Abstract
Voriconazole is a potent second generation triazole antifungal agent with broad-spectrum activity against clinically important fungi. It is cleared predominantly via metabolism in all species tested including humans. N-oxidation of the fluoropyrimidine ring, its hydroxylation, and hydroxylation of the adjacent methyl group are the known pathways of voriconazole oxidative metabolism, with the N-oxide being the major circulating metabolite in human. In vitro studies have shown that CYP2C19, CYP3A4, and to a lesser extent CYP2C9 contribute to the oxidative metabolism of voriconazole. When CYP-specific inhibitors and antibodies were used to evaluate the oxidative metabolism of voriconazole by human liver microsomes (HLM), the results suggested that CYP-mediated metabolism accounted for ~75% of the total oxidative metabolism. The studies presented here provide evidence that the remaining ~25% of the metabolic transformations are catalyzed by flavin-containing monooxygenase (FMO). This conclusion was based on the evidence that the NADPH-dependent metabolism of voriconazole was sensitive to heat (45 °C for 5 min), a condition known to selectively inactivate FMO without affecting CYP activity. The role of FMO in the metabolic formation of voriconazole N-oxide was confirmed by the use of recombinant FMO enzymes. Kinetic analysis of voriconazole metabolism by FMO1 and FMO3 yielded Km values of 3.0 mM and 3.4 mM and Vmax values of 0.025 pmol/min/pmol and 0.044 pmol/min/pmol, respectively. FMO5 did not metabolize voriconazole effectively. This is the first report of the role of FMO in the oxidative metabolism of voriconazole.
Introduction
Voriconazole (Vfend™), a second generation triazole, is a potent antifungal agent with activity against a broad spectrum of fungal pathogens (Boucher et al., 2004; Patterson, 2002). Its pharmacokinetic properties after intravenous (IV) and oral administration have been thoroughly investigated in healthy volunteers (Purkins et al., 2002; Purkins et al., 2003a; Purkins et al., 2003b) and in patients at risk of fungal infections (Lazarus et al., 2002); the pharmacokinetic / pharmacodynamic profile has been recently reviewed recently (Theuretzbacher et al., 2006). These studies show that voriconazole is readily absorbed upon oral administration (oral bioavailability > 90%), and that it is eliminated with a terminal elimination half-life of approximately 6 h. Studies with radioisotope-labeled voriconazole have demonstrated that it is cleared via extensive hepatic metabolism in preclinical species and in humans; less than 2% of the administered dose is excreted as the parent drug in humans and a slightly higher percentage of the dose (<10%) appears as voriconazole in the excreta of other preclinical species - mouse, rat, guinea pig, and dog (Roffey et al., 2003). After either IV or oral administration to humans, nearly 80% of the voriconazole dose is excreted renally, mostly as metabolites (Purkins et al., 2003b). Multiple dose studies have revealed non-linear pharmacokinetics with both Cmax and AUCτ (area under the plasma concentration-time curve during dosage intervals τ) increasing more than dose-proportionately. For example, a 2-fold increase in oral dose (200 mg to 400 mg) caused 2.8-fold and 3.9-fold increase in Cmax and AUCτ, respectively (Purkins et al., 2002). Since voriconazole is predominantly cleared by metabolism, it is reasonable to conclude that the non-linear pharmacokinetics is likely due to saturation of metabolism.
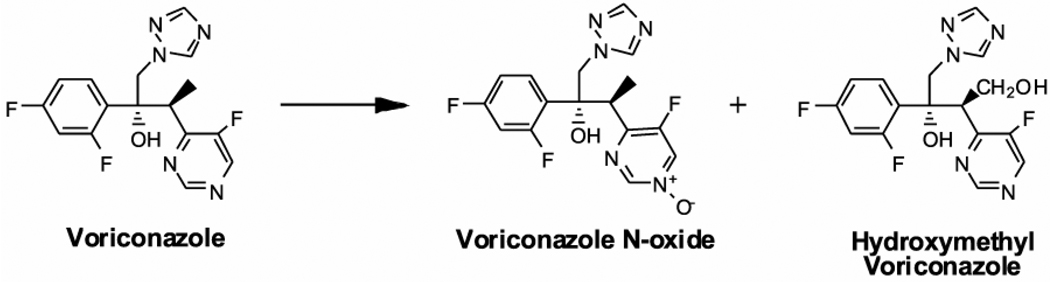
Voriconazole is metabolized to several oxidative metabolites, with N-oxidation of the fluoropyrimidine ring and hydroxylation of the adjacent methyl group being the major pathways in humans (Roffey et al., 2003; Murayama, et al., 2007). Of note, the N-oxide (Figure 1) is a major circulating metabolite in humans and in preclinical species, such as rat and dog (Roffey et al., 2003). In vitro studies with human liver microsomes (HLM) and expressed enzymes have indicated that the N-oxide metabolite is formed predominantly by CYP3A4, CYP2C19, and to a smaller extent by CYP2C9 (Hyland et al., 2003). These studies suggested that at low µM concentrations of voriconazole, CYP2C19 plays a major role in the formation of the N-oxide, whereas at low mM concentrations, CYP3A4 appears to be the major contributor. Recently, in vitro studies have shown that oxidative metabolism of voriconazole to the hydroxymethyl metabolite (Figure 1) by human and rat liver microsomes is catalyzed exclusively by CYP3A (Murayama et al., 2007). Significant alteration of voriconazole clearance in CYP2C19 poor metabolizers, resulting in approximately 4-fold greater exposure to voriconazole, provides evidence for a substantial contribution of CYP2C19 to the clearance of voriconazole in vivo (cf. Theuretzbacher et al., 2006). Because two key CYP enzymes, CYP3A4 and CYP2C19, play a pivotal role in its metabolic clearance, significant drug interactions involving voriconazole are likely when co-administered with inducers or inhibitors of CYP3A4 and CYP2C19 or with drugs that are predominantly cleared by these enzymes (reviewed by Theuretzbacher et al., 2006).
Figure 1.
Structures of Voriconazole and Major Oxidative Metabolites Formed by Human Liver Microsomes
The NADPH-dependent oxygenation of functional groups containing soft nucleophiles is often catalyzed by another class of oxidative enzymes, flavin-containing monooxygenase (FMO) (Rodriguez et al., 1999; Krueger and Williams, 2005). In this study, we demonstrate that, in addition to CYP enzymes, certain FMO isoforms can metabolize voriconazole to N-oxide; we further show that these FMO isoforms contribute significantly to the formation of the N-oxide by HLM, a finding that may significantly affect our understanding of the drug interactions involving voriconazole metabolism.
Materials and Methods
Chemicals and Reagents
Voriconazole was generously supplied by Pfizer Central Research (Groton, CT). Testosterone and 6β-hydroxy testosterone were purchased from Steraloids, Inc. (Newport, RI). CYP substrates and their respective hydroxylated metabolites, mephenytoin, 4-hydroxy-mephenytoin, diclofenac, and 4-hydroxy-diclofenac were purchased from BD Gentest (Woburn, MA). Benzydamine (FMO substrate), ketoconazole, sulfaphenazole, and fluvoxamine (CYP inhibitors), and 3-chloroperoxybenzoic acid were purchased from Sigma-Aldrich (St. Louis, MO). Human FMO1, FMO3, and FMO5 enzymes, expressed in the baculovirus system, were purchased from BD Gentest (Woburn, MA). Inhibitory antibodies for CYP3A4, CYP2C19, and CYP2C9, and pooled human liver microsomes (HLM) were purchased from Xenotech PLC (Lenexa, KS). HLM from CYP2C19 poor metabolizers (2*/2*) were purchased from BD Gentest (Woburn, MA), while the CYP2C19-competent (WT) HLM were purchased from CellzDirect (Austin, TX).
Voriconazole N-oxide (Figure 1) was synthesized by reacting voriconazole with 3-chloroperoxy benzoic acid following the procedure for the synthesis of pyridine N-oxide as described by Luning et al. (1998). The identity of the N-oxide was verified by high performance liquid chromatography/mass spectrometry (HPLC/MS) (1100 MSD-Trap, Agilant, Santa Clara, CA), m/z transition 366→224, (Figure 5A) and by 1H-NMR1 and 13C-NMR spectra in acetone-d6 at 300 MHz and 75 MHz, respectively (Varian 300 MHz-Palo Alto, CA). 1H NMR (300 MHz, acetone-d6) ppm 8.88 (d, J = 1.6 Hz, 1H, H2 pyrimidine), 8.72 (dd, J = 5.5 Hz, 1.6 Hz, 1H, H4 pyrimidine), 8.18 (s, 1H), 7.59 (s, 1H), 7.56 (dt, J = 9.0, 6.7 Hz, 1H), 7.03 (ddd, J = 12.3, 9.0, 2.6 Hz, 1H), 6.89 (dddd, J = 9.0, 8.2, 2.6, 0.9 Hz, 1H), 5.43 (s, 1H), 4.97 (d, J = 14.3 Hz, 1H), 4.53 (d, J = 14.3 Hz, 1H), 4.01 (dq, J = 7.1, 1.3 Hz, 1H), 1.19 (d, J = 7.1 Hz, 1H); 13C NMR (75 MHz, acetone-d6) ppm 156.8, 153.3, 150.2, 145.2, 143.6, 143.4, 130.3, 110.6, 110.4, 103.7, 103.4, 103.0, 77.2, 55.6, 38.4, 13.1. The chemical shifts for the H2 and H4 pyrimidine protons in voriconazole were 9.00 and 8.81 ppm, respectively. Purity of the N-oxide was determined by HPLC.
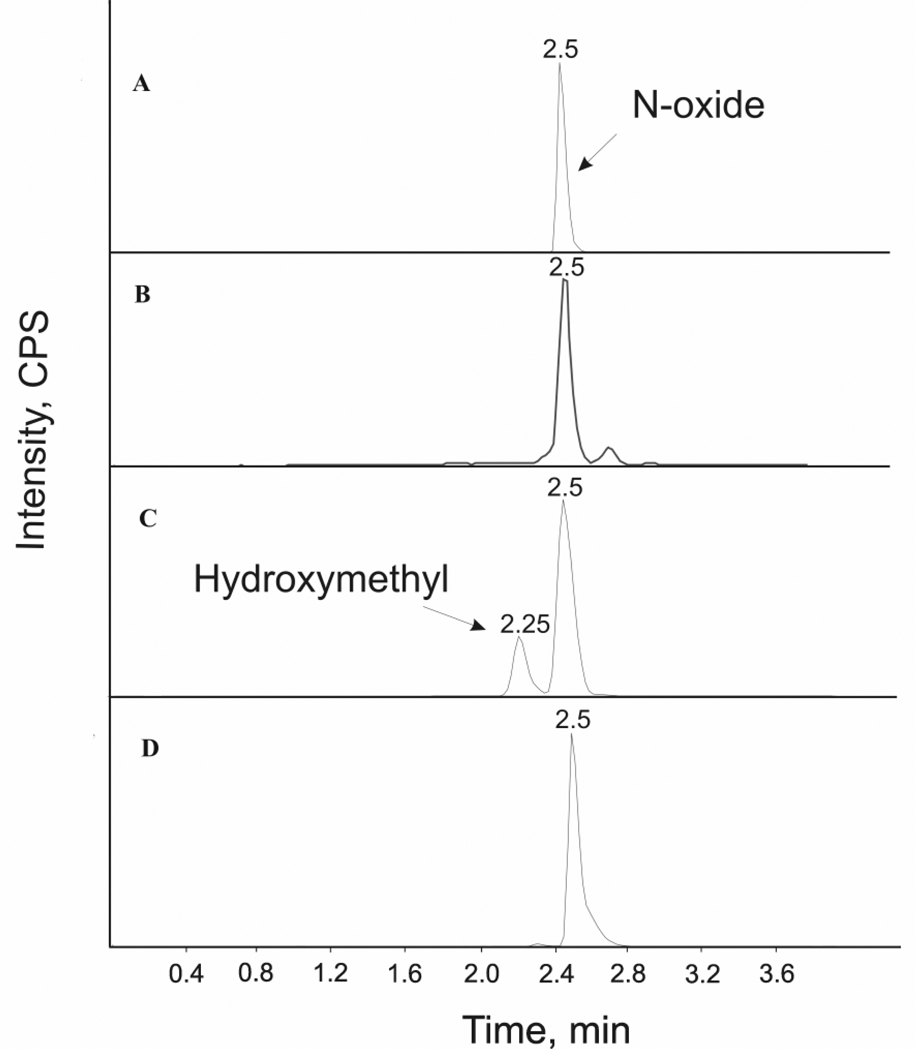
Figure 5. HPLC-MS/MS Profiles of Voriconazole Metabolites formed by Human Liver Microsomes and Flavin-containing Monooxygenase.
A- The HPLC-MS/MS chromatogram of authentic N-oxide standard (366→224; retention time, 2.5 min). B- The HPLC-MS/MS chromatogram of the products formed by recombinant human FMO3 in the presence of NADPH; the N-oxide (366→224; retention time, 2.5 min) was the only metabolite detected. C- The HPLC-MS/MS chromatogram of metabolites formed by incubating voriconazole and HLM in the presence of NADPH; two metabolites were formed - the hydroxymethyl (366→224; retention time, 2.25 min) and the N-oxide (366→224; retention time, 2.5 min). D- Chromatogram of metabolites formed upon co-incubation of voriconazole (25 µM) and ketoconazole (3µM) with HLM. Formation of hydroxymethyl (366→224; retention time, 2.25 min), but not the N-oxide (366→224; retention time, 2.5 min) was diminished by inhibition of CYP3A activity. ALL traces represent extracted ion chromatograms and the Y scales are not identical.
Metabolism of Voriconazole by Human Liver Microsomes
Incubations of voriconazole (2 µM) with HLM (1.0 mg/mL) were carried out in phosphate buffer (0.1 M, pH 7.4) containing MgCl2 (3 mM) at 37 °C. The reactions were initiated by addition of NADPH (2 mM); 200 µL aliquots were removed at defined time points and mixed with 600 µL of ice cold methanol (containing internal standard-1 µM diclofenac) to terminate the reactions. After centrifugation at 10,000×g for 10 min, the supernatants were evaporated to dryness under a stream of nitrogen gas; and the dry residues were dissolved in 10% acetonitrile in water. Quantitative determination of voriconazole was achieved using HPLC with ultraviolet (UV) detection (see the HPLC-UV analysis section). The conditions for initial velocity with respect of time and protein (<20% depletion of initial voriconazole concentration) were determined, and voriconazole metabolism rate and half life (T1/2) were then calculated as described by Obach and Reed-Hagen (2002).
To assess the role of the three putative CYP enzymes in voriconazole metabolism, incubations of voriconazole with HLM were carried out as described above in the presence of selective chemical inhibitors of CYP3A4 (ketoconazole, 3 µM), CYP2C9 (sulfaphenazole, 30 µM), CYP2C19 (fluvoxamine, 3 µM), or a mixture of all three inhibitors to assess the role of the three CYP enzymes, individually and collectively, in its metabolism. The role of CYP enzymes in the metabolism of voriconazole by HLM was also assessed by selective inhibition of CYP-mediated voriconazole metabolism using CYP-specific inhibitory antibodies. Aliquots of HLM (0.1 mg) were pre-incubated at 4°C with the inhibitory antibody against CYP2C19 (10µl), CYP3A4 (20 µl), CYP2C9 (20 µl), or with a mixture of the three antibodies. Pre-incubation of HLM with non-specific rabbit IgG (20 µL) for 15 min was conducted to serve as no inhibition control. The ratio of microsomal protein to antibody volume was chosen to give a maximal inhibitory effect. Voriconazole (2 µM) was then added and mixed with other reaction constituents as described above, and incubations were carried out for 20 minutes.
To investigate the potential role of FMO in voriconazole metabolism, thermal inactivation condition that selectively diminish FMO activity and not CYP activity was applied by preheated HLM (20 mg/mL) at 45 °C for 5 minutes (without NADPH); pre-incubated HLM (20 mg/mL) at 37 °C for 5 minutes served as control. Aliquots of preheated or control HLM were then incubated with voriconazole and NADPH, with or without a cocktail of CYP chemical inhibitors to determine the contribution of FMO, CYP, or both in voriconazole metabolism by HLM. Effect of thermal inactivation condition on FMO activity was confirmed by the incubation of benzydamine (a selective FMO substrate) with preheated HLM and determining the attenuation of its metabolism by the heat treatment.
Metabolism of Voriconazole with Recombinant Human Flavin-containing monooxygenase
To determine the metabolic products formed by metabolism of voriconazole by FMO enzymes, FMO1 (60 pmol/mL), FMO3 (60 pmol/mL), or FMO5 (150 pmol/mL) was incubated with voriconazole (100 µM) in the presence of NADPH (2 mM) for 60 minutes. The voriconazole N-oxide formed in the incubation samples was measured by LC-MS/MS (see the LC-MS/MS analysis section) against a calibration curve of authentic N-oxide standard. The rates of voriconazole N-oxide formed were calculated and normalized to protein concentration in the samples. For kinetic analysis, voriconazole was incubated with 60 pmol/mL of FMO1 or FMO3 at several concentrations ranging from 1 µM - 8000 µM as described above. Reactions were initiated by addition of NADPH and terminated after 30 min by addition of ice cold methanol-containing internal standard, centrifuged, and supernatants were analyzed by HPLC-MS/MS for the formed N-oxide. The rates of N-oxide formation, expressed as pmol of N-oxide formed/min/mg protein or as pmol of N-oxide formed/min/pmol FMO1 or FMO3 were calculated and plotted against voriconazole concentration. Non-linear regression analysis (WinNonLin, Pharsight, Cary, NC) was performed using the Michaels-Menten kinetic equation for the determination of Km and Vmax.
HPLC-UV and HPLC-MS/MS Analysis
The HPLC-UV method was used in the metabolism studies involving the measurement of substrate consumption (see Section “Metabolism of Voriconazole by Human Liver Microsomes”). For HPLC-UV analysis, voriconazole and its N-oxide in the incubation samples were chromatographically separated using an Agilent 1100 HPLC system (Santa Clara, CA) and C18 Zorbax Eclipse column (150 × 4.6 mm, 5μ - Agilent). These two analytes were eluted with a linear gradient of mobile phase A:B (v/v) starting from 95:5 at 0 min to 30:70 in 10 minutes at a flow rate 1.1 mL/min. The mobile phase A was 5 mM ammonium acetate with 0.1% formic acid, pH 4.0, and mobile phase B was acetonitrile with 0.1% formic acid. Voriconazole and the N-oxide were detected by UV at their respective λ max of 254nm and 265nm; the retention time was 6.9 min for N-oxide and 8.2 min for voriconazole. The HPLC-MS/MS method was used for the analysis of voriconazole N-oxide in samples from the metabolism studies with recombinant FMO enzymes. An Applied Biosystems, API 4000-triple quadrupole mass spectrometer (MDS Sciex Instruments - San Francisco, CA) fitted with TurbolonSpray® interface was used in conjunction with Shimadzu solvent delivery system (Kyoto, Japan) and a CTC PAL auto-sampler (Carborro, NC). The HPLC separation was achieved on an Aquasil C18 analytical column (50 × 2.1, 5μ - Thermo Scientific, Waltham, MA) using gradient elution of mobile phases A:B (v/v), where mobile phase A was 0.1% formic acid and mobile phase B was methanol with 0.1% formic acid. The starting condition of 90:10 (A:B) was held as the eluent was diverted to waste for 0.5 min. From 0.5 min to 3 min, the mobile phase composition changed linearly increased 5:95 (A:B). The flow rate was 0.7 mL/min, and the injection volume was 5 µL. Multiple reaction monitoring (MRM) was used to monitor voriconazole (m/z transition 350→127), the N-oxide (m/z transition 366→224), and the internal standard (diclofenac, m/z transition 296→215) in positive ion mode. The retention time for the N-oxide and voriconazole were 2.5 min and 2.8 min, respectively. For quantitative determination of the N-oxide formed, a calibration curve was constructed using the peak area ratio of authentic standard of the N-oxide:internal standard over a concentration range of 1 nM - 5 µM (R2>0.99).
The quantitative determination of hydroxy metabolites of CYP probe substrates was achieved by a simultaneous HPLC-MS/MS method with similar HPLC conditions as used in voriconazole separation and monitored at m/z transitions of 235→150, 312→230, 305→269 by positive mode MRM for 4-hydroxy mephenytoin, 4-hydroxy diclofenac, and 6-hydroxy testosterone, respectively. Quantitative determination was based on a constructed calibration curve of corresponding hydroxyl metabolite standards as previously reported (Yanni, 2004).
Statistical Analysis
All data represent a minimum of three experimental determinations and were expressed as mean ± standard deviation (S.D.). The statistical significance for the difference of mean values of treated and control was evaluated by unpaired t- test and was also evaluated using non-parametric method as Wilcoxon rank sum with *p < 0.05; ** p <0.01; *** p <0.001 compared to the control.
Results
Metabolism of Voriconazole by Human Liver Cytochrome P450 enzymes
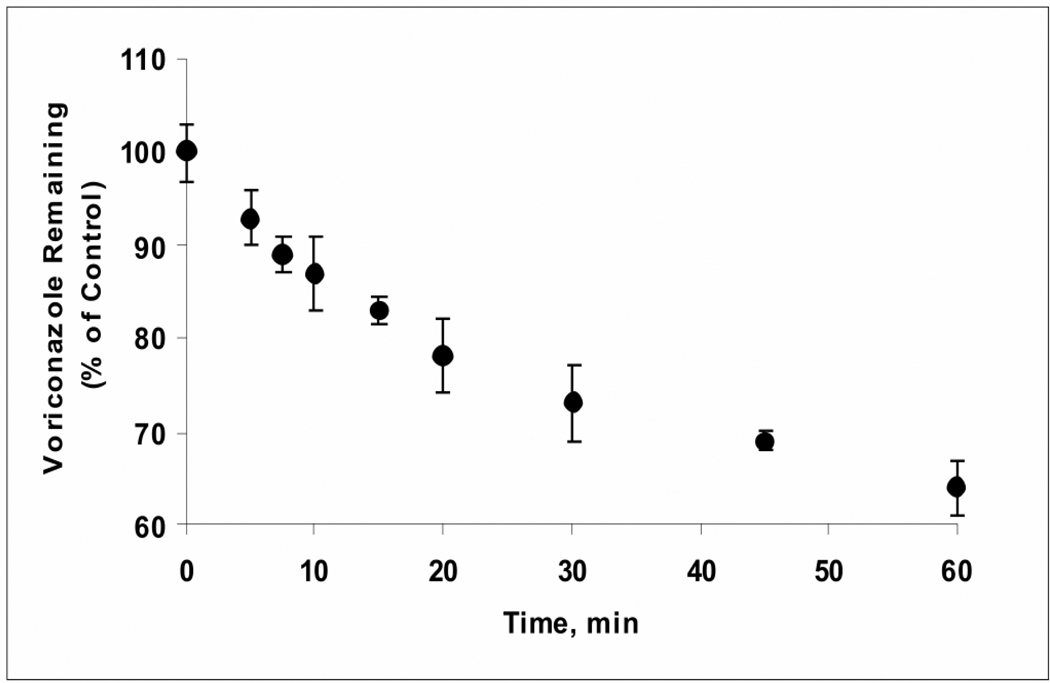
Voriconazole (2 µM) was metabolized by HLM in the presence of NADPH as determined by the loss of the substrate as a function of time (Figure 2); no loss of the substrate was observed in the absence of NADPH (data not shown). The rate of metabolism over the linear range (up to 20 min incubation and 20% loss of voriconazole) was 54 pmol/min/mg protein and T1/2 was estimated to be 57 min.
Figure 2. Oxidative Metabolism of Voriconazole by Human Liver Microsomes.
Voriconazole (2 µM) was metabolized by HLM in the presence of NADPH (2 mM) as determined by the loss of the substrate as a function of time; the rate of metabolism over the linear range (up to 20 min incubation and 20% loss of voriconazole) was 54 pmol/min/mg protein and T1/2 was estimated to be 57 min. The data points represent the mean of three determinations ± S.D.
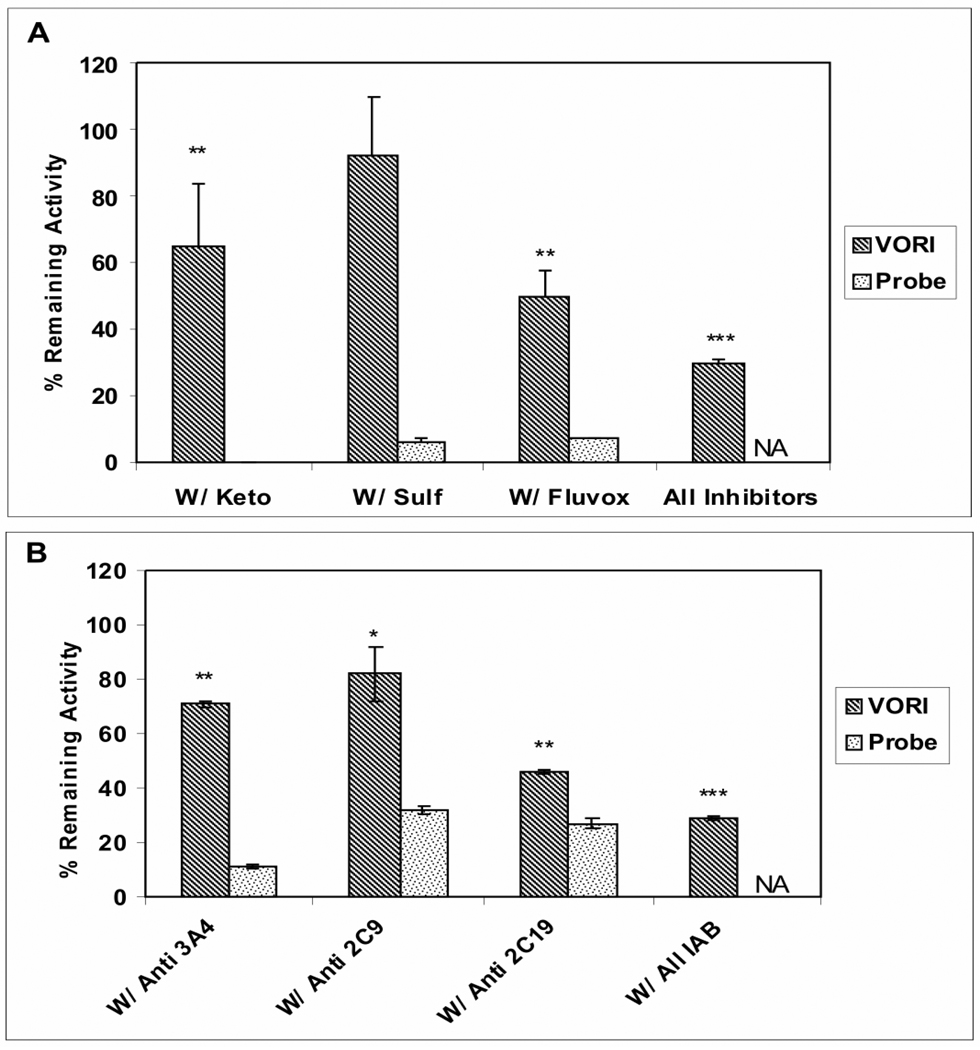
Metabolism of voriconazole (2 µM) by HLM was measured in the presence of ketoconazole (3µM), sulfaphenazole (30 µM), or fluvoxamine (3µM), which selectively inhibit CYP3A4, CYP2C9, and CYP2C19, respectively, to determine the relative contributions of these enzymes toward voriconazole metabolism. Approximately 35%, <5%, and 50% of voriconazole metabolism was inhibited by ketoconazole, sulfaphenazole, and fluvoxamine, respectively. A mixture of the three CYP inhibitors (at respective concentrations indicated above) inhibited the metabolism by 75% (Figure 3A); thus approximately 25% of the oxidative metabolism of voriconazole by HLM persisted following chemical inhibition of CYP3A4, CYP2C9, and CYP2C19. Other CYP enzymes were not considered because Hyland et al. (2003) have previously reported that none of the other CYP enzymes, e.g. CYP1A2, CYP2D6, CYP2B6, and CYP2A6, played a role in voriconazole metabolism. The CYP-selective inhibitors, at the concentrations employed, nearly completely inhibited the respective CYP enzyme activities: CYP3A4, (testosterone 6-β hydroxylase), CYP2C9 (diclofenac 4-hydroxylase), and CYP2C19 (S-mephenytoine 4- hydroxylase) activities in HLM were inhibited by 100 %, 93 %, and 93 % by ketoconazole (3µM), sulfaphenazole (30 µM), and fluvoxamine (3µM), respectively (Figure 3A).
Figure 3. Role of CYP3A4, CYP2C9, and CYP2C19 in the Metabolism of Voriconazole by Human Liver Microsomes.
A- The metabolism of voriconazole (VORI) measured by loss of parent following co-treatment with CYP inhibitors ketoconazole (Keto, 3 uM), sulfaphenazole (Sulf, 30 uM), fluvoxamine (Fluvox, 3 uM), and a mixture of three inhibitors was determined in incubations with voriconazole (2 µM) and HLM (1 mg/mL) in presence of NADPH (2mM) for 20 minutes (Striped bar); the results are presented as percent of “no inhibition” control (100%, 54 pomol/min/mg). Under the same experimental conditions, the effect of these inhibitors on the CYP activities was measured using the probe substrates (testosterone for CYP3A4, diclofenac for CYP2C9, and S-mephenytoin for CYP2C19) (dotted bar). B- Voriconazole metabolism by HLM was determined upon co-incubation with CYP3A4-, CYP2C9-, or CYP2C19-specific inhibitory antibody (Anti 3A4, Anti 2C9, Anti 2C19, respectively) and with a mixture of the three CYP inhibitory antibody (ALL IAB) (striped bar); the results are presented as percent of “no inhibition” control as explained in the Methods Section, where the “no inhibition” control was the metabolism resulting from co-incubation with rabbit IgG serum (100%). The effect of IAB on corresponding CYP probe substrates was also measured (dotted bar). The data are the mean of three separate experiments ± S.D. Effect Unpaired t-test analysis were reported as *p < 0.05; ** p <0.01; *** p <0.001 compared to no inhibition control. The statistical analysis by non-parametric method like Wilcoxon rank sum yielded the same outcome as unpaired t test. NA, non applicable.
The contributions of CYP3A4, CYP2C9, and CYP2C19 to voriconazole metabolism by HLM were also assessed using commercially available selective inhibitory antibodies against CYP3A4, CYP2C9, and CYP2C19. As shown in Figure 3B, inhibitory antibodies against CYP3A4, CYP2C9, and CYP2C19 inhibited voriconazole metabolism by 30%, 20% and 52%, respectively. Similarly to the chemical inhibitors, the three inhibitory antibodies together, caused approximately 70% reduction in voriconazole metabolism (Figure 3B). Under the experimental conditions employed, the respective CYP inhibitory antibodies inhibited CYP3A4, CYP2C9, and CYP2C19 activities (determined using their probe substrates) by 90%, 70%, and 78%. Evaluation of the cross-reactivity of the three inhibitory antibodies showed that anti CYP2C9 inhibited CYP2C19 and CYP3A4 by 40% and 45%, respectively, anti CYP2C19 inhibited CYP3A4 by 17%, and anti CYP3A4 inhibited CYP2C9 by 20%. The cross-reactivity of anti CYP2C9 for CYP2C19 could explain the difference between the estimated contribution of CYP2C9 to voriconazole metabolism determined by the chemical inhibitor, sulfaphenazole, (5%, Figure 3A) and by anti CYP2C9 (20%, Figure 3B). The information supplied by Xenotech in the package insert for anti CYP2C9 indicated that it inhibited 20% of CYP2C19 activity.
Evidence for Voriconazole Metabolism by Human Liver Flavin-containing Monooxygenase
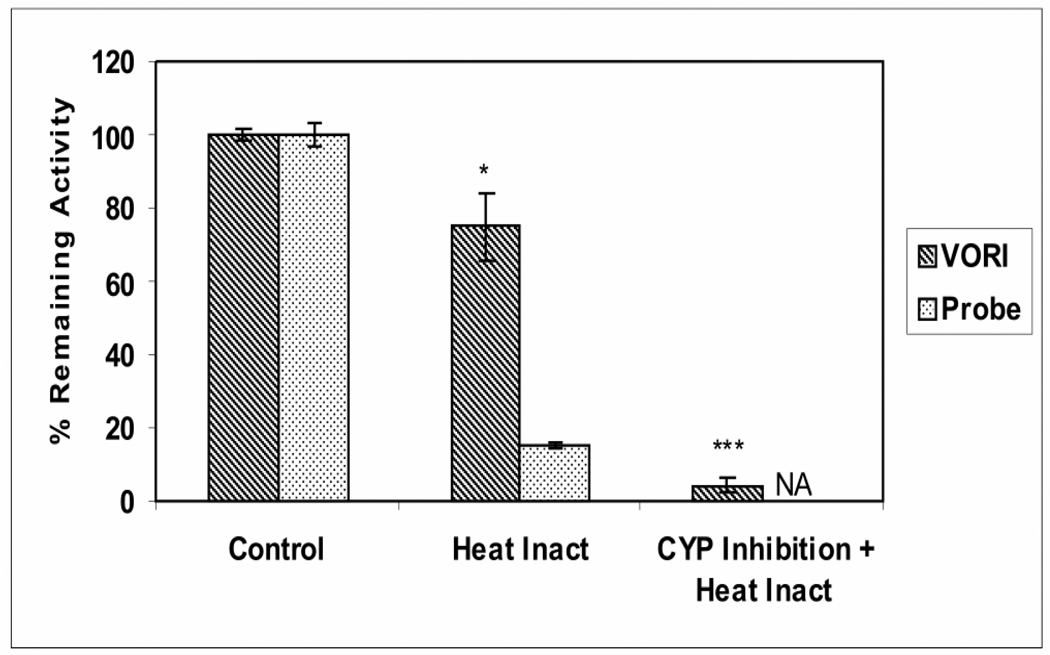
The results in Figure 3 indicate that 25–30% of the oxidative metabolism of voriconazole by HLM persisted following inhibition of CYP3A4, CYP2C9, and CYP2C19, thus suggesting a role for another CYP enzyme(s) or for a different (non-CYP) oxidative enzyme family. A role for FMO enzymes was anticipated based on their known ability to catalyze oxidation of hetero atoms (Ziegler DM, 1993) including N-oxidation (Cashman, 1995), and on previous identification of an N-oxide (Figure 1) as the major microsomal metabolite of voriconazole (Roffey et al., 2003; Hyland et al., 2003; Murayama et al., 2007). This hypothesis was first investigated using the lability of FMO toward mild heat, a condition which does not affect CYP enzymes. The metabolism of voriconazole by HLM, preheated to 45 °C for 5 minutes, was reduced to 75% of control (Figure 4); under these conditions, metabolism by HLM of the FMO probe substrate, benzydamine, was inhibited by 85% (Figure 4). The metabolism of the CYP probes by HLM was not affected by the heat treatment (data not shown). Further, nearly complete (95%) inhibition of voriconazole metabolism by HLM was observed when the heat-treatment of the HLM and the treatment with the cocktail of inhibitors for CYP3A, CYP2C9, and CYP2C19 were combined in a single experiment (Figure 4). These results strongly suggest that 25–30% of the human microsomal metabolism of voriconazole can be catalyzed by FMO with the remaining 70–75% of the metabolism catalyzed by CYP enzymes, mostly CYP3A4 and CYP2C19.
Figure 4. Effect of Heat Treatment on the Metabolism of Voriconazole by Human Liver Microsomes.
Voriconazole (2 µM) metabolism measured by loss of parent by HLM, preheated to 45 ºC for 5 minutes (striped bar), was determined and compared with the metabolism by HLM preheated to 37 ºC for 5 minutes (100% control). The effect of heat treatment on the metabolism of the FMO probe substrate benzydamine (60 µM) was also measured by the loss of parent (dotted bar) (Stormer et al., 2000). Incubation of voriconazole with HLM, preheated to 45 ºC for 5 minutes, was conducted in the presence of a mixture of CYP inhibitors to determine the combined effect of heat pretreatment and CYP inhibition on voriconazole metabolism by HLM. Voriconazole metabolism by control HLM, preheated to 37 ºC for 5 minutes, co-incubated without the CYP inhibitors served as a control. NA, indicated not applicable. The data are the mean of three separate experiments ± S.D. Unpaired t-test analysis were reported as *p < 0.05; ** p <0.01; *** p <0.001 compared to control. VORI, voriconazole.
Metabolism of Voriconazole to N-oxide by Recombinant Human Flavin Monoxygenase
To confirm the role of FMO in the metabolism of voriconazole, its metabolism by recombinant FMO enzymes (FMO1, FMO3, and FMO5) was examined. The HPLC-MS/MS chromatogram of the products generated from the incubation of voriconazole with recombinant FMO enzymes showed that voriconazole was metabolized to a product that co-eluted with the authentic N-oxide (N1-oxide of the 5-fluoropyrimidine moiety) (Figure 5 panels A and B); ion trap mass spectrometry analysis of this metabolite further confirmed its identity. Interestingly, the HPLC-MS chromatogram of voriconazole metabolites produced by HLM showed two metabolites – the N-oxide and a more polar metabolite (Figure 5 panel C), presumably hydroxymethyl voriconazole, reported as a CYP3A4-specific metabolite by Murayama et al., 2007. The formation of the more polar (hydroxymethyl) metabolite was abolished by ketoconazole (3µM) treatment of the HLM (Figure 5 panel D), confirming the finding of Murayama, et al., 2007 that it is selectively formed by CYP3A4. Evaluation of the cross reactivity of CYP inhibitors, ketoconazole (3µM) and fluvoxamine (3µM), for voriconazole N-oxidation by FMO1- and FMO3 indicated that these inhibitors diminished FMO3-catalyzed voriconazole N-oxidation by 25% and 40%, respectively, but had negligible effect on the FMO1 activity.
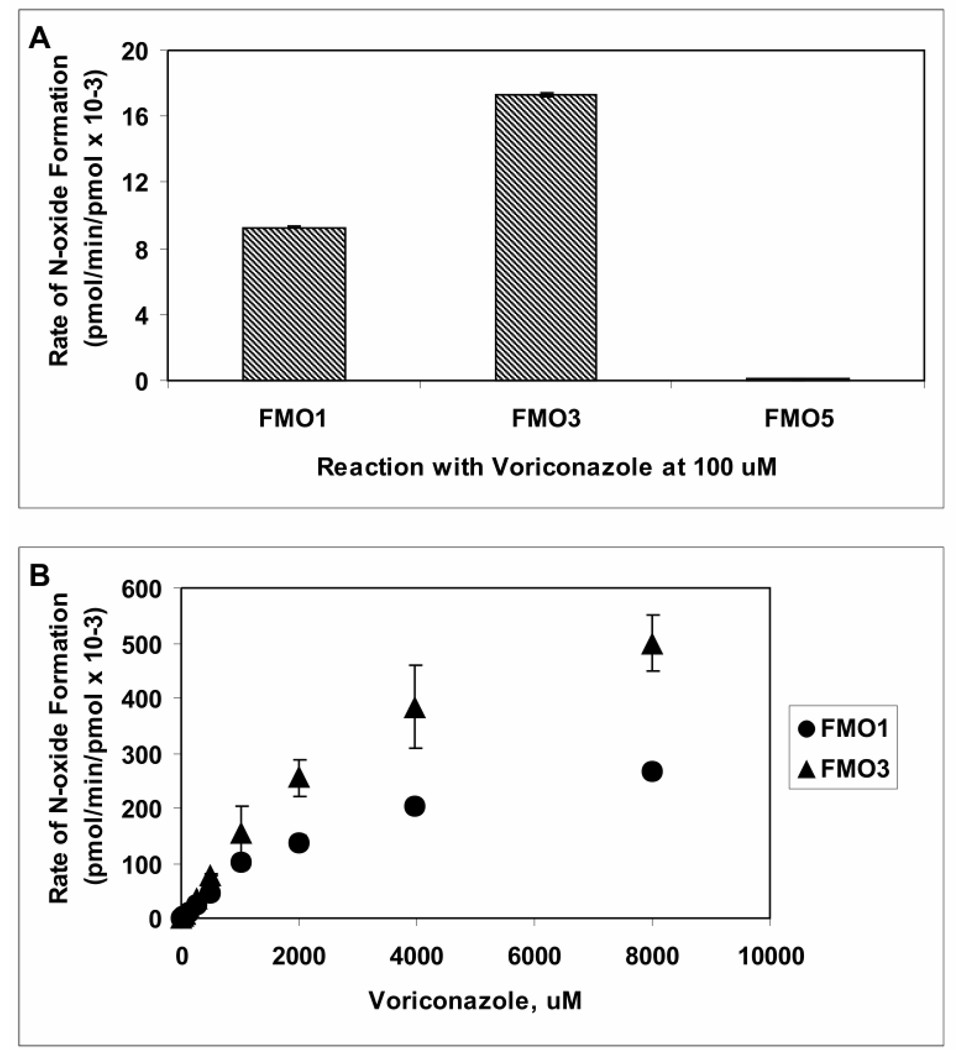
The rate at which voriconazole (100 µM) was metabolized by the three commercially available recombinant FMO1, FMO3, or FMO5 enzymes is shown in Figure 6A. FMO3 metabolized voriconazole at a greater velocity than did FMO1, and metabolism by FMO5 was too low to detect any product. The rate of N-oxide formation by either FMO1 or FMO3 as a function of voriconazole concentrations (1 µM - 8000 µM) followed Michaelis-Menten kinetics (Figure 6B), yielding Km values of 3.0 mM and 3.3 mM, and Vmax values of 0.025 and 0.044 pmol/min/pmol, respectively (Table 1).
Figure 6. Catalytic Activities of Recombinant Human Flavin-containing Monooxygenase 1, 3, and 5 toward Voriconazole.
A- Voriconazole (100 µM) was metabolized by recombinant human FMO enzymes FMO1 (60 pmol/mL), FMO3 (60 pmol/mL) and FMO5 (150 pmol/mL) in presence of NADPH for 60 minutes, and rates of metabolism (pmol/min/pmol enzyme) were calculated from a calibration curve constructed using the authentic N-oxide standard. B- Initial velocity of voriconazole N-oxide formation by FMO1 (triangle) and FMO3 (circle) was determined as a function of voriconazole concentration; data are reported as mean (n=3) ± S.D. The N-oxide formation by FMO5, was undetectable.
Table 1.
Kinetic Constants for Voriconazole Metabolism by Recombinant Human Flavin-containing Monooxygenases
| Parameter | FMO1 | FMO3 |
|---|---|---|
| Km, mM | 3.0 ± 0.5 | 3.4 ± 0.8 |
| Vmax, pmol/min/mg | 21.3 ± 2.0 | 42.5 ± 5.9 |
| Vmax, pmol/min/pmol | 0.025 ± 0.002 | 0.044 ± 0.006 |
Kinetic constants were determined based on metabolism experiments with voriconazole (1 µM-8000 µM) and recombinant human FMO enzymes (60 pmol/mL) for 30 minutes in the presence of NADPH. The Michaelis-Menten equation was fitted to the rates of N-oxide formation using non-linear regression approach in WinNonLin software v.4.
Metabolism of Voriconazole to N-oxide by Liver Microsomes from CYP2C19 Poor Metabolizers
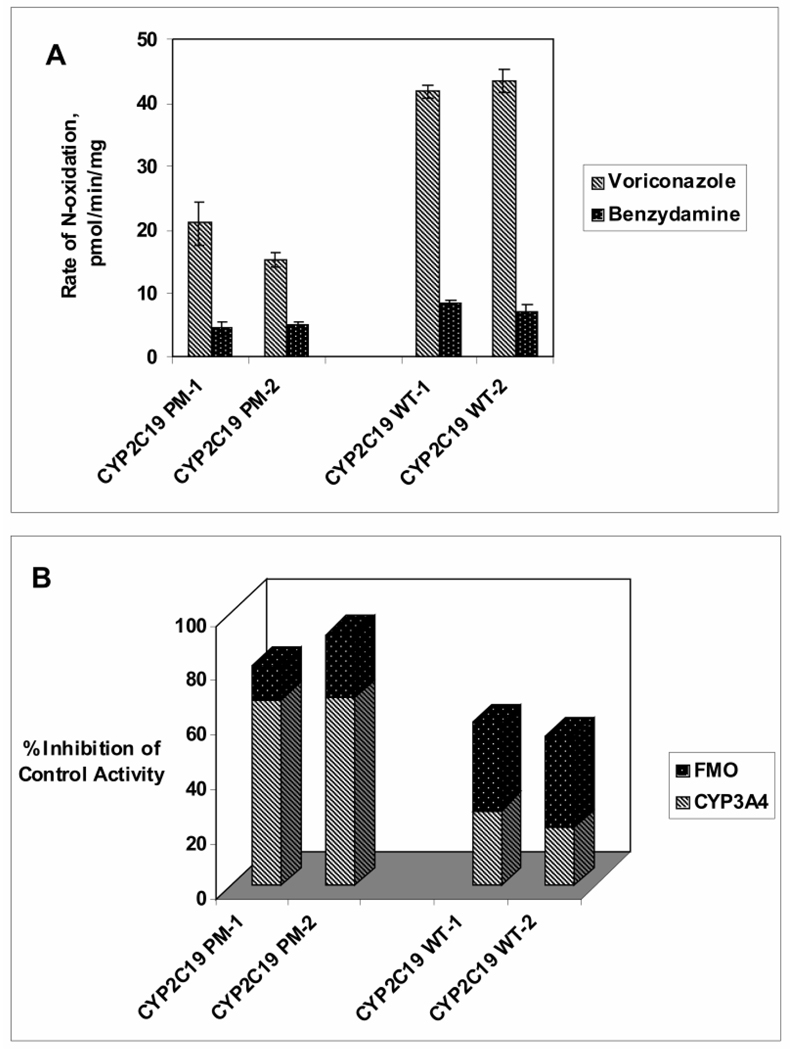
If CYP3A4 and FMO are the two most important contributors, next to CYP2C19, toward voriconazole N-oxidation, it is important to assess their relative role in CYP2C19 poor metabolizers. Thus, inhibition of voriconazole N-oxidation by ketoconazole (3 µM) and by heat-inactivation (45 ºC for 5 minutes) was assessed using 2 samples of HLM obtained from CYP2C19 poor metabolizers (2*/2*) in comparison to 2 samples of CYP2C19-competent HLM (WT). As expected, the total voriconazole N-oxidation by CYP2C19-deficient HLM (~20 pmol/min/mg) was less than 50% of that by CYP2C19-competent HLM (>40 pmol/min/mg) (Figure 7A). Interestingly, FMO activity (as determined by metabolism of the probe substrate, benzydamine) appeared to be somewhat lower (< 50%) in CYP2C19-deficient HLM than in CYP2C19-competent HLM (Figure 7A). CYP3A4 contributed to over 60% of the total voriconazole N-oxidation in CYP2C19-deficient HLM, whereas contribution of FMO was <20%. This may reflect lower FMO expression in CYP2C19-deficient individuals. In contrast, FMO appeared to contribute as much as or greater than CYP3A4 to voriconazole N-oxidation by CYP2C19-competent HLM, together the two enzymes contributing to >50% of the total voriconazole N-oxidation.
Figure 7. Voriconazole N-oxidation by Human Liver Microsomes from CYP2C19 Poor Metabolizers and from Normal Subjects.
A- Voriconazole (5 µM) N-oxidation by HLM (1.0 mg/mL) from two CYP2C19 poor metabolizers (2*/2*) (PM-1, PM-2) and by HLM (1.0 mg/mL) from two CYP2C19 wild type subjects (1*/1*) (WT-1, WT-2) was expressed as pmol/min/mg (striped bar) and compared to the maximal rate of benzydamine (250 µM) metabolism expressed as nmol/min/mg (dotted bar). Data for each sample are reported as mean (n=3) ± S.D. B- Contribution of CYP3A4 and FMO toward voriconazole N-oxidation was determined by measuring the N-oxide formation in the presence of ketoconazole (3 µM) or by heat-treatment (45 °C for 5 minutes), respectively, and comparing it to no-inhibition controls. Percent Inhibition by ketoconazole and by heat-inactivation is shown as striped bar and dotted bar, respectively; data for each sample are reported as mean (n=3).
Discussion
Voriconazole is an antifungal agent that is known to be cleared predominantly via metabolism in all species tested; for example, only 2% of the administrated dose is recovered in excreta as the parent drug in humans. Its elimination is capacity-limited in preclinical species (Roffey et al., 2003) and in humans (Purkins et al., 2002), presumably due to saturation of metabolic transformations. Thus in humans, with a 2-fold increase in the oral dose (200 to 400 mg) of voriconazole, a 2.8-fold increase in Cmax and a 3.9-fold increase in AUCτ was observed (Purkins et al., 2002). The major metabolic pathways in humans (Roffey et al., 2003) appear to involve N-oxidation of the fluoropyrimidine ring, hydroxylation of the methyl group, and glucuronidation of the hydroxylated metabolites. An interesting metabolic step that appears to be stimulated by the formation of the N-oxide is the cleavage of voriconazole, resulting in the loss of the fluoropyrimidine ring (Roffey et al., 2003). Thus, the N-oxide may contribute to even greater percentage of the total metabolites than would be apparent from the percentages of the isolable oxide. The N-oxide is also the major circulating metabolite in humans, rats, and dogs. In vitro studies have implicated CYP2C19, CYP3A4, and to a lesser extent CYP2C9 in the oxidative metabolism of voriconazole (Hyland et al., 2003; Murayama et al., 2007), and their role in the formation of N-oxide, seems to depend on the concentration of voriconazole in the reaction (Hyland et al., 2003).
The results reported here provide unequivocal evidence that in addition to the CYP enzymes, FMO can metabolize voriconazole to its major circulating metabolite, the N-oxide. Studies with chemical inhibitors and with CYP-specific antibodies clearly established that CYP3A4, CYP2C9, and CYP2C19 could not account for more than approximately 70% of voriconazole metabolism by HLM.
Because of the known selectivity of this enzyme family to oxidize compounds at the hetero atoms, such as sulfur and nitrogen (reviewed in Krueger and Williams, 2005), the role of FMO in the oxidation of voriconazole at the ring nitrogen of the fluoropyrimidine ring was investigated. Initial evidence for the involvement of FMO in the oxidation of voriconazole was obtained when the results showed that about 20% of the total enzyme activity contributing to oxidative metabolism of voriconazole was heat-labile; it has been established previously that short incubation (2–5 min) at 45–50 ºC (without NADPH) destroys FMO activity without significantly affecting CYP activity (Krueger and Williams, 2005; Ziegler, 1980). Further studies with recombinant enzymes (FMO1, FMO3, FMO5) definitively established that enzymes of the FMO family oxidized voriconazole to the N-oxide metabolite. The kinetic analysis yielded Km values in the low millimolar range for FMO1 and FMO3, whereas FMO5 appeared to exhibit only a very low level of catalytic activity toward voriconazole. Taken together, these results suggest that certain FMO isoforms contribute significantly to oxidative metabolism of voriconazole. We recognize that the estimation of the relative contribution of FMO toward voriconazole metabolism by HLM is solely based on the heat-sensitivity of a fraction of the metabolism. Unfortunately, the lack of inhibitory antibodies and FMO-selective inhibitors do not allow us to estimate the contribution of individual FMO enzymes in the overall metabolism of voriconazole. When the kinetic parameters for voriconazole metabolism by FMO1/FMO3 (Table 1) vs. CYP2C19/CYP3A4 (Hyland et al., 2003) are scaled to a typical human liver, containing 80 (FMO3), 15 (CYP2C9), and 100 (CYP3A4) pmol enzyme/mg of microsomal protein (Fisher et al., 2002; Koukouritaki et al., 2004; Hines 2006), it is expected that FMO would contribute to less than 2% of the voriconazole metabolic clearance by CYP3A4 and even smaller percentage of the metabolic clearance by CYP2C19. However, the in vitro (heat inactivation) studies with human liver microsomes (Figure 4) suggest much greater (~20%) contribution of FMO toward voriconazole N-oxidation. This is not surprising since a similar scaling exercise suggests a much smaller contribution of CYP3A4 (<4%) compared to CYP2C19 toward voriconazole metabolic clearance than has been found (~35%) in clinical studies (Mikus et al., 2006).
It is important to note that both CYP3A4 and FMO play a lesser role in comparison to CYP2C19 toward the metabolic clearance of voriconazole. In two separate studies, inhibitors of CYP3A4, erythromycin (1 g twice daily for 7 days, Purkins et al., 2003c) and indinavir (800 mg three times daily for 7 days, Purkins et al., 2003d), did not alter Cmax or AUCτ of co-administered voriconazole (200 mg twice daily). These studies led to the conclusion that while CYP3A4 might contribute to metabolic clearance, it did not govern the pharmacokinetics of voriconazole in humans. However, these conclusions should be considered with caution since these studies did not include appropriate positive controls. In fact, Mikus et al. (2006) contradicted this conclusion, and reported that the CYP3A4 inhibitor, ritonavir (300 mg twice daily), reduced the oral clearance of voriconazole (400 mg) by ~35%. Not surprisingly, the effect of ritonavir in reducing the oral clearance of voriconazole was much larger (>80%) in CYP2C19 poor metabolizers. Thus, it appears that CYP3A4 plays a much more significant role in CYP2C19 poor metabolizers; the in vitro data in Figure 7, in fact, confirm this. Further clinical and in vitro studies should be conducted to investigate the role of FMO in CYP2C19 poor metabolizers and in normal subjects.
This is the first report of the role of FMO in the metabolism of voriconazole in an in vitro system. Clearly, it would be important to assess the role of FMO in the metabolic clearance of this antifungal agent in preclinical species and in humans. There is compelling evidence that the metabolic clearance of voriconazole is saturated over the therapeutic dose range of this drug, since doubling the dose leads to an almost four-fold increase in the systemic exposure of the parent drug in adult patients. Interestingly, the metabolic clearance does not show evidence of saturation in children (Walsh et al, 2004). Whether this difference in dose-dependence observed in adults vs. children is due to different relative contributions of FMO, CYP3A4, and CYP2C19 in these two patient populations remains to be determined.
Acknowledgement
The authors would like to thank Pfizer for providing voriconazole, and Dr. Ralph Raasch for valuable discussions.
Financial support: Financial support by NICHD (NCC-PPRU 5U10 HD045962-04) is acknowledged.
Abbreviation
- CYP
cytochrome P450
- FMO
flavin-containing monooxygenase
- HLM
human liver microsomes
- Km
apparent Michaelis constant
- Vmax
maximum reaction velocity
- HPLC-MS/MS
liquid chromatography coupled with tandem mass spectrometry
Footnotes
The 1H-NMR is consistent with the partial 1H-NMR data published by Murayama et al. (2007).
References
- Allerston CK, Shimizu M, Fujieda M, Shephard EA, Yamazaki H, Phillips IR. Molecular evolution and balancing selection in the flavin-containing monooxygenase 3 gene (FMO3) Pharmacogenet Genomics. 2007;17:827–839. doi: 10.1097/FPC.0b013e328256b198. [DOI] [PubMed] [Google Scholar]
- Boucher HW, Groll AH, Chiou CC, Walsh TJ. Newer systemic antifungal agents : pharmacokinetics, safety and efficacy. Drug. 2004;64:1997–2020. doi: 10.2165/00003495-200464180-00001. [DOI] [PubMed] [Google Scholar]
- Cashman JR. Structural and catalytic properties of mammalian flavin-containing monooxygenase. Chem Res Toxicol. 1995;8:165–181. doi: 10.1021/tx00044a001. [DOI] [PubMed] [Google Scholar]
- Fisher MB, Yoon K, Vaughn ML, Strelevitz TJ, Foti RS. Flavin-containing monooxygenase activity in hepatocytes and microsomes: in vitro characterization and in vivo scaling of benzydamine clearance. Drug Metab Dispos. 2002;30:1087–1093. doi: 10.1124/dmd.30.10.1087. [DOI] [PubMed] [Google Scholar]
- Hines RN. Developmental and tissue-specific expression of human flavin-containing monooxygenases 1 and 3. Expert Opin Drug Metab Toxicol. 2006;2:41–49. doi: 10.1517/17425255.2.1.41. [DOI] [PubMed] [Google Scholar]
- Hyland R, Jones BC, Smith DA. Identification of the cytochrome P450 enzymes involved in the N-oxidation of voriconazole. Drug Metab Dispos. 2003;31:540–547. doi: 10.1124/dmd.31.5.540. [DOI] [PubMed] [Google Scholar]
- Koukouritaki SB, Manro JR, Marsh SA, Stevens JC, Rettie AE, McCarver DG, Hines RN. Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther. 2004;308:965–974. doi: 10.1124/jpet.103.060137. [DOI] [PubMed] [Google Scholar]
- Krueger SK, Williams DE. Mammalian-Flavin Containg Monooexygenase: structure/function, genetic polymorphism and role in drug metabolism. Pharmacol Therap. 2005;106:357–387. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus HM, Blumer JL, Yanovich S, Schlamm H, Romero A. Safety and pharmacokinetics of oral voriconazole in patients at risk of fungal infection: a dose escalation study. J Clin. Pharmacol. 2002;42:395–402. [PubMed] [Google Scholar]
- Luning U, Ross H, Thondorf I. Steric requirements of intraannular substituents in A,D-bridged calyx[6]arenes. J. Chem. Soc. Perkin Trans. 1998;2:1313–1317. [Google Scholar]
- Mikus G, Schowel V, Drzewinska M, Rengelshausen J, Ding R, Riedel KD, Burhenne J, Weiss J, Thomsen T, Haefeli WE. Potent cytochrome P450 2C19 genotype-related interaction between voriconazole and the cytochrome P450 3A4 inhibitor ritonavir. Clin Pharmacol Ther. 2006;80:126–135. doi: 10.1016/j.clpt.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Murayama N, Imai N, Nakane T, Shimizu M, Yamazaki H. Roles of CYP3A4 and CYP2C19 in methyl hydroxylated and N-oxidized metabolite formation from voriconazole, a new anti-fungal agent, in human liver microsomes. Biochem Pharmacol. 2007;73:2020–2026. doi: 10.1016/j.bcp.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Obach RS, Reed-Hagen AE. Measurement of michaelis constants for cytochrome P450-mediated biotransformation reactions using a substrate depletion approach. Drug Metab Dispos. 2002;30:831–837. doi: 10.1124/dmd.30.7.831. [DOI] [PubMed] [Google Scholar]
- Patterson TF. Early use of antifungal therapy in high-risk patients. Curr Opin Infect Dis. 2002;15:561–563. doi: 10.1097/00001432-200212000-00001. [DOI] [PubMed] [Google Scholar]
- Purkins L, Wood N, Ghahramani P, Greenhalgh K, Allen MJ, Kleinermans D. Pharmacokinetics and safety of voriconazole following intravenous- to oral-dose escalation regimens. Antimicrob Agents Chemother. 2002;46:2546–2553. doi: 10.1128/AAC.46.8.2546-2553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkins LN, Wood NK, Greenhalgh K, Eve SD, MD, Oliver SD. British Journal of Clinical Pharmacology. Vol. 56. 2003a. The pharmacokinetics and safety of intravenous voriconazole- a novel wide-spectrum antifungal agent; pp. 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkins Lynn, Purkins L, Wood N, Greenhalgh K, Allen MJ, Oliver SD. Voriconazole, a novel wide-spectrum triazole: oral pharmacokinetics and safety. British Journal of Clinical Pharmacology. 2003b;56:10–16. doi: 10.1046/j.1365-2125.2003.01993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkins L, Wood N, Ghahramani P, Kleinermans D, Layton G, Nichols D. No clinically significant effect of erythromycin or azithromycin on the pharmacokinetics of voriconazole in healthy male volunteers. Br J Clin Pharmacol. 2003c;56:30–36. doi: 10.1046/j.1365-2125.2003.01996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkins L, Wood N, Kleinermans D, Love ER. No clinically significant pharmacokinetic interactions between voriconazole and indinavir in healthy volunteers. Br J Clin Pharmacol. 2003d;56:62–68. doi: 10.1046/j.1365-2125.2003.02001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RJ, Proteau PJ, Marquez BL, Hetherington CL, Buckholz CJ, O'Connell KL. Flavin-containing monooxygenase-mediated metabolism of N-deacetyl ketoconazole by rat hepatic microsomes. Drug metab Dispos. 1999;27:880–886. [PubMed] [Google Scholar]
- Roffey SJ, Cole S, Comby P, Gibson D, Jezequel SG, Nedderman AN, Smith DA, Walker DK, Wood N. The disposition of voriconazole in mouse, rat, rabbit, guinea pig, dog, and human. Drug Metab Dispos. 2003;31:731–741. doi: 10.1124/dmd.31.6.731. [DOI] [PubMed] [Google Scholar]
- Stormer E, Roots I, Brockmoller J. Benzydamine N-oxidation as an index reaction reflecting FMO activity in human liver microsomes and impact of FMO3 polymorphisms on enzyme activity. Br J Clin Pharmacol. 2000;50:553–561. doi: 10.1046/j.1365-2125.2000.00296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuretzbacher U, Ihle F, Derendorf H. Pharmacokinetic/pharmacodynamic profile of voriconazole. Clin Pharmacokinet. 2006;45:649–663. doi: 10.2165/00003088-200645070-00002. [DOI] [PubMed] [Google Scholar]
- Walsh TJ, Karlsson MO, Driscoll T, Arguedas AG, Adamson P, Saez-Llorens X, Vora AJ, Arrieta AC, Blumer J, Lutsar I, Milligan P, Wood N. Pharmacokinetics and safety of intravenous voriconazole in children after single- or multiple-dose administration. Antimicrob Agents Chemother. 2004;48:2166–2172. doi: 10.1128/AAC.48.6.2166-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanni SB, Thiel P, Haroldsen P, Samara E. ISSX. Canada: Vancouver; 2004. High Throughput Evaluation of Clinical Drug Interaction using LCMSMS. [Google Scholar]
- Ziegler DM. Microsomal flavin-containing monooxygenase: Oxygenation of nucleophilic nitrogen and sulfur compounds. Enzymatic Basis of Detoxification1. 1980:201–217. [Google Scholar]
- Ziegler DM. Recent studies on the structure and function of multisubstrate flavin-containing monooxygenases. Annu Rev Pharmacol Toxicol. 1993;33:179–199. doi: 10.1146/annurev.pa.33.040193.001143. [DOI] [PubMed] [Google Scholar]