA damage inducible genes, poly-ADP-ribose polymerase
Introduction
15-methoxypinusolidic acid (15-MPA) is a pinusolide derivative isolated from leaves of Biota orientalis (Cupressaceae). Since we first reported 15-MPA as a neuroprotective agent against glutamate-induced neurotoxicity in primary cultures of rat cortical cells (Koo et al., 2002), 15-MPA has been investigated for the possibility as a therapeutic drug for neurodegenerative disorders. We found that 15-MPA was neuroprotective against staurosporine-induced apoptosis by preventing the increase in intracellular Ca2+ concentration and cellular oxidation in rat cortical cells (Koo et al., 2007). In contrast, Shults et al. (2006) reported that pinusolide, a precursor of 15-MPA, decreased proliferation and induced apoptosis in tumour cells.
Microglia are resident immune cells in the CNS and are also recognized as key cellular mediators of neurodegenerative processes (Cuadros and Navascués, 1998; Kaur et al., 2001; Streit et al., 2004; Kim and Joh, 2006). They readily become activated in response to infection or injury. Activated microglia secrete a number of pro-inflammatory and neurotoxic factors such as interleukin-1β, tumour necrosis factor α, nitric oxide and prostanoids, which cause neuronal damage (Stoll and Jander, 1999; Liu and Hong, 2003; Lai and Todd, 2006). Microglial activation and proliferation after CNS injury such as ischaemia, trauma and spinal cord injury, have been reported to be associated with up-regulation of cell cycle components (Kato et al., 2003; Koguchi et al., 2003; Cernak et al., 2005; Di Giovanni et al., 2005; Tian et al., 2007). In an earlier study, 15-MPA inhibited lipopolysaccharide-induced inflammation in murine microglial BV2 cells (Choi et al., 2008), which was independent of nuclear factor-κB. Apart from its anti-inflammatory effect, 15-MPS also activated extracellular signal-regulated kinase 1/2, which plays key regulatory roles in many biological pathways.
High-density DNA microarray technology has been proven to be a powerful tool for the analysis of gene expression profiles and, therefore, for assessing biological responses (Gene Ontology Consortium, 2001; Hamadeh et al., 2002; Zhong and Sternberg, 2006). In the present study, we took a systematic approach to exploring the biological effects of 15-MPA and conducted a genome wide gene expression analysis in microglial BV2 cells. The results revealed that a number of genes related to cell cycle regulation, DNA replication, cell proliferation and signal transduction were differentially regulated by 15-MPA in the microglial transcriptome.
Methods
Preparation of 15-MPA
15-MPA was isolated from the leaves of B. orientalis as previously reported, and its purity was higher than 95% (Koo et al., 2002). For each experiment, 15-MPA was dissolved in dimethyl sulphoxide (DMSO) (final culture concentration, 0.05%). Preliminary studies indicated that the solvent had no effect on cell viability at the concentration used.
Cell line and culture conditions
Murine microglial BV2 cells, originally developed by Dr V Bocchini at University of Perugia (Perugia, Italy; Blasi et al., 1990), were generously provided by Dr Sun Yeou Kim at Kyunghee University (Suwon, Korea). The cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen Corporation, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen Corporation), 100 units·mL−1 penicillin G, 100 µg·mL−1 streptomycin and 0.25 µg·mL−1 amphotericin B at 37°C in a humidified atmosphere of 95% air/5% CO2.
RNA isolation and DNA microarray
BV2 cells were plated overnight in six-well plates at a density of 5 × 106 cells per plate in the maintenance medium, starved for 4 h in the medium without serum, and then treated with 50 µmol·L−1 15-MPA for 6 h. Serum-free media were used as a vehicle control. Total RNA was prepared from BV2 cells by using the TRIzol® reagent (Invitrogen Corporation) according to the manufacturer's protocol. Quantification and purity analysis (OD260/OD280 ratio) of RNA were performed by using a ND-1000 UV/VIS spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). A commercial mouse genome survey array (Applied Biosystems, Foster City, CA, USA) containing 33 015 probes of 60-mer oligonucleotides representing a set of 30 988 individual mouse genes and more than 1000 control probes was used for differential gene expression profiling. Microarray experiments were performed according to the manufacturer's instructions. Digoxigenin (DIG)-UTP-labelled cRNA was generated from 2 µg of total RNA and amplified by using chemiluminescent reverse transcription in vitro transcription labelling kit (Applied Biosystems). Briefly, each microarray was prehybridized in hybridization buffer with blocking reagent at 55°C for 1 h. DIG-labelled cRNA targets (10 µg) were fragmented to 100–400 bps and hybridized with each prehybridized microarray at 55°C for 16 h. The arrays were washed with hybridization wash buffer and then with chemiluminescence rinse buffer. Chemiluminescent signals were generated by incubating the arrays with anti-DIG alkaline phosphatase and chemiluminescence substrate. Images were collected for each microarray by using the Model 1700 Chemiluminescent Microarray Analyzer (Applied Biosystems). Microarray images were auto-gridded and the chemiluminescent signals were quantified, corrected for background, spatially normalized and exported for quality report. Microarray data with quality reports above the manufacturer's threshold were used for further analysis.
Analysis of microarray expression data
Signal intensities were imported into GenPlex software (ISTECH, Inc., Goyang, Korea), where inter-array quantile normalization was performed to minimize the effect of external variables that might be introduced into the data. Quality filtering of unreliable spots was performed before normalization, and 14 961 probes (flag value <5000 and S/N ≥3) were taken into account. Then, the expression intensities were log2 transformed. We took the average value from the gene expression ratio obtained in three biological replicates. Differentially expressed genes (DEGs) were selected on the basis of ratios (fold change ≥2) and Welch's t-test (P < 0.05). For further analysis, DEGs were categorized according to their biological function by using the PANTHER (Protein ANalysis THrough Evolutionary Relationships) protein classification system (http://www.pantherdb.org/) and to their biological pathway using the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (http://www.genome.jp/kegg/) (Trajkovski et al., 2008).
Reverse transcription-polymerase chain reaction (RT-PCR) analysis
BV2 cells were seeded, starved, treated, and then total RNA of the cells was extracted as described above (RNA isolation and DNA microarray). Total RNA was reverse-transcribed using the AccuPower® RT Premix (Bioneer Corporation, Daejeon, Korea) with 2 µg total RNA and oligo dT. Primer sequences were as follows: gadd45α, sense: 5′-GCTCAACGTAGACCCCGATA-3′, antisense: 5′-GTTCGTCACCAGCACACAGT-3′; DNA damage-inducible transcript 3 (ddit3), sense: 5′-GCATGAAGGAGAAGGAGCAG-3′, antisense: 5′-ACTGTTCATGCTTGGTGCAG-3′; cyclin-dependent kinase inhibitor 2B (cdkn2b), sense: 5′-TTACCAGACCTGTGCACGAC-3′, antisense: 5′-GCAGATACCTCGCAATGTCA-3′; fos, sense: 5′-CCAGTCAAGAGCATCAGCAA-3′, antisense: 5′-AAGTAGTGCAGCCCGGAGTA-3′; orc1l, sense: 5′-AACCCAGCTATGACGACCAC-3′, antisense: 5′-ACGGATGGGTTCTTTGTGAG-3′; ccnd1, sense: 5′-AGTGCGTGCAGAAGGAGATT-3′, antisense: 5′-CACAACTTCTCGGCAGTCAA-3′; cell division cycle 6 homologue (cdc6), sense: 5′-AGGAGCCAGACAGTCCTCAA-3′, antisense: 5′-GGGTCAAAAGCAGCAAAGAG-3′; gapdh, sense: 5′-TCGTGGAGTCTACTGGCGT-3′, antisense: 5′-GCCTGCTTCACCACCTTCT-3′. PCR amplification of the resulting cDNA template was conducted by using the following conditions for 30 cycles: denaturation at 94°C for 30 s, annealing at 57°C for 45 s and extension at 72°C for 30 s. The amplified DNA products were visualized on 1.2% agarose gels and photographed under UV light.
Western blot analysis
BV2 cells were plated, starved and treated as described above (RNA isolation and DNA microarray). Cells were harvested with ice-cold phosphate-buffered saline (PBS) and centrifuged at 110×g for 5 min at 4°C. The pellet was lysed in 60 µL PRO-PREP™ protein extraction solution (iNtRON Biotechnology, Seongnam, Korea) containing 4 mmol·L−1 sodium orthovanadate and incubated at −20°C for 20 min. Cell lysates were centrifuged at 20 000×g for 5 min at 4°C, then the supernatants were collected. Protein content was determined by using the BCA protein assay (Pierce, Rockford, IL, USA). Equal amounts of protein (50 µg) were loaded per lane onto 12% SDS-PAGE gel. Proteins were transferred to nitrocellulose membranes (Invitrogen Corporation) and subsequently blocked in 4% bovine serum albumin (BSA)-TBST (100 mmol·L−1 Tris, pH 8.0, 150 mmol·L−1 NaCl and 0.1% Tween 20) for 2 h at room temperature. Antibody to poly-ADP-ribose polymerase (PARP; 1:500 dilution; Cell Signaling Technology, Danvers, MA, USA) was employed in 4% BSA-TBST. The membrane was incubated with the primary antibody at 4°C overnight. After washing three times with TBST, the immunoreactive band was visualized by using secondary antibody conjugated to horseradish peroxidase (1:10 000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and ECL chemiluminescence detection kit (Amersham Biosciences, Piscataway, NJ, USA). The membrane was stripped with Restore™ Western Blot Stripping Buffer (Pierce) for 30–40 min at room temperature and reprobed with anti-actin antibody (1:1000 dilution in 4% BSA-TBST, overnight, 4°C; Santa Cruz Biotechnology).
Bromodeoxyuridine (BrdU) incorporation assay
BV2 cells were plated overnight in 96-well plates at a density of 1 × 104 cells per well in the maintenance medium, starved for 4 h in the medium without serum, and then treated with 12.5, 25 or 50 µmol·L−1 of 15-MPA for 6 h. Serum-free media were used as a vehicle control for 15-MPA. The assay was performed by using the Calbiochem® BrdU Cell Proliferation Assay (EMD Biosciences, Inc., Darmstadt, Germany) according to the instruction manual. In brief, after labelling the cells with BrdU for 16 h, the cells were fixed in the 96-well plate, and then anti-BrdU antibody was added to the wells, allowing it to bind to the incorporated BrdU. Unbound antibody was washed away, and horseradish peroxidase-conjugated goat anti-mouse IgG antibody was added. The chromogenic substrate, tetramethylbenzidine, was added to each well, and the absorbance at 450 nm was measured by using a microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
Cell viability assay
BV2 cells were plated overnight in 96-well plates at a density of 8 × 104 cells per well in the maintenance medium and starved for 3 h in the medium without serum. Cells were then treated with 15-MPA (12.5, 25 or 50 µmol·L−1) for 6 or 22 h before the addition of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) followed by further incubation for 2.5 h. The medium was displaced by 130 µL DMSO, and the amount of MTT formazan product was determined by measuring the absorbance at 562 nm using a microplate reader (BioTek Instruments, Inc.).
Flow cytometry
BV2 cells were plated overnight in 100 mm culture dishes at 90% confluency in the maintenance medium, incubated for 4 h in the serum-free medium and then treated with 15-MPA (50 µmol·L−1) for 6 or 22 h. Serum-free media were used as a vehicle control. Cells were harvested and washed with ice-cold PBS and then incubated with fluorescein isothiocyanate (FITC)-conjugated anti-Annexin V antibody (BD Biosciences, San Jose, CA, USA) at 4°C for 20–30 min in the dark room. After washing with ice-cold PBS, cells were stained with propidium iodide (PI) (BD Biosciences) at room temperature for 15 min, followed by flow cytometry using FACscan flow cytometer (BD Biosciences). Percentages of apoptotic cells were determined by using Cell Quest software (BD Biosciences).
Statistical analysis
Each experiment was performed at least in triplicate. All data are presented as the mean ± standard deviation (SD). Statistical analyses were performed by using SigmaStat® 3.1 (Systat Software, Point Richmond, CA, USA). The differences among groups were analysed by one way anova (analysis of variance) followed by Dunnett's test for comparing a control group with all other groups. The differences between two groups were evaluated by unpaired t-test. Values of P < 0.05 were considered to be statistically significant.
Results
Identification and analysis of genes differentially expressed by 15-MPA
Microarray analysis was performed to examine the effect of 15-MPA on expression of genes in microglial BV2 cells. 15-MPA treatment distinctly induced alterations in the expression pattern of a number of genes in BV2 cells. After comparison of fold change and statistical analysis between 15-MPA-treated and untreated cells, 514 genes were identified as DEGs (fold change ≥2 and P < 0.05), 254 genes were up-regulated, and 260 genes were down-regulated by 15-MPA.
To understand the mechanisms underlying the cellular response to 15-MPA, we carried out an ontological study on 514 DEGs. For this purpose, gene enrichment analyses were performed by using expression analysis tool provided on the PANTHER protein classification system. The biological functions of DEGs showing significant enrichment were obtained by comparing the lists of DEGs by 15-MPA with a reference list from total genes on ABI 1700 gene survey array. By comparing the number of the ‘expected’ and the ‘observed’ genes and taking consideration of the P-value, the biological functions that are statistically over- or under-represented by 15-MPA were extracted. The analysis showed significant enrichment of DEGs involved in biological functions including amino acid metabolism, cell cycle, cell cycle control, cell proliferation and differentiation, chemosensory perception, DNA metabolism, DNA replication and immunity and defence (Figure 1).
Figure 1.
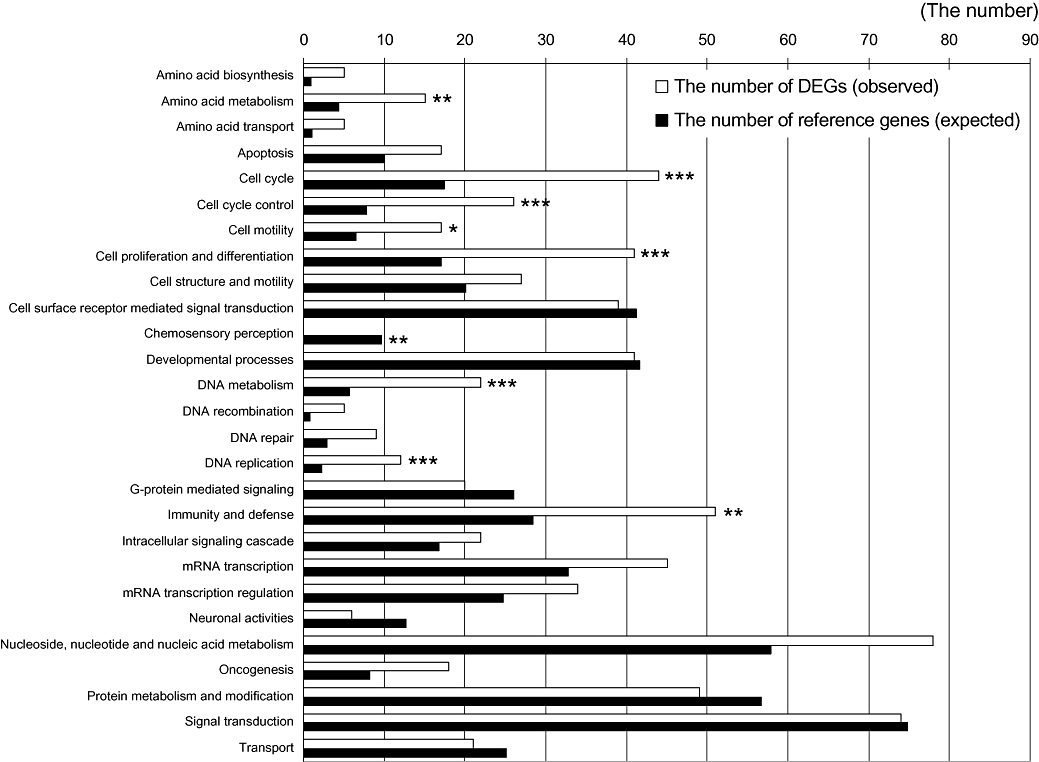
List of enriched gene sets in the differentially expressed genes (DEGs) altered by 15-methoxypinusolidic acid (15-MPA) in BV2 cells. The lists of DEGs altered by 15-MPA were compared with a reference list from total genes on ABI 1700 gene survey array by using the Biological Process of the PANTHER classification system. The ‘expected’ number means the number of genes for the specific biological process when the genes are randomly selected from a reference list. The ‘observed’ number means the number of DEGs for the specific biological process. The over- and under-represented biological processes with high significance by Fisher's exact test with the Bonferroni correction for multiple testing were marked with * (*P < 0.05, **P < 0.01, ***P < 0.001).
Pathway analysis of 15-MPA based on DEG analysis
To obtain further information on the mechanism of 15-MPA in BV2 cells, 514 DEGs were used again for pathway analysis using KEGG database. The results of pathway analysis using the KEGG database were shown in Tables 1–8. We selected biological pathways that had more than five DEGs (fold change ≥2 and P < 0.05) for further consideration in order to focus on the relationship among the DEGs in the pathway (Table 1). Some of the genes in Table 1 were listed in several groups as they contributed to more than one biological function. The biological pathways in which functionally related genes were repeatedly involved were as follows: MAPK signalling pathway (Table 2), cell cycle (Table 3), Jak-STAT signalling pathway (Table 4), focal adhesion (Table 5), B cell receptor signalling pathway (Table 6), T cell receptor signalling pathway (Table 7) and Wnt signalling pathway (Table 8). Genes involved in cytokine–cytokine receptor interaction (ccl2, ccl6, csf2rβ2, cxcl2, il1β, il10rα, osm, tnfrsf19l and tnfrsf9), neuroactive ligand–receptor interaction (c3ar1, c5ar1, fpr1, gpr35, p2ry1, p2ry5 and ptger4) and complement and coagulation cascades (c1qα, c3ar1, c5ar1, f7, f10 and thbd) were excluded from consideration. They were almost certainly different from each other (except c3ar1 and c5ar1) and different from those in Tables 2–8 (sharing only csf2rβ2, il1β, il10rα and osm).
Table 1.
Classification of genes differentially expressed by 15-methoxypinusolidic acid in BV2 cells using the KEGG database
| Biological pathway | Number of genes |
|---|---|
| MAPK signalling pathway | 13 |
| Cell cycle | 10 |
| Cytokine–cytokine receptor interaction | 9 |
| Jak-STAT signalling pathway | 8 |
| Neuroactive ligand–receptor interaction | 7 |
| Complement and coagulation cascades | 6 |
| Focal adhesion | 6 |
| B cell receptor signalling pathway | 5 |
| Colorectal cancer | 5 |
| T cell receptor signalling pathway | 5 |
| Wnt signalling pathway | 5 |
Table 8.
Genes that changed expression significantly (P < 0.05) upon 15-methoxypinusolidic acid treatment were functionally grouped (see Table 1)
| Gene ID | Gene symbol | Gene title | Fold change | P-value |
|---|---|---|---|---|
| 595490 | ppp3cc | Protein phosphatase 3, catalytic subunit, γ isoforms | 2.74 | 1.78E-02 |
| 463565 | fos | FBJ osteosarcoma oncogene | 0.49 | 3.48E-03 |
| 534427 | jun | Jun oncogene | 0.49 | 3.26E-02 |
| 352483 | ccnd3 | Cyclin D3 | 0.35 | 3.40E-02 |
| 684806 | ccnd1 | Cyclin D1 | 0.31 | 9.48E-03 |
These are the genes from the Wnt signalling pathway.
Table 2.
Genes that changed expression significantly (P < 0.05) upon 15-methoxypinusolidic acid treatment were functionally grouped (see Table 1)
| Gene ID | Gene symbol | Gene title | Fold change | P-value |
|---|---|---|---|---|
| 496423 | gadd45α | Growth arrest and DNA damage-inducible 45 α | 14.62 | 2.39E-03 |
| 422947 | ddit3 | DNA damage-inducible transcript 3 | 7.39 | 5.50E-04 |
| 595490 | ppp3cc | Protein phosphatase 3, catalytic subunit, γ isoform | 2.74 | 1.78E-02 |
| 371178 | atf4 | Activating transcription factor 4 | 2.36 | 1.32E-02 |
| 303843 | dusp1 | Dual specificity phosphatase 1 | 2.28 | 4.91E-02 |
| 930147 | gadd45γ | Growth arrest and DNA damage-inducible 45 γ | 2.07 | 3.42E-02 |
| 825386 | sos2 | Son of sevenless homologue 2 | 2.05 | 6.29E-03 |
| 341967 | map3k6 | Mitogen-activated protein kinase kinase kinase 6 | 0.5 | 1.27E-02 |
| 548588 | dusp7 | Dual specificity phosphatase 7 | 0.49 | 1.41E-02 |
| 463565 | fos | FBJ osteosarcoma oncogene | 0.49 | 3.48E-03 |
| 534427 | jun | Jun oncogene | 0.49 | 3.26E-02 |
| 920514 | grb2 | Growth factor receptor-bound protein 2 | 0.46 | 1.37E-02 |
| 734612 | il1β | Interleukin-1 β | 0.44 | 2.05E-02 |
These are the genes from the MAPK signalling pathway.
Table 3.
Genes that changed expression significantly (P < 0.05) upon 15-methoxypinusolidic acid treatment were functionally grouped (see Table 1)
| Gene ID | Gene symbol | Gene title | Fold change | P-value |
|---|---|---|---|---|
| 496423 | gadd45α | Growth arrest and DNA damage-inducible 45 α | 14.62 | 2.39E-03 |
| 807657 | cdkn2b | Cyclin-dependent kinase inhibitor 2B (p15, inhibits cyclin-dependent kinase 4) | 6.77 | 2.88E-07 |
| 930147 | gadd45γ | Growth arrest and DNA damage-inducible 45 γ | 2.07 | 3.42E-02 |
| 911031 | mcm4 | Minichromosome maintenance deficient 4 homologue | 0.49 | 5.96E-03 |
| 580590 | orc1l | Origin–recognition complex, subunit 1-like | 0.44 | 1.52E-02 |
| 466841 | ccne1 | Cyclin E1 | 0.37 | 3.10E-03 |
| 352483 | ccnd3 | Cyclin D3 | 0.35 | 3.40E-02 |
| 930704 | cdkn1c | Cyclin-dependent kinase inhibitor 1C (P57) | 0.34 | 4.41E-03 |
| 684806 | ccnd1 | Cyclin D1 | 0.31 | 9.48E-03 |
| 579729 | cdc6 | Cell division cycle 6 homologue | 0.24 | 3.47E-03 |
These are the genes from the cell cycle pathway.
Table 4.
Genes that changed expression significantly (P < 0.05) upon 15-methoxypinusolidic acid treatment were functionally grouped (see Table 1)
| Gene ID | Gene symbol | Gene title | Fold change | P-value |
|---|---|---|---|---|
| 563182 | osm | Oncostatin M | 2.74 | 1.43E-02 |
| 825386 | sos2 | Son of sevenless homologue 2 | 2.05 | 6.29E-03 |
| 920514 | grb2 | Growth factor receptor-bound protein 2 | 0.46 | 1.37E-02 |
| 352483 | ccnd3 | Cyclin D3 | 0.35 | 3.40E-02 |
| 486066 | csf2rβ2 | Colony-stimulating factor 2 receptor, β 2, low-affinity (granulocyte-macrophage) | 0.33 | 1.21E-02 |
| 684806 | ccnd1 | Cyclin D1 | 0.31 | 9.48E-03 |
| 743607 | socs3 | Suppressor of cytokine signalling 3 | 0.26 | 6.89E-03 |
| 340636 | il10rα | Interleukin-10 receptor, α | 0.25 | 8.05E-03 |
These are the genes from the Jak-STAT signalling pathway.
Table 5.
Genes that changed expression significantly (P < 0.05) upon 15-methoxypinusolidic acid treatment were functionally grouped (see Table 1)
| Gene ID | Gene symbol | Gene title | Fold change | P-value |
|---|---|---|---|---|
| 825386 | sos2 | Son of sevenless homologue 2 | 2.05 | 6.29E-03 |
| 534427 | jun | Jun oncogene | 0.49 | 3.26E-02 |
| 920514 | grb2 | Growth factor receptor-bound protein 2 | 0.46 | 1.37E-02 |
| 926460 | zyx | Zyxin | 0.43 | 9.88E-03 |
| 352483 | ccnd3 | Cyclin D3 | 0.35 | 3.40E-02 |
| 684806 | ccnd1 | Cyclin D1 | 0.31 | 9.48E-03 |
These are the genes from the focal adhesion pathway.
Table 6.
Genes that changed expression significantly (P < 0.05) upon 15-methoxypinusolidic acid treatment were functionally grouped (see Table 1)
| Gene ID | Gene symbol | Gene title | Fold change | P-value |
|---|---|---|---|---|
| 595490 | ppp3cc | Protein phosphatase 3, catalytic subunit, γ isoforms | 2.74 | 1.78E-02 |
| 463565 | fos | FBJ osteosarcoma oncogene | 0.49 | 3.48E-03 |
| 534427 | jun | Jun oncogene | 0.49 | 3.26E-02 |
| 920514 | grb2 | Growth factor receptor-bound protein 2 | 0.46 | 1.37E-02 |
| 496390 | card11 | Caspase recruitment domain family, member 11 | 0.43 | 2.54E-03 |
These are the genes from the B cell receptor signalling pathway.
Table 7.
Genes that changed expression significantly (P < 0.05) upon 15-methoxypinusolidic acid treatment were functionally grouped (see Table 1)
| Gene ID | Gene symbol | Gene title | Fold change | P-value |
|---|---|---|---|---|
| 595490 | ppp3cc | Protein phosphatase 3, catalytic subunit, γ isoforms | 2.74 | 1.78E-02 |
| 463565 | fos | FBJ osteosarcoma oncogene | 0.49 | 3.48E-03 |
| 534427 | jun | Jun oncogene | 0.49 | 3.26E-02 |
| 496390 | card11 | Caspase recruitment domain family, member 11 | 0.43 | 2.54E-03 |
| 880995 | cd72 | CD72 antigen | 0.39 | 2.49E-02 |
These are the genes from the T cell receptor signalling pathway.
Pathway analysis revealed that the most prominent changes in gene expression occurred in the cell cycle pathway (P-value 5.26E-04). The cell cycle pathway is regulated via various upstream signalling pathways including MAPK signalling pathway, Jak-STAT signalling pathway, Wnt signalling pathway and focal adhesion, and the exact pathway by which 15-MPA affects the cell cycle remains to be determined. Among genes involved in B and T cell receptor signalling pathway, we were also interested in the transcription factors, jun and fos, because of their biological function of cell proliferation and differentiation.
Putting together the functional relationship among pathways and the properties of genes belonging to the pathways, we could formulate a hypothesis that 15-MPA might induce cell cycle inhibition and growth arrest in BV2 cells. We have summarized the genes related to those biological effects in Table 9. Genes involved in growth arrest and DNA damage (GADD) (gadd45α, gadd45γ and ddit3) and cyclin-dependent kinase inhibitor (cdkn2b) were up-regulated. However, genes involved in cell cycle progression, DNA replication and cell proliferation were down-regulated: genes encoding cyclins, which contributed to cell cycle progression (ccnd1 for cyclin D1, ccnd3 for cyclin D3 and ccne1 for cyclin E1), genes involved in DNA replication (mcm4, orc1l and cdc6) and the early-response genes for transcription factors jun and fos.
Table 9.
List of selected genes that are functionally related to cell cycle regulation and DNA damage and were changed by 15-methoxypinusolidic acid treatment
| Gene ID | Gene symbol | Protein name | Fold change | Function |
|---|---|---|---|---|
| 496423 | gadd45α | Growth arrest and DNA damage-inducible protein GADD45 α | 14.62 | Binds to proliferating cell nuclear antigen; might affect PCNA interaction with some CDK (cell division protein kinase) complexes; stimulates DNA excision repair in vitro and inhibits entry of cells into S phase |
| 422947 | ddit3 | DNA damage-inducible transcript 3 | 7.39 | Inhibits the DNA-binding activity of C/EBP and LAP by forming heterodimers that cannot bind DNA |
| 807657 | cdkn2b | Cyclin-dependent kinase 4 inhibitor B (p14-INK4b, p15-INK4b) | 6.77 | Interacts strongly with CDK4 and CDK6; potent inhibitor; potential effector of TGF-β-induced cell cycle arrest |
| 930147 | gadd45γ | Growth arrest and DNA damage-inducible protein GADD45 γ | 2.07 | Involved in the regulation of growth and apoptosis; mediates activation of stress-responsive MTK1/MEKK4 MAPKKK |
| 463565 | fos | Proto-oncogene protein c-fos | 0.49 | Nuclear phosphoprotein that forms a tight but non-covalently linked complex with the JUN/AP-1 transcription factor; has a critical function in regulating the development of cells destined to form and maintain the skeleton; it is thought to have an important role in signal transduction, cell proliferation and differentiation |
| 534427 | jun | Transcription factor AP-1 | 0.49 | Transcription factor that recognizes and binds to the enhancer heptamer motif 5′-TGA[CG]TCA-3′ |
| 911031 | mcm4 | DNA replication licensing factor MCM4 | 0.49 | Involved in the control of DNA replication |
| 580590 | orc1l | Origin–recognition complex subunit 1 | 0.44 | Component of the origin–recognition complex that binds origins of replication; it has a role in both chromosomal replication and mating type transcriptional silencing; binds to the ARS consensus sequence of origins of replication in an ATP-dependent manner |
| 466841 | ccne1 | G1/S-specific cyclin E1 | 0.37 | Essential for the control of the cell cycle at the G1/S (start) transition |
| 352483 | ccnd3 | G1/S-specific cyclin D3 | 0.35 | Essential for the control of the cell cycle at the G1/S (start) transition; potentiates the transcriptional activity of ATF5 |
| 684806 | ccnd1 | G1/S-specific cyclin D1 | 0.31 | Essential for the control of the cell cycle at the G1/S (start) transition |
| 579729 | cdc6 | Cell division control protein 6 homologue | 0.24 | Involved in the initiation of DNA replication; also participates in checkpoint controls that ensure DNA replication is completed before mitosis is initiated |
RT-PCR analysis confirms the expression of representative genes listed in Table 9
We performed RT-PCR analysis for representative genes in Table 9 in order to confirm the expression levels: gadd45α and ddit3 (genes involved in GADD), cdkn2b, ccnd1 (cyclins), orc1l and cdc6 (genes involved in DNA replication), and fos. All of the genes showed significant changes in expression levels corresponding to the results of microarray expression data (Figure 2). Treatment of BV2 cells with 15-MPA (50 µmol·L−1) for 6 h that is the same condition as in the microarray experiment significantly increased the mRNA expressions of gadd45α (493%), ddit3 (176%) and cdkn2b (463%), as compared with untreated control. The mRNA expression of ccnd1, orc1l, cdc6 and fos was significantly inhibited in 15-MPA-treated BV2 cells, compared with values in untreated controls.
Figure 2.
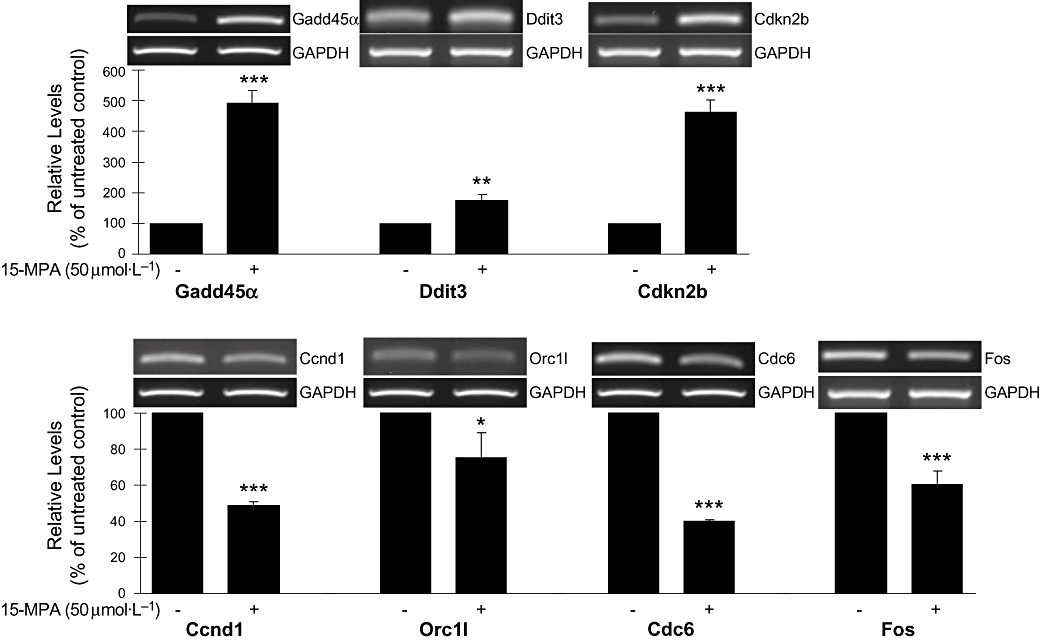
RT-PCR analysis of genes showing altered expression in microarray analysis. The level of mRNA transcript was assessed by semi-quantitative RT-PCR analysis. PCR product for each gene was resolved on ethidium-containing agarose gels, quantified by scanning densitometry and normalized to GAPDH level. The values shown are mean ± SD of data from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 versus untreated control. 15-MPA, 15-methoxypinusolidic acid; ccnd1, cyclin D1; cdc, cell division cycle; cdkn2b, cyclin-dependent kinase inhibitor 2B; ddit3, DNA damage-inducible transcript 3; GADD, growth arrest and DNA damage; ORC, origin–recognition complex; RT-PCR, reverse transcription-polymerase chain reaction.
Inhibition of cell proliferation and induction of apoptosis by 15-MPA
We examined the effect of 15-MPA on microglial proliferation. Microglial BV2 cells were proliferated during overnight incubation with 10% FBS, and 15-MPA was added to the medium without serum at the concentrations of 12.5, 25 or 50 µmol·L−1. Cell proliferation was determined by measuring the incorporation of BrdU. 15-MPA significantly inhibited cell proliferation in a concentration-dependent manner, compared with untreated control (Figure 3). We also determined the cytotoxicity of 15-MPA on BV2 cells by MTT assay. BV2 cells grown to confluence were treated with 15-MPA at concentrations of 12.5, 25 or 50 µmol·L−1, and cell viability was determined after 6 and 22 h. The results, however, did not show any significant decreased cell viability at either time point (data not shown).
Figure 3.
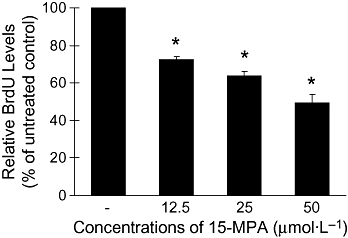
Effects of 15-methoxypinusolidic acid (15-MPA) on bromodeoxyuridine (BrdU) incorporation in BV2 cells. BV2 cells were treated with 15-MPA at various concentrations for 6 h and incubated for 16 h in the presence of BrdU. BrdU incorporated in cells was determined as described in the Methods. *P < 0.05 versus untreated control.
Inhibition of the cell cycle by 15-MPA through down-regulating the expression of cyclin genes and inhibition of BrdU incorporation indicate that 15-MPA inhibits entry of BV2 cells into S phase. Therefore, we assessed the pro-apoptotic potential of 15-MPA. BV2 cells were treated with 15-MPA (50 µmol·L−1) for 6 h and the expression of PARP, a nuclear enzyme activated by DNA damage was detected by Western blot analysis. As shown in Figure 4, 15-MPA significantly increased the expression of cleaved PARP protein over untreated control BV2 cells. However, the actual amounts of cleaved PARP were small, and that of uncleaved PARP was hardly changed. These results might indicate that the 15-MPA-treated BV2 cells are at a very early stage of apoptosis in this experimental condition.
Figure 4.
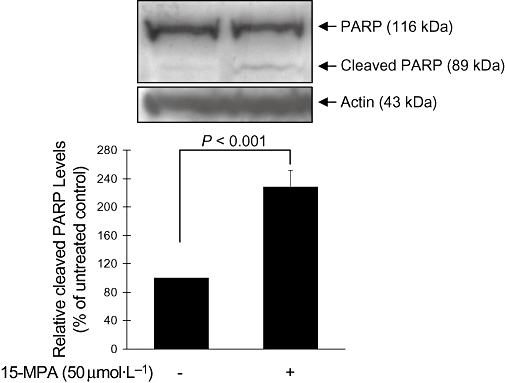
Western blot analysis of the poly-ADP-ribose polymerase (PARP) protein. BV2 cells were treated with 15-methoxypinusolidic acid (15-MPA) (50 µmol·L−1) for 6 h, and cell lysates were prepared and subjected to Western blot analysis using an antibody specific for PARP. Equivalent loading of cell lysates was determined by reprobing the blots with anti-actin antibody. The relative protein levels were quantified by scanning densitometry and normalized to actin. The values shown are the mean ± SD of data from three independent experiments.
In order to confirm the apoptosis-inducing activity of 15-MPA, we performed double staining of BV2 cells treated with 15-MPA using PI and anti-Annexin V antibody conjugated with FITC followed by flow cytometry. As shown in Figure 5, the proportion of apoptotic cells (Annexin V-FITC and PI-double positive) was significantly higher in 15-MPA-treated cells than in control cells at 22 h (P < 0.01), indicating the induction of apoptosis by 15-MPA in BV-2 cells. The number of early apoptotic cells (Annexin V-FITC positive and PI negative) was also significantly increased by treatment of 15-MPA at 6 h (P < 0.05), although the proportion of apoptotic cells was low at this time.
Figure 5.
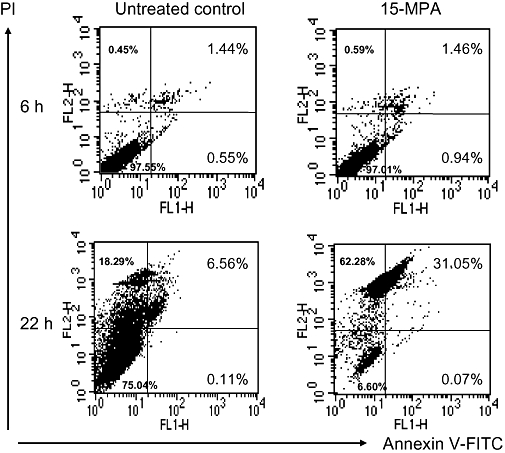
Flow cytometric analysis of BV2 cells treated with 15-methoxypinusolidic acid (15-MPA). BV2 cells were treated with 15-MPA (50 µmol·L−1) for 6 or 22 h. Apoptotic cells were detected by staining cells with propidium iodide and anti-annexin V antibody-conjugated with fluorescein isothiocyanate (FITC) followed by flow cytometry. The percentage of the number of cells for each quadrant is shown (mean ± SD of data from three independent experiments).
Discussion
Microarray analysis of microglial gene expression response to 15-MPA revealed that a group of genes were regulated by 15-MPA in a functionally related manner, which led to inhibition of cell cycle progression and suppression of cell proliferation in murine microglial BV2 cells. Up-regulation of cell cycle proteins was reported to induce proliferation in mitotic cells such as microglia, whereas such up-regulation initiates caspase-related apoptosis in post-mitotic cells such as neurons (Kato et al., 2003; Koguchi et al., 2003; Cernak et al., 2005; Di Giovanni et al., 2005; Tian et al., 2007). Cyclin D1, which triggers the progression of the cell cycle by activating cyclin-dependent kinases (CDKs), may contribute to the onset of microglial proliferation (Wiessner et al., 1996). Thus, cell cycle inhibition has been shown to be neuroprotective by attenuating neuronal cell death and microglial proliferation/activation after CNS injury (Yang et al., 2000; Di Giovanni et al., 2005; Tian et al., 2007).
Our data show that 15-MPA affects cell cycle regulation systematically. Cell cycle progression is regulated by the sequential activation and inactivation of CDKs. In somatic cells, movement through G1 and into S phase is driven by the active form of the cyclin D1, 2, 3/CDK4, 6 complex and the subsequent phosphorylation of retinoblastoma (Rb) protein in mid G1. Once cyclin E and CDK2 are induced by phosphorylation of some Rb, the resulting cyclin E/CDK2 further phosphorylates Rb in late G1, leading to a rapid rise in the expression and activity of E2F and the transcription of multiple other genes essential for entry into S phase and DNA synthesis (Lodish et al., 2004a). 15-MPA suppressed the expression of genes encoding cyclin D1, D3 and cyclin E1. In contrast, the gene encoding INK4 (inhibitors of kinase 4) protein, p15INK4b, which blocks passage through G1 by specifically inhibiting CDK4 and CDK6, was up-regulated by 15-MPA. These results indicate that 15-MPA may effectively inhibit the transition of microglial cells from G1 to S phase and attenuate proliferation of microglia. These results are supported by others suggesting microglia are the source of cyclin D1 mRNA in the post-ischaemic brain and cyclin D1 contributes to microglia proliferation (Wiessner et al., 1996). The 15-MPA-induced G1 arrest was also confirmed by its inhibition on BrdU incorporation, which is incorporated in the DNA of cells in the S phase.
15-MPA increased the expression of the family of GADD-inducible genes, gadd45α, gadd45γ and ddit3. The genes are induced by various genotoxic and non-genotoxic stresses such as UV exposure, redox imbalance, nutrient deprivation, perturbations of endoplasmic reticulum homeostasis and drug therapies (Hollander and Fornace, 2002; Lawrence et al., 2007; Siafakas and Richardson, 2008). Up-regulation of gadd genes results in cell cycle arrest, DNA repair and growth inhibition and/or apoptosis (Kearsey et al., 1995; Sheikh et al., 2000; Oyadomari and Mori, 2004; Zerbini et al., 2004; Bhattacharjee et al., 2005; Pan et al., 2007; Li et al., 2008). The gadd45α is regulated by both p53-dependent and independent mechanisms, although it is the first stress-inducible gene activated by p53 (Kastan et al., 1992; Zhan et al., 1998; Zerbini et al., 2005). In our microarray data, p53 mRNA was not differentially expressed by 15-MPA. Gadd45 isoforms, Gadd45α, β and γ are known to be implicated in the G2/M checkpoint as well as the G1/S arrest (Hollander and Fornace, 2002; Mak and Kültz, 2004). Gadd45γ is found to be epigenetically repressed in some cancers including pituitary tumours by methylation of CpG islands and suggested to be a functional tumour suppressor (Bahar et al., 2004; Ying et al., 2005). Ddit, also known as Gadd153 or Chop (C/EBP homologous protein) can be induced by inflammatory cytokines or endoplasmic reticulum stress signals, which are implicated in a variety of diseases such as diabetes, ischaemia and neurodegenerative disorders (Oyadomari and Mori, 2004; Cardozo et al., 2005; Lawrence et al., 2007). Therefore, 15-MPA could have the potential to protect cells from stress-induced alterations, which might be mediated by an induction of the above gadd genes through a p53-independent G1 or G2 arrest.
The cell cycle arrest induced by 15-MPA leads the cells to the apoptotic pathway, although the apoptotic pathway did not show a significant enrichment in the microarray analysis by using the PANTHER ontological terms. Two genes belonging to the apoptotic pathway were differentially expressed, ppp3cc (up-regulated by 2.74-fold) and csf2rβ2 (down-regulated by 0.33-fold), but, the expression of genes such as the bcl2 family showed no significant differences in the pathway analysis by KEGG database. This could be related to the short treatment time of 15-MPA. In this study, we treated cells with 15-MPA for 6 h in order to determine the molecular mechanisms underlying cellular response to 15-MPA. In this condition, the 15-MPA-treated cells appeared to be at an early stage of apoptosis. This was indicated by the results that the number of early apoptotic cells and the expression of cleaved PARP were significantly increased by 15-MPA, whereas the proportion of each was very small as compared with total cell number and uncleaved PARP respectively. PARP is cleaved by caspases, the effector proteins in the apoptotic pathway (Oliver et al., 1998; Kauppinen and Swanson, 2007). If the cells are treated longer (up to 24 h), changes in the gene expression commonly observed in the late stage of apoptosis (Kroemer et al., 2007) would be expected to be seen. The FACS data at 22 h clearly indicate that 15-MPA induces apoptosis in BV2 cells, after a longer exposure time.
The MTT assay was performed to determine whether the inhibitory effect of 15-MPA on BrdU incorporation was due to its cytotoxicity. Thus, we measured the cytotoxicity after 22 h as well as after 6 h that was the total incubation time of 15-MPA including BrdU labelling time. 15-MPA showed no cytotoxicity at either time point, and this could be due to the fact that the cells might be committed to apoptosis by treatment of 15-MPA but which were not yet (22 h) undergoing cell death. However, apoptotic cell death was observed at 22 h in the FACS analysis. We consider that PARP cleavage and membrane leakage (resulting in PI staining of cells) give more accurate information on cell status. In contrast, the cytotoxicity test using MTT assay often does not reflect the real situation of cells in terms of ‘viability’ because it measures mitochondrial enzyme activity, the reduction of which is often observed in the late stage of cell death. Such differences should explain the discrepancy between the results of the MTT assays and the FACS analysis.
15-MPA could also regulate DNA replication in BV2 cells by controlling the initiation of transcription of genes. Synthesis of eukaryotic DNA is regulated by controlling the activity of the hexameric MCM helicases (MCM2-7) that initiate DNA replication at multiple origins spaced along chromosomal DNA (Lodish et al., 2004b). cdc6 is an essential regulator of DNA replication that cooperates with ORC (origin–recognition complex) to load MCM proteins and also of the activation and maintenance of the checkpoint mechanisms that coordinate S phase and mitosis. Over-expression of cdc6 showed an oncogenic potential via interfering with INK4/ARF tumour suppressor genes and increasing DNA replication (Borlado and Méndez, 2008; Boronat and Campbell, 2008). 15-MPA might effectively coordinate BV2 cells to inhibit DNA replication by down-regulating the expression of mcm4, orc1l and cdc6 genes.
Early-response genes encoding the transcription factors, Fos and Jun induce expression of many genes encoding proteins necessary for cells to progress through the cell cycle G1 to S transition. These genes are over-expressed in some human tumours and encode proteins that sometimes associate to form a heterodimeric transcription factor, called AP-1. The growth factors in serum are strong stimulators of these genes (Lodish et al., 2004c). 15-MPA inhibited the expression of fos and jun genes.
Overall, our results indicated that 15-MPA might induce cell cycle inhibition and growth arrest in murine microglial BV2 cells, based on the ontological analysis of the microglial transcriptome. The treatment with 15-MPA suppressed the expression of early-response genes, fos and jun, which appeared to make the cells remain quiescent and not to initiate DNA replication. The decreased expression of genes responsible for DNA replication by 15-MPA could potently inhibit DNA synthesis in S phase: 15-MPA-mediated suppression of cdc6 gene could down-regulate the assembly of DNA pre-replication complexes at origins during G1 phase, and the suppression of mcm4 and orc1l gene could synergize to inhibit DNA replication. 15-MPA induced a G1/S arrest by suppressing G1 and early S cyclin expression and by inducing expression of ink4 gene. In addition, 15-MPA probably induced a G2/M arrest by up-regulating gadd genes. RT-PCR analysis for representative genes confirmed the expression levels. The results of the BrdU incorporation assay, Western blot of PARP and FACS analysis indicate that 15-MPA induced apoptosis in BV2 cells, presumably via inhibition of the cell cycle progression.
Considering that microglial activation and proliferation are detrimental in CNS injuries including neurodegenerative disorders (Dheen et al., 2007), the outcomes of our experiments suggest that 15-MPA might have strong therapeutic potential for the treatment of such injuries by using the cell cycle machinery to induce arrest through a combination of related components. Further, as 15-MPA showed a neuroprotective effect against staurosporine-induced apoptosis by inhibiting caspase 3/7 activation in rat cortical cells (Koo et al., 2007), 15-MPA could play both roles, pro-apoptotic or pro-survival, depending on the cell type (mitotic or post-mitotic), and would be likely to induce decreased neuronal cell death and inhibition of microglial proliferation. We think this study provides meaningful results, but further study is needed in order to demonstrate the effectiveness of 15-MPA against neurodegenerative diseases. Work to confirm the expression of the corresponding proteins and to evaluate their biological activity using appropriate disease models will provide much useful information.
Acknowledgments
We thank Professor Na Gyong Lee (Sejong University, Seoul) for comments on the manuscript. This research was supported by a grant (M103KV010027-08K2201-02710) from Brain Research Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology, the Republic of Korea.
Glossary
Abbreviations:
- 15-MPA
15-methoxypinusolidic acid
- BrdU
bromodeoxyuridine
- ccnd1
cyclin D1
- ccnd3
cyclin D3
- ccne1
cyclin E1
- cdc
cell division cycle
- CDK
cyclin-dependent kinase
- cdkn2b
cyclin-dependent kinase inhibitor 2B
- ddit3
DNA damage-inducible transcript 3
- DEG
differentially expressed gene
- GADD
growth arrest and DNA damage
- ORC
origin–recognition complex
- PARP
poly-ADP-ribose polymerase
- RT-PCR
reverse transcription-polymerase chain reaction
Conflict of interest
The authors state no conflict of interest.
References
- Bahar A, Bicknell JE, Simpson DJ, Clayton RN, Farrell WE. Loss of expression of the growth inhibitory gene GADD45γ, in human pituitary adenomas, is associated with CpG island methylation. Oncogene. 2004;23:936–944. doi: 10.1038/sj.onc.1207193. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee RN, Park KS, Uematsu S, Okada K, Hoshino K, Takeda K, et al. Escherichia coli verotoxin 1 mediates apoptosis in human HCT116 colon cancer cells by inducing overexpression of the GADD family of genes and S phase arrest. FEBS Lett. 2005;579:6604–6610. doi: 10.1016/j.febslet.2005.10.053. [DOI] [PubMed] [Google Scholar]
- Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F. Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J Neuroimmunol. 1990;27:229–237. doi: 10.1016/0165-5728(90)90073-v. [DOI] [PubMed] [Google Scholar]
- Borlado LR, Méndez J. CDC6: from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis. 2008;29:237–243. doi: 10.1093/carcin/bgm268. [DOI] [PubMed] [Google Scholar]
- Boronat S, Campbell JL. Linking mitosis with S-phase: Cdc6 at play. Cell Cycle. 2008;7:597–601. doi: 10.4161/cc.7.5.5519. [DOI] [PubMed] [Google Scholar]
- Cardozo AK, Ortis F, Storling J, Feng YM, Rasschaert J, Tonnesen M, et al. Cytokines downregulate the sarcoendoplasmic reticulum pump Ca2+ ATPase 2b and deplete endoplasmic reticulum Ca2+, leading to induction of endoplasmic reticulum stress in pancreatic β-cells. Diabetes. 2005;54:452–461. doi: 10.2337/diabetes.54.2.452. [DOI] [PubMed] [Google Scholar]
- Cernak I, Stoica B, Byrnes KR, Di Giovanni S, Faden AI. Role of the cell cycle in the pathobiology of central nervous system trauma. Cell Cycle. 2005;4:1286–1293. doi: 10.4161/cc.4.9.1996. [DOI] [PubMed] [Google Scholar]
- Choi Y, Moon A, Kim YC. A pinusolide derivative, 15-methoxypinusolidic acid from Biota orientalis inhibits inducible nitric oxide synthase in microglial cells: implication for a potential anti-inflammatory effect. Int Immunopharmacol. 2008;8:548–555. doi: 10.1016/j.intimp.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Cuadros MA, Navascués J. The origin and differentiation of microglial cells during development. Prog Neurobiol. 1998;56:173–189. doi: 10.1016/s0301-0082(98)00035-5. [DOI] [PubMed] [Google Scholar]
- Dheen ST, Kaur C, Ling EA. Microglial activation and its implications in the brain diseases. Curr Med Chem. 2007;14:1189–1197. doi: 10.2174/092986707780597961. [DOI] [PubMed] [Google Scholar]
- Di Giovanni S, Movsesyan V, Ahmed F, Cernak I, Schinelli S, Stoica B, et al. Cell cycle inhibition provides neuroprotection and reduces glial proliferation and scar formation after traumatic brain injury. Proc Natl Acad Sci USA. 2005;102:8333–8338. doi: 10.1073/pnas.0500989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamadeh HK, Bushel PR, Jayadev S, Martin K, DiSorbo O, Sieber S, et al. Gene expression analysis reveals chemical-specific profiles. Toxicol Sci. 2002;67:219–231. doi: 10.1093/toxsci/67.2.219. [DOI] [PubMed] [Google Scholar]
- Hollander MC, Fornace AJ., Jr Genomic instability, centrosome amplification, cell cycle checkpoints and Gadd45a. Oncogene. 2002;21:6228–6233. doi: 10.1038/sj.onc.1205774. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Kato H, Takahashi A, Itoyama Y. Cell cycle protein expression in proliferating microglia and astrocytes following transient global cerebral ischemia in the rat. Brain Res Bull. 2003;60:215–221. doi: 10.1016/s0361-9230(03)00036-4. [DOI] [PubMed] [Google Scholar]
- Kauppinen TM, Swanson RA. The role of poly(ADP-ribose) polymerase-1 in CNS disease. Neuroscience. 2007;145:1267–1272. doi: 10.1016/j.neuroscience.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Kaur C, Hao AJ, Wu CH, Ling EA. Origin of microglia. Microsc Res Tech. 2001;54:2–9. doi: 10.1002/jemt.1114. [DOI] [PubMed] [Google Scholar]
- Kearsey JM, Coates PJ, Prescott AR, Warbrick E, Hall PA. Gadd45 is a nuclear cell cycle regulated protein which interacts with p21Cip1. Oncogene. 1995;11:1675–1683. [PubMed] [Google Scholar]
- Kim YS, Joh TH. Microglia, major player in the brain inflammation: their roles in the pathogenesis of Parkinson's disease. Exp Mol Med. 2006;38:333–347. doi: 10.1038/emm.2006.40. [DOI] [PubMed] [Google Scholar]
- Koguchi K, Nakatsuji Y, Okuno T, Sawada M, Sakoda S. Microglial cell cycle-associated proteins control microglial proliferation in vivo and in vitro and are regulated by GM-CSF and density-dependent inhibition. J Neurosci Res. 2003;74:898–905. doi: 10.1002/jnr.10829. [DOI] [PubMed] [Google Scholar]
- Koo KA, Sung SH, Kim YC. A new neuroprotective pinusolide derivative from the leaves of Biota orientalis. Chem Pharm Bull. 2002;50:834–836. doi: 10.1248/cpb.50.834. [DOI] [PubMed] [Google Scholar]
- Koo KA, Lee MK, Kim SH, Jeong EJ, Kim SY, Oh TH, et al. Pinusolide and 15-methoxypinusolidic acid attenuate the neurotoxic effect of staurosporine in primary cultures of rat cortical cells. Br J Pharmacol. 2007;150:65–71. doi: 10.1038/sj.bjp.0706944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- Lai AY, Todd KG. Microglia in cerebral ischemia: molecular actions and interactions. Can J Physiol Pharmacol. 2006;84:49–59. doi: 10.1139/Y05-143. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, McGlynn K, Naziruddin B, Levy MF, Cobb MH. Differential regulation of CHOP-10/GADD153 gene expression by MAPK signaling in pancreatic β-cells. Proc Natl Acad Sci USA. 2007;104:11518–11525. doi: 10.1073/pnas.0704618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Qian H, Li X, Wang H, Yu J, Liu Y, et al. Adenoviral-mediated gene transfer of Gadd45a results in suppression by inducing apoptosis and cell cycle arrest in pancreatic cancer cell. J Gene Med. 2008;11:3–13. doi: 10.1002/jgm.1270. [DOI] [PubMed] [Google Scholar]
- Liu B, Hong JS. Role of Microglia in inflammation-mediated neurodegenerative diseases: mechanisms and strategies for therapeutic intervention. J Pharmacol Exp Ther. 2003;304:1–7. doi: 10.1124/jpet.102.035048. [DOI] [PubMed] [Google Scholar]
- Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, et al. Regulating the eukaryotic cell cycle: cell-cycle control in mammalian cells. In: Byrd ML, editor. Molecular Cell Biology. New York: W. H. Freeman and Company; 2004a. pp. 881–886. [Google Scholar]
- Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, et al. Basic molecular genetic mechanisms: DNA replication. In: Byrd ML, editor. Molecular Cell Biology. New York: W. H. Freeman and Company; 2004b. pp. 131–137. [Google Scholar]
- Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, et al. Cancer: oncogenic mutations in growth-promoting proteins. In: Byrd ML, editor. Molecular Cell Biology. New York: W. H. Freeman and Company; 2004c. pp. 951–956. [Google Scholar]
- Mak SK, Kültz D. Gadd45 proteins induce G2/M arrest and modulate apoptosis in kidney cells exposed to hyperosmotic stress. J Biol Chem. 2004;279:39075–39084. doi: 10.1074/jbc.M406643200. [DOI] [PubMed] [Google Scholar]
- Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Murcia JM. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J Biol Chem. 1998;273:33533–33539. doi: 10.1074/jbc.273.50.33533. [DOI] [PubMed] [Google Scholar]
- Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- Pan MH, Chang YH, Badmaev V, Nagabhushanam K, Ho CT. Pterostilbene induces apoptosis and cell cycle arrest in human gastric carcinoma cells. J Agric Food Chem. 2007;55:7777–7785. doi: 10.1021/jf071520h. [DOI] [PubMed] [Google Scholar]
- Sheikh MS, Hollander MC, Fornance AJ., Jr Role of Gadd45 in apoptosis. Biochem Pharmacol. 2000;59:43–45. doi: 10.1016/s0006-2952(99)00291-9. [DOI] [PubMed] [Google Scholar]
- Shults EE, Velder J, Schmalz HG, Chernov SV, Rubalova TV, Gatilov YV, et al. Gram-scale synthesis of pinusolide and evaluation of its antileukemic potential. Bioorg Med Chem Lett. 2006;16:4228–4232. doi: 10.1016/j.bmcl.2006.05.077. [DOI] [PubMed] [Google Scholar]
- Siafakas AR, Richardson DR. Growth arrest and DNA damage-45 alpha (GADD45α) Int J Biochem Cell Biol. 2008;41:986–989. doi: 10.1016/j.biocel.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Stoll G, Jander S. The role of microglia and macrophages in the pathophysiology of the CNS. Prog Neurobiol. 1999;58:233–247. doi: 10.1016/s0301-0082(98)00083-5. [DOI] [PubMed] [Google Scholar]
- Streit WJ, Mrak RE, Griffin WS. Microglia and neuroinflammation: a pathological perspective. J Neuroinflammation. 2004;1:14. doi: 10.1186/1742-2094-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian DS, Xie MJ, Yu ZY, Zhang Q, Wang YH, Chen B, et al. Cell cycle inhibition attenuates microglia induced inflammatory response and alleviates neuronal cell death after spinal cord injury in rats. Brain Res. 2007;1135:177–185. doi: 10.1016/j.brainres.2006.11.085. [DOI] [PubMed] [Google Scholar]
- Trajkovski I, Lavrac N, Tolar J. SEGS: search for enriched gene sets in microarray data. J Biomed Inform. 2008;41:588–601. doi: 10.1016/j.jbi.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wiessner C, Brink I, Lorenz P, Neumann-Haefelin T, Vogel P, Yamashita K. Cyclin D1 messenger RNA is induced in microglia rather than neurons following transient forebrain ischaemia. Neuroscience. 1996;72:947–958. doi: 10.1016/0306-4522(95)00601-x. [DOI] [PubMed] [Google Scholar]
- Yang NC, Jeng KC, Ho WM, Chou SJ, Hu ML. DHEA inhibits cell growth and induces apoptosis in BV-2 cells and the effects are inversely associated with glucose concentration in the medium. J Steroid Biochem Mol Biol. 2000;75:159–166. doi: 10.1016/s0960-0760(00)00180-1. [DOI] [PubMed] [Google Scholar]
- Ying J, Srivastava G, Hsieh WS, Gao Z, Murray P, Liao SK, et al. The stress-responsive gene GADD45G is a functional tumor suppressor, with its response to environmental stresses frequently disrupted epigenetically in multiple tumors. Clin Cancer Res. 2005;11:6442–6449. doi: 10.1158/1078-0432.CCR-05-0267. [DOI] [PubMed] [Google Scholar]
- Zerbini LF, Wang Y, Czibere A, Correa RG, Cho JY, Ijiri K, et al. NF-κB-mediated repression of growth arrest- and DNA-damage-inducible proteins 45α and γ is essential for cancer cell survival. Proc Natl Acad Sci USA. 2004;101:13618–13623. doi: 10.1073/pnas.0402069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbini LF, Wang Y, Correa RG, Cho JY, Libermann TA. Blockage of NF-κB induces serine 15 phosphorylation of mutant p53 by JNK kinase in prostate cancer cells. Cell Cycle. 2005;4:1247–1253. doi: 10.4161/cc.4.9.1966. [DOI] [PubMed] [Google Scholar]
- Zhan Q, Chen IT, Antinore MJ, Fornace AJ., Jr Tumor suppressor p53 can participate in transcriptional induction of the GADD45 promoter in the absence of direct DNA binding. Mol Cell Biol. 1998;18:2768–2778. doi: 10.1128/mcb.18.5.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]


