Abstract
Historically, microglia/macrophages are quantified in the pathological central nervous system (CNS) by counting cell profiles then expressing the data as cells/mm2. However, because it is difficult to visualize individual cells in dense clusters and in most cases it is unimportant to know the absolute number of macrophages within lesioned tissue, alternative methods may be more efficient for quantifying the magnitude of the macrophage response in the context of different experimental variables (e.g., therapeutic intervention or time post-injury/infection). The present study provides the first in-depth comparison of different techniques commonly used to quantify microglial/macrophage reactions in the pathological spinal cord. Individuals from the same and different laboratories applied techniques of digital image analysis (DIA), standard cell profile counting and a computer-assisted cell counting method with unbiased sampling to quantify macrophages in focal inflammatory lesions, disseminated lesions caused by autoimmune inflammation or at sites of spinal trauma. Our goal was to find a simple, rapid and sensitive method with minimal variability between trials and users. DIA was consistently the least variable and most time-efficient method for assessing the magnitude of macrophage responses across lesions and between users. When used to evaluate the efficacy of an anti-inflammatory treatment, DIA was 5–35x faster than cell counting and was sensitive enough to detect group differences while eliminating inter-user variability. Since lesions are clearly defined and single profiles of microglia/macrophages are difficult to discern in most pathological specimens of brain or spinal cord, DIA offers significant advantages over other techniques for quantifying activated macrophages.
Keywords: image analysis, macrophages, microglia, inflammation, CNS, immunohistochemistry
Introduction
The accumulation and activation of macrophages is a ubiquitous consequence of trauma or disease in any tissue and serves as a reliable index of injury or disease severity (Carson, 2002; Stoll and Bendszus, 2006; Szekanecz and Koch, 2007; Donnelly and Popovich, 2008). As such, the ability to reliably quantify then compare differences in these responses is useful for documenting disease progression, tissue healing or the efficacy of different therapies. In the central nervous system (CNS), resident microglia and infiltrating monocyte-derived macrophages (collectively referred to as CNS macrophages) respond to injury or infection by proliferating and undergoing dramatic morphological changes. These parameters vary as a function of tissue pathology and directly impact the accuracy and sensitivity of different quantitative techniques. Currently, there is no standardized approach for quantifying CNS macrophages; some prefer to manually count cellular profiles through the microscope (Roggendorf, Strupp et al., 1996; Felts, Woolston et al., 2005) while others use semi-quantitative techniques (e.g., categorical/subjective rating scales) (Tak, Thurkow et al., 1995; Colburn, DeLeo et al., 1997; Deininger and Schluesener, 1999) or digital image analysis (DIA) (Popovich, Wei et al., 1997; Furness, Rogers-Wheatley et al., 1997). Indeed, an informal survey of >30 primary research articles involving quantitation of CNS macrophages revealed that profile counting (standard or computer-assisted) was used most of the time, followed by DIA, categorical analyses, and lastly stereology (citations not shown).
Under ideal conditions, counting cell profiles or applying unbiased stereology (e.g., optical dissector) yields total numbers of CNS macrophages within a lesion. Indeed, Beggs and Salter found a significant difference in spinal microglial numbers between naïve mice and mice that received peripheral nerve injury using the optical fractionator technique (Beggs and Salter, 2007). However, in regions of hemorrhage and necrosis, where phagocytic microglia and blood monocytes cluster together, it is difficult or impossible to distinguish individual cell nuclei making it unlikely that either technique will yield consistent or reliable data. Moreover, in most cases it may not be useful to know how many CNS macrophages are present. Indeed, the function of CNS macrophages is not always predicted from cell numbers. Instead, function may be better predicted by quantifying how much a select surface or cytoplasmic antigen is up- or down-regulated. These changes can occur without increasing absolute cell number (McCann, O’Callaghan et al., 1996; McPherson, Kubik et al., 2003; Yasuda, Shinagawa et al., 2007).
In some circumstances, it may be feasible to categorically rank the magnitude of a CNS macrophage response as high, intermediate or low. This can be completed quickly and does not require that individual cell nuclei be discernible nor does it require one to distinguish between subtle changes in cell morphology. However, this type of analysis is prone to user bias and may not be sensitive enough to detect small changes in the magnitude of the CNS macrophage response. Because an increase in tissue surveillance can be inferred by the presence of more or larger (e.g., phagocytic) CNS macrophages (Popovich, Wei et al., 1997), the process of measuring the area occupied by labeled cells within a region of interest may be ideal for quantifying the magnitude of the CNS macrophage response, especially when individual cell profiles cannot be distinguished. Moreover, since all software programs that are capable of performing DIA recognize labeled targets in similar ways (e.g., based on densitometric scanning of thresholded targets), semi-automated methods can be universally applied without introducing significant user error.
The need to develop sensitive and reproducible methods for quantifying macrophage reactions in tissue sections has been recognized within (Blackbeard, O’Dea et al., 2007) and outside (Youssef, Smeets et al., 1998; Kraan, Haringman et al., 2000) the neuroscience community. Kraan et al. found DIA to be a more sensitive and time-efficient method than profile counting or categorical rankings for quantifying macrophages in inflamed synovial tissue (Kraan, Haringman et al., 2000). Recently, Blackbeard et al. reported similar advantages of using DIA to quantify intraspinal microglial activation after peripheral nerve injury (Blackbeard, O’Dea et al., 2007). However, to date, there have been no attempts to determine which of the most commonly used techniques should be used when analyzing specimens that involve graded levels of microglial activation with concomitant recruitment of blood monocytes. This question is relevant across a spectrum of experimental and clinical neurological disorders including stroke/cerebral ischemia, brain or spinal trauma, Wallerian degeneration, glioma, viral encephalitis and multiple sclerosis.
Here, different models of spinal cord pathology with distinct types of CNS macrophage activation were analyzed: 1) acute focal non-traumatic inflammation (zymosan microinjection), 2) chronic disseminated neuroinflammation (experimental autoimmune encephalomyelitis; EAE) and 3) inflammation caused by traumatic spinal cord injury (SCI). To measure the magnitude of the response in each case, we compared DIA, standard profile counting (PC) and profile counting with an unbiased sampling technique (UST). The sensitivity and consistency of each approach was assessed through intra-user and inter-user comparisons. Our results show that DIA yields the most sensitive and consistent data. In follow-up studies, when two users applied the three techniques to quantify traumatized spinal cord sections prepared from rodents that had or had not received an anti-inflammatory therapy, only DIA detected a treatment effect without introducing inter-user variability. Moreover, DIA was 5–35x faster to use than the PC method.
Materials and Methods
Animal models
All housing, surgical and postoperative care procedures were performed in accordance with The Ohio State University Institutional Animal Care and Use Committee and have been described previously (Jakeman, Guan et al., 2000; Kigerl, McGaughy et al., 2006). All tissues, except those from rats with cervical SCI (see Figure 6), were generated in previous studies. Non-traumatic focal inflammatory lesions were created by nanoinjecting zymosan (50 nl; 12.5 mg/ml) into the lateral funiculus of the mid-thoracic spinal cord of female Sprague-Dawley rats and lesions were analyzed at 7 days post-injection (Popovich, Guan et al., 2002). Experimental autoimmune encephalomyelitis (EAE) was induced by immunizing female C57BL/6 mice with myelin oligodendrocyte protein (400 μg; emulsified in complete Freund’s adjuvant containing M. tuberculosis at 0.5 mg/ml) (Ankeny and Popovich, 2007). Pertussis toxin (0.2 ml; i.p.) was injected at the time of immunization then again 48 hours later (Papenfuss, Rogers et al., 2004). Inflammatory lesions were analyzed in sections of cervical spinal cord 42 days post-immunization.
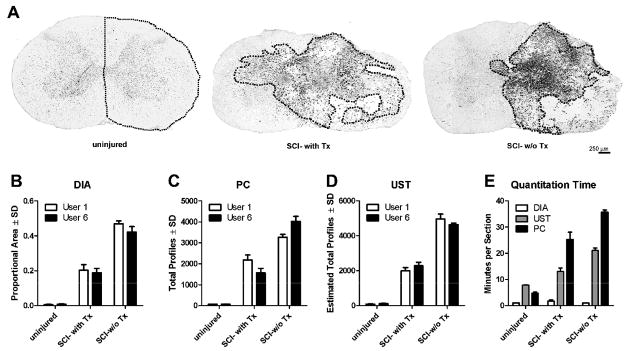
Figure 6.
Comparative analysis of quantitative techniques in uninjured or injured spinal cord with and without treatment to reduce the CNS macrophage response. (A) Representative sections from each condition; dotted line indicates areas quantified in (B–E). (B) DIA was not significantly different between users (P=0.12) but detected significant differences between treatment conditions (P<0.001). (C–D) Although profile counts [either using standard counting (C) or estimated using UST (D)] significantly differentiated between experimental conditions (P<0.001 for both methods), there were significant user by condition interactions for UST (p=0.043, Df=2, F=4.726) and PC (p=0.029, Df=2, F=5.679) signifying that these measure were more variable between users. (E) PC and UST counting methods take significantly more time per section than DIA (P<0.01 among all methods for naïve, SCI w-Tx and SCI-w/o-Tx).
Two models of spinal contusion injuries were used in this study. In the mouse model, CNS macrophages were analyzed in sections from female C57BL/6 mice that received a mid-thoracic spinal contusion injury (OSU device; 0.5 mm displacement) 42 days earlier (Jakeman, Guan et al., 2000; Jones, Basso et al., 2002; Sroga, Jones et al., 2003). In the rat model, cervical spinal cord contusion injuries were generated as described previously (Gensel, Tovar et al., 2006). Briefly, CNS macrophages were analyzed in sections from female Sprague-Dawley rats that received a unilateral cervical (C5) spinal contusion injury (Infinite Horizons device; probe=1.5 mm, force=175 kdyn) 7 days earlier. To deplete macrophages systemically and limit intraspinal macrophage accumulation, a subset of injured rats was injected with phosphatidylcholine liposomes containing clodronate on days 1, 3, and 6 post-injury (2 ml/day via the tail vein) (Popovich, Guan et al., 1999). Controls received identical PBS injections. For the zymosan, EAE, and mouse contusion lesions, one individual section from one representative animal was analyzed. For the rat contusion lesions with/without macrophage depletion, one section each from n = 3–4 animals were analyzed in each group. Clodronate was a gift of Roche Diagnostics GmbH (Mannheim, Germany) and was encapsulated into liposomes using methods that were described previously (van Rooijen and Sanders, 1994).
Immunohistochemistry
All tissues were prepared and stained using procedures described previously (Popovich, Guan et al., 2002; Kigerl, McGaughy et al., 2006). Briefly, frozen tissue sections were cut (10 μm) on a cryostat then stained with macrophage-specific antibodies: 1) OX-42 (1:4000; Serotec; zymosan and rat contusion), 2) anti-Iba1 (1:750; Wako; EAE), and 3) M1/70 (1:200; Serotec; mouse contusion). OX-42 and M1/70 bind membrane-bound CD11b (complement type 3 receptor) on rat and mouse macrophages, respectively (Springer, Galfre et al., 1979; Robinson, White et al., 1986). Iba1 is Ionized calcium-Binding Adaptor molecule 1, an intracellular signaling protein expressed by myeloid cells (Imai, Ibata et al., 1996). CD11b and Iba1 are constitutively expressed but are upregulated on activated cells (Popovich, Wei et al., 1997; Ito, Tanaka et al., 2001). Bound antibody was visualized using the Elite ABC reagent (Vector Laboratories, Burlingame, CA) and diaminobenzidine (DAB) as a substrate (Vector Laboratories). Antibody specificity has been previously documented for all antibodies used in this study by Western blot or immunoprecipitation assays (Kigerl, McGaughy et al., 2006). Once DIA was completed, coverslips were removed and sections were counterstained with cresyl violet. Cresyl violet stains nuclei and Nissl bodies (i.e., ribosomal RNA), and was used to facilitate identification of individual cell nuclei. Cresyl violet staining was performed after DIA to prevent interference with digital thresholding of stained tissue.
Lesion area selection
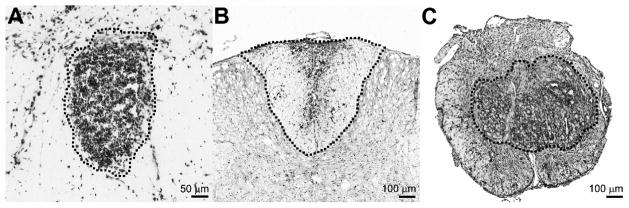
All analyses were performed on a Zeiss Axioplan 2 imaging microscope equipped with a motorized stage (Ludl Electronic Products Inc., Hawthorne, NY) and an MCID 6.0 Elite image analysis station (Interfocus, Linton, England). The lesion area for the focal inflammation (zymosan) model was a well-demarcated cluster of activated microglia/macrophages in the lateral funiculus (Figure 1a). Given the heterogeneity of EAE lesions, the sample area needed to be standardized and easily interpreted by each user. Consequently, an entire dorsal funiculus containing a focus of active inflammation was chosen since it provided clearly-defined anatomical boundaries in which to restrict quantitative analyses (Figure 1b). For contused spinal cord, a section from the necrotic lesion core was analyzed (Sroga, Jones et al., 2003) (Figures 1c, 6a).
Figure 1.
Examples of activated intraspinal microglia/macrophages in zymosan microinjection lesions (A), dorsal funiculus of mouse spinal cord with EAE (B) and the impact site of contused mouse spinal cord (C). Tissues are stained with OX-42 (A; mouse anti-rat anti-CD11b), anti-Iba1 (B) or the M1/70 clone (C; mouse anti-mouse CD11b). Dashed lines delineate the boundaries of the lesions that were quantified.
Users
Six individuals participated in this study. Users 1, 2, and 3 were trained in the Popovich lab while Users 4, 5 and 6 were trained in collaborating labs in OSU’s Center for Brain and Spinal Cord Repair. Users 1, 2, 4 and 6 were experienced with macrophage quantitation while Users 3 and 5 were familiar with microscopy but inexperienced at macrophage quantitation. User 2 trained User 1. User 1 then trained Users 3 and 5. Users 2, 4, and 6 were trained by their respective principal investigators. Therefore, Users 1, 2, 3, and 5 were all trained in a similar way. Inter-user comparisons showed no significant user effect for any technique (p = 0.2392 for PC, p = 0.1384 for UST, p = 0.4134 for DIA) meaning that all users applied each technique in the same way.
Before counterstaining, User 1 analyzed each lesion using DIA and PC four times each. However, in order to minimize any effect of “training” or “familiarity” with the tissue, none of the different analyses were conducted consecutively and > 4 hours passed before a lesion was quantified again. Users 2, 3, 4 and 5 then performed DIA once for each lesion. After counterstaining, User 1 analyzed each lesion using PC and UST four times each, while Users 2, 3, 4 and 5 used these techniques once for each lesion. Replicate data created by User 1 were used to determine intra-user variability. Inter-user comparisons were completed using data from all users. As an additional test of sensitivity, Users 1 and 6 compared the ability of DIA, UST and PC to distinguish the effects of a treatment designed to attenuate the CNS macrophage response to trauma.
Digital image analysis (DIA)
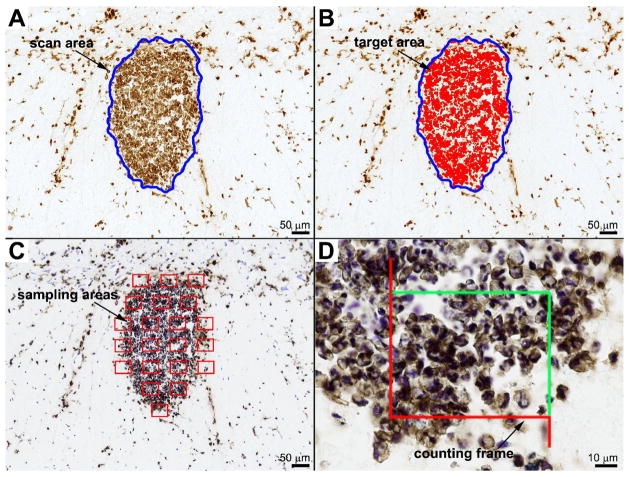
DIA was performed using the basic densitometric thresholding features of an MCID™ Elite Imaging Station (InterFocus Imaging Ltd, England). The strength of this approach is its ease of use, the need for minimal training and the wide-availability of similar software and shareware programs that can be downloaded from the Internet free of charge (e.g., ImageJ, NIH Image, Scion Image). Indeed, most image analysis programs are capable of quantifying pixel areas based on color thresholds. Previously, we and others have used the DIA technique to quantify microglia and macrophages in different forms of spinal cord pathology (Popovich, Wei et al., 1997; Popovich, Yu et al., 1997; Popovich and Hickey, 2001; Popovich, Guan et al., 2002; Olson and McKeon, 2004; Tsutsui, Noorbakhsh et al., 2005; Kigerl, McGaughy et al., 2006) for variations on applying the DIA technique). For the present study, DIA was performed by first capturing images at the highest magnification that allowed the entire lesion area (i.e., scan area) to be visualized within a single image frame. For the zymosan lesion, a 10X objective was used; for the EAE and mouse contusion lesions, a 5X objective; and for the rat contusion lesion, a 2.5X objective. The perimeter of each lesion was digitally outlined by each user and for each replicate to define the region of interest (i.e., scan area) (Figure 2a). Next, background labeling was subtracted from all images then the contrast was enhanced slightly to improve resolution. A threshold value was obtained for each image ensuring that all labeled cells were selected (i.e., target area) (Figure 2b). Each user determined the detection threshold independently. User 1 determined independent threshold levels for each replicate trial. The magnitude of macrophage activation was reported as the proportional area (PA), i.e., ratio of target to scan area (Table 1).
Figure 2.
Micrographs and overlays to illustrate digital image analysis (DIA) and the unbiased sampling technique (UST) used to quantify microglia/macrophages. The lesion area was digitally outlined (blue line delineates zymosan lesion) and the area calculated automatically (A). Within the region of interest, a digital threshold was obtained to identify areas of positive staining (red fill) (B). Proportional area is the ratio of target area/lesion area. For the UST, the lesion area (A) was randomly sampled using MCID’s 3-D fractionator program. After selecting a counting frame size to yield ~5–30 cells/frame, the program randomly generated enough counting frames to sample 25% of the lesion area (C). Cells were counted within the counting frame (enlarged in D) using specific inclusion/exclusion criteria. Note: sample area and counting frames are not to scale and the image that would be quantified would be live on screen and ~4X larger than that shown in (D).
Table 1.
Quantitation of macrophage accumulation within each lesion using PC, UST, and DIA
Intra-user comparisons were derived from 4 replicate data sets obtained by User 1. Inter-user comparisons were derived from the data sets obtained by Users 1–5 using User 1’s first replicate only. Counting was performed using standard profile counting (PC) and an unbiased sampling technique (UST). The outcome of digital image analysis (DIA) was a proportional area measurement.
| Comparison | Lesion | Trial | Lesion area (mm2) | PC (count) | PC (cells/mm2) | UST (estimated count) | UST (estimated cells/mm2) | DIA (proportional area) |
|---|---|---|---|---|---|---|---|---|
| Intra-user | Zymosan | 1 | 0.083 | 877 | 10594 | 848 | 10244 | 0.51 |
| 2 | 0.082 | 768 | 9339 | 796 | 9679 | 0.50 | ||
| 3 | 0.081 | 818 | 10042 | 612 | 7513 | 0.52 | ||
| 4 | 0.084 | 680 | 8063 | 764 | 9059 | 0.50 | ||
| Mean ± SD | 0.083 ± 0.0012 | 786 ± 83 | 9509 ± 1093 | 755 ± 101 | 9124 ± 1178 | 0.51 ± 0.008 | ||
| EAE | 1 | 0.366 | 438 | 1196 | 512 | 1398 | 0.19 | |
| 2 | 0.353 | 410 | 1162 | 460 | 1303 | 0.19 | ||
| 3 | 0.353 | 394 | 1117 | 460 | 1304 | 0.18 | ||
| 4 | 0.353 | 370 | 1048 | 604 | 1711 | 0.19 | ||
| Mean ± SD | 0.356 ± 0.0067 | 403 ± 29 | 1131 ± 64 | 509 ± 68 | 1429 ± 193 | 0.19 ± 0.008 | ||
| Contusion | 1 | 0.317 | 910 | 2870 | 724 | 2283 | 0.48 | |
| 2 | 0.320 | 1211 | 3786 | 820 | 2564 | 0.39 | ||
| 3 | 0.304 | 975 | 3212 | 1012 | 3334 | 0.42 | ||
| 4 | 0.315 | 1364 | 4332 | 1020 | 3239 | 0.46 | ||
| Mean ± SD | 0.314 ± 0.0072 | 1115 ± 210 | 3550 ± 644 | 894 ± 146 | 2855 ± 513 | 0.44 ± 0.038 | ||
| Inter-user | Zymosan | 1 | 0.083 | 877 | 10594 | 848 | 10244 | 0.51 |
| 2 | 0.075 | 912 | 12115 | 1552 | 20616 | 0.56 | ||
| 3 | 0.079 | 669 | 8457 | 772 | 9760 | 0.73 | ||
| 4 | 0.074 | 578 | 7763 | 824 | 11067 | 0.52 | ||
| 5 | 0.081 | 1004 | 12370 | 1532 | 18875 | 0.47 | ||
| Mean ± SD | 0.078 ± 0.0032 | 808 ± 178 | 10260 ± 2091 | 1106 ± 399 | 14113 ± 5200 | 0.56 ± 0.101 | ||
| EAE | 1 | 0.366 | 438 | 1196 | 512 | 1398 | 0.19 | |
| 2 | 0.327 | 593 | 1816 | 616 | 1886 | 0.19 | ||
| 3 | 0.339 | 635 | 1875 | 692 | 2043 | 0.22 | ||
| 4 | 0.361 | 265 | 734 | 532 | 1474 | 0.19 | ||
| 5 | 0.365 | 230 | 630 | 692 | 1895 | 0.18 | ||
| Mean ± SD | 0.348 ± 0.0183 | 432 ± 184 | 1250 ± 584 | 609 ± 85 | 1739 ± 289 | 0.20 ± 0.014 | ||
| Contusion | 1 | 0.317 | 910 | 2870 | 724 | 2283 | 0.48 | |
| 2 | 0.317 | 614 | 1935 | 1584 | 4992 | 0.40 | ||
| 3 | 0.317 | 954 | 3013 | 1404 | 4434 | 0.39 | ||
| 4 | 0.346 | 292 | 844 | 1012 | 2926 | 0.35 | ||
| 5 | 0.335 | 745 | 2226 | 1076 | 3215 | 0.43 | ||
| Mean ± SD | 0.329 ± 0.0142 | 703 ± 267 | 2178 ± 868 | 1160 ± 339 | 3570 ± 1114 | 0.41 ± 0.049 |
To ensure that changes in tissue volume did not skew PA measures, we used Cavalieri’s method to analyze the reference volume of our specimens (Gundersen and Jensen, 1987; Kigerl, McGaughy et al., 2006). Briefly, digital images were obtained from transverse sections spanning the rostro-caudal extent of the spinal cord containing the lesion of interest. Using ImageJ software, a digital point grid of known area was randomly overlaid onto each image and the number of points falling within the tissue area was tallied. Total tissue volume was calculated according to the formula: V = T · a/p · nΣp, where T equals the distance between sections, a/p equals calculated area per point, and nΣp equals the sum of points counted across all sections.
Standard profile counting (PC)
With a 63X oil immersion objective, microglia and macrophages were counted in a raster-like fashion throughout the entire lesion using an ocular objective with a 10×10 grid overlay. All cell profiles with clearly demarcated cell membranes that were within a single plane of focus were counted (Table 1). For the zymosan, EAE, and mouse contusion lesions, User 1 quantified profiles before and after counterstaining to assess differences in accuracy when the counterstain was applied. Users 2–5 then counted profiles in these same sections with the addition of counterstain (i.e., cresyl violet). When counterstain was used, the presence of a nuclear profile was required for a cell to be counted. Only Users 1 and 6 analyzed rat cervical contusion lesions (with and without treatment; see Figure 6). Profile counts are expressed as total number of profiles within a region of interest or as profiles/area (cell density).
Unbiased sampling technique (UST)
The stereology module on the MCID system was used to perform unbiased sampling of each lesion. Briefly, a digital image of each lesion was obtained at low power after which the lesion area was digitally outlined (Figure 2a). A counting frame (digital overlay) was sized to yield 5–30 cells per frame. These sizes were determined through a preliminary study performed by User 1. For the zymosan lesion, the counting frame was ~8300 μm2; for the EAE lesion, ~45,000 μm2; for the mouse contusion lesion, ~30,000 μm2; and for the rat contusion lesions, ~80,000 μm2. For each lesion, 25% of the lesion area was randomly sampled. Based on this size counting frame and the sampling frequency, a variable number of sample areas were randomly generated for each lesion (Figure 2c). With a 63X oil immersion objective, a live image was captured of each sample area and displayed on a color monitor. With the counting frame superimposed on the live image, profiles in each frame were counted using pre-determined inclusion and exclusion criteria (Figure 2d). Specifically, profiles were counted when the top of the nucleus came into focus concurrent with surrounding positive immunostaining. Profiles contained within the frame or touching green lines were counted while those touching red lines were excluded. Once each sample area was quantified, the field of view was automatically moved by the motorized stage to the next sample area. After all sample areas were analyzed, the total number of profiles counted was multiplied by four to obtain an estimate of total profile number (Table 1). This number was then divided by the scan area determined by each user, yielding a profiles/area measurement.
Variability analysis
The DIA technique yields area measurements while the other techniques yield profile counts or measures of cell density. As such, data from the three techniques must be normalized before statistical comparisons can be made. Percent deviations were calculated using the formula:
Percent deviation provides a measure of the degree of precision or variability among a group of values when all values are normalized to the group average. As variability decreases, so does the percent deviation. Unlike standard deviations, the percent deviation is unaffected by the total number of values. Intra-user variability was calculated from the replicate trials conducted by User 1. Inter-user variability was calculated from individual scores from all five users (note: only the first trial from User 1 was used for inter-user comparisons).
Statistics
All tests were performed using GraphPad Prism version 4.03 (GraphPad Software Inc., San Diego, CA). The Pearson correlation was used to determine if the data produced by the various techniques were related. The ability of each technique to detect differences in macrophage activation was compared using one-way analyses of variance (ANOVAs). Average percent deviations between user analysis paradigms and techniques were assessed with two-way ANOVA. The utility of counterstain was evaluated with a two-way ANOVA. The interaction of user by condition for the rat contusion lesions was analyzed by repeated measures 2-way ANOVA. Post-hoc analyses were done using Bonferroni’s test. Significance was set at p ≤ 0.05.
Results
Data obtained via DIA correlates with profile counting techniques
Data produced using each technique correlated with each other for each lesion type (p ≤ 0.0002 for all correlations), suggesting that the outcomes of the three techniques can be described as functions of one another. However, the presence of positive correlations does not imply that the different techniques possess equal sensitivity or consistency. Evaluation of these latter parameters is described below.
Each technique can distinguish between graded levels of macrophage activation
Qualitative microscopic inspection of each lesion predicted that zymosan lesions would have the most macrophages per unit area and EAE lesions the least. Our quantitative analyses confirmed this prediction showing that macrophage density was always greatest in the zymosan lesion (Table 1). Moreover, whether one or multiple users performed an analysis, each technique could reproducibly detect overall differences in the magnitude of the CNS macrophage response (p < 0.0001 via ANOVA; shown for inter-user comparisons only in Figure 3). However, pair-wise comparisons reveal that only DIA was sensitive enough to detect subtle differences in microglia/macrophage activation between lesions (Figure 3). These findings suggest that counting cell profiles (i.e., total cells or cells/mm2) yields data that are more variable than those produced by DIA.
Figure 3.
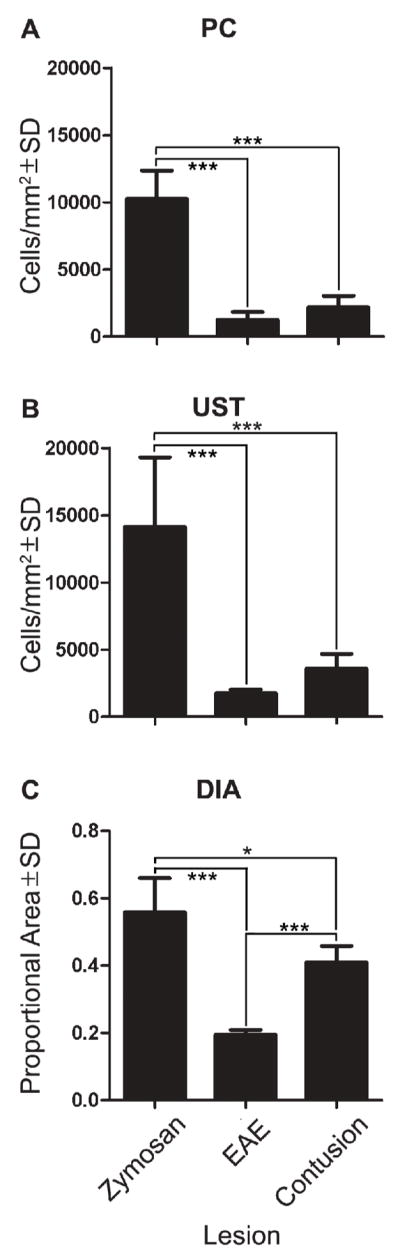
Data from standard profile counting (PC; A), unbiased sampling technique (UST; B) and digital image analysis (DIA; C). For PC and the UST, macrophage accumulation is expressed as cells/mm2; for DIA, accumulation is expressed as a proportional area. All three techniques could differentiate the level of activation in the zymosan lesion from the EAE and contusion lesion. However, only DIA could differentiate between the EAE and contusion lesions.
Cresyl violet counterstaining aids in cell profile identification
Counterstains (e.g., cresyl violet) help reveal individual cell nuclei and should minimize variability and improve sensitivity of profile counting techniques. Unfortunately, a random survey of 20 different primary research articles revealed that when macrophage profiles are counted, counterstains are only used ~30% of the time (citations not shown). To determine if counterstaining reduces variability and improves sensitivity, the results of the PC technique were compared before and after counterstaining for each lesion (Figure 4). Only single user comparisons were made. Interestingly, we found that without counterstain, the total number of CNS macrophages was underestimated in contusion lesions but not in the smaller more defined lesions like those found in zymosan-injected or EAE tissue (Figure 4).
Figure 4.
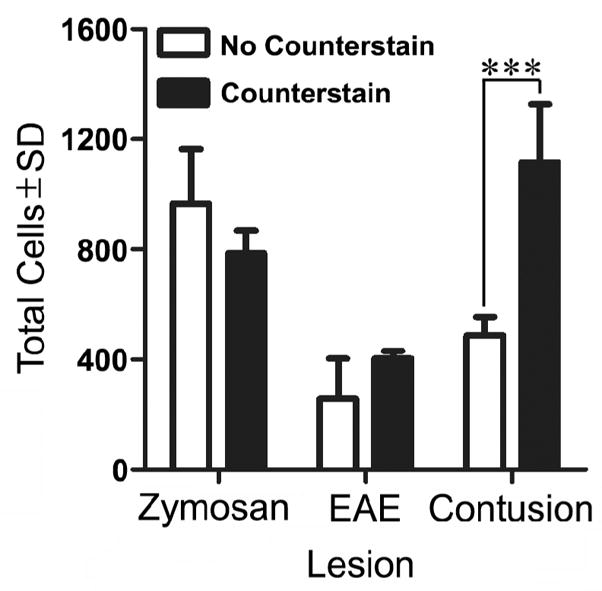
Quantitation of macrophage accumulation within each lesion using standard profile counting (PC) with and without a counterstain. When total cells were analyzed, the presence of counterstain allowed significantly more cells to be detected in the contusion lesion, resulting in a significant increase in the cell count. The presence of counterstain had no effect on the variability of the technique.
Digital image analysis minimizes variability
Differences in the variability of data produced by profile counting (PC and UST) or DIA are influenced by the number of users, the technique being used and the type of lesion being analyzed. To evaluate the influence of single or multiple users on the inherent variability of the three techniques, a 2-way ANOVA was performed with user analysis (intra- or inter-user) and quantitative technique as the two factors (data from Table 1). No significant differences were noted when a single user applied the different techniques to different lesions. This suggests that any technique can be used reliably to analyze a given lesion if a single user completes the analysis. This may be useful for documenting the magnitude of a microglia/macrophage reaction for a single research report; however, this is not practical if comparisons are to be made between different laboratories (e.g., multiple users participating in a multi-center trial).
When multiple users generated data, the variability increased markedly and was influenced by lesion type and technique (Table 2 and Figure 5). Profile counts (PC and UST) were more variable than DIA. DIA consistently produced low, insignificant variability in all lesions and for all users (Figure 5).
Table 2.
Effects of user analysis and technique on variability
Both intra-user and inter-user comparisons were performed. Intra-user comparisons showed less variability than inter-user comparisons. Digital image analysis (DIA) consistently had less variability than both standard profile counting (PC) and the unbiased sampling technique (UST). Variability was significantly different (p < 0.05) for inter-user comparisons for both the zymosan and EAE lesions.
| Lesion | Source of variation | P value |
|---|---|---|
| Zymosan | Interaction | 0.1625 |
| User Analysis | 0.0002* | |
| Technique | 0.0069* | |
| EAE | Interaction | 0.0026* |
| User Analysis | 0.0015* | |
| Technique | 0.0035* | |
| Contusion | Interaction | 0.5534 |
| User Analysis | 0.0739 | |
| Technique | 0.0568 |
Figure 5.
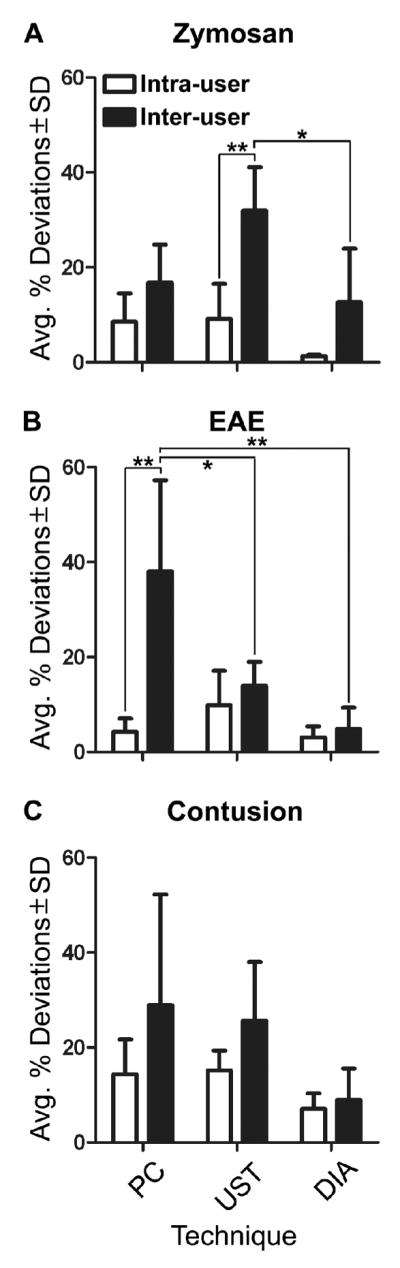
Average percent deviations of profile counting (PC), unbiased sampling technique (UST) and digital image analysis (DIA) data obtained from zymosan (A), EAE (B) and contusion (C) lesions. Comparisons were made between intra-user data (n = 4 replicates) and inter-user data (n = 5 users; only User 1’s first score was used). A) Intra-user and inter-user comparisons show that DIA is consistently the least variable approach with and between users.
DIA rapidly detects treatment effects with minimal inter-user variability
We next tested the sensitivity of DIA, UST and PC for distinguishing a treatment effect. Specifically, these techniques were used to determine if systemic macrophage depletion would reduce the magnitude of the CNS macrophage response elicited by spinal contusion trauma. Using Cavalieri’s method (Gundersen and Jensen, 1987; Kigerl, McGaughy et al., 2006), the volume of injured spinal cord was found to be similar between macrophage-depleted and non-depleted subjects (P=0.91). Thus, any changes in cell density or proportional area of tissue occupied by microglia/macrophages can be attributed to the treatment.
All three techniques detected significant reductions in the CNS macrophage response with equal sensitivity (Figure 6b–d). However, analysis of the UST and PC data revealed a significant user by condition interaction. This indicates that the counting methods cannot be reliably compared between users. Also, quantitation times for DIA were significantly faster (~5–35x) then PC and UST (Figure 6e). Indeed, the time required to implement DIA is independent of lesion severity, whereas the time needed to distinguish and count individual cells for PC and UST increased with increasing severity of pathology and inflammation.
Discussion
The ability to deplete macrophages or modify their effects on surrounding cells is a goal for treating various pathologies including atherosclerosis, rheumatoid arthritis and neurological disease (Blight, 1994; Smith, Trogan et al., 1995; Popovich, Guan et al., 1999; Barrera, Blom et al., 2000). To document the efficacy of these treatments, macrophage responses are often quantified in tissue specimens using microscopy-based techniques. In this report we show that the consistency and sensitivity of these quantitative measures vary as a function of the lesion (type and severity) and the technique that is used. Moreover, the variability introduced by these factors will be further amplified if different investigators fail to apply the quantitative assays in an identical manner. For these reasons, it can be difficult to compare indices of macrophage activation between investigators in the same or different laboratories. These are important considerations given the recent emphasis on manipulating CNS macrophages in pre-clinical and clinical trials across a range of neurological diseases (Popovich and Longbrake, 2008).
In this report, we compared the sensitivity and reproducibility of three common quantitative techniques in spinal cord specimens with distinct types of pathology and graded levels of involvement of resident and recruited macrophages (i.e., microglia and monocyte-derived macrophages). In general, all techniques were able to differentiate between different degrees of macrophage accumulation/density. However, DIA was consistently the most sensitive technique and yielded data with the lowest variability when applied by multiple users. When applied to identical lesions with or without anti-inflammatory therapy (i.e., acute spinal cord trauma with/without macrophage depletion), DIA remained the most sensitive and time-efficient technique. This finding confirms previous data from our laboratory showing that DIA can differentiate between macrophage responses in different experimental groups and between genetically distinct animal strains (Popovich, Guan et al., 1999; Kigerl, McGaughy et al., 2006; Kigerl, Lai et al., 2007). Similar findings by Blackbeard et al. show DIA to be a sensitive technique for revealing changes in microglial activation in a model of spinal nerve transection (Blackbeard, O’Dea et al., 2007). The present data extend validity of this approach by showing that it simplifies and reduces variability when compared with different techniques, different users and across lesions of different etiology and complexity.
The decreased sensitivity and increased variability of data obtained by cell counting is likely explained by increased user error due to fatigue and the inability to consistently identify individual cells in a complex lesion. Indeed, all microscopy-based counting techniques dictate that investigators make a choice about whether a cell profile should be counted. Since CNS macrophages overlap or fuse together forming densely packed layers of cells, it is difficult or impossible to identify individual cell profiles in specimens with marked pathology. For these reasons, neither fractal analyses nor stereological counting techniques can be used reliably. Indeed, to complete fractal analyses, which yield quantitative indices of different morphological features of individual cells (e.g., form factor, convexity, ramification factor, solidity), each cell must be easily distinguished from others in the environment (Soltys, Ziaja et al., 2001; Soltys, Orzylowska-Sliwinska et al., 2005). Similarly, stereological techniques like the optical dissector or fractionator require that users identify individual cell nuclei within a reference volume of tissue and that individual objects have a near uniform distribution within the region of interest (i.e., no clumping of cells in clusters with varying cell densities) (West, Slomianka et al., 1991).
Although the ability to resolve single profiles can be improved using counterstains, our current data indicate this is only true in a subset of lesions (see Figure 4). Regardless, it is still tedious and time consuming to quantify thousands of macrophages (Jander, Sitzer et al., 1998; Kigerl, McGaughy et al., 2006). In our hands, it took ~5–35 times longer to count cells with PC than to complete DIA (see Figure 6e). Even when PC quantification time was reduced by sub-sampling using UST method, DIA remained ~7–21 times faster (see Figure 6e). Still, when multiple factors are needed to define a given macrophage population, it may be necessary to count cell profiles. For example, if the goal is to double label tissue sections then discriminate between microglia/macrophages with unique membrane and/or cytoplasmic proteins, one may need to tally the number of cell profiles that define each phenotype. As our present data show, these data are prone to user error and may under- or overestimate the actual contribution of CNS macrophages in a pathological context. This makes comparisons difficult between individual users or laboratories.
In most cases, meaningful quantitative comparisons can be made without the tedium and variability inherent in counting cell profiles. Indeed, DIA can detect the magnitude of the microglia/macrophage response regardless of whether it results from enhanced cell infiltration, local hyperplasia or cell hypertrophy (Popovich, Wei et al., 1997). Digital thresholding also compensates for sub-optimal staining or variable staining between sections. Thus, two different sections containing equivalent microglia/macrophage reactions with unequal staining intensities will yield identical results. This helps overcome the problems with object identification that are inherent with standard counting techniques (see above). Despite the considerable strengths of DIA, tissue processing artifacts (e.g., shrinking, swelling) can influence any type of histological analysis. These confounding variables may increase or decrease brain or spinal cord volume, thereby altering the density or proportional area of tissue occupied by microglia/macrophages. Ensuring that the volume of tissue being analyzed does not vary between groups can minimize the impact of these variables. This is easily accomplished using Cavalieri’s method which integrates area measurements from systematically random sampled reference sections throughout a 3-dimensional structure (Contreras, Ragan et al., 1991; Mayhew and Olsen, 1991; Costa, Mandarim-de-Lacerda et al., 1991). Should tissue volume vary between experimental groups, then microglia/macrophage cell counts or area measurements can be normalized to their respective reference volumes. For all of these reasons, DIA offers significant advantages over other techniques for quantifying microglia/macrophages in various forms of experimental or clinical neuropathology.
Acknowledgments
This work was supported by the NIH (NINDS NS37846) and the Paralysis Project of America.
The authors thank Dr. Kurt Lucin, David Schonberg, and Amy Tovar for their involvement in the quantitation study. We thank Dr. Dana McTigue for reviewing the manuscript. This work was supported by the NIH (NINDS NS37846) and the Paralysis Project of America.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Ankeny DP, Popovich PG. Central nervous system and non-central nervous system antigen vaccines exacerbate neuropathology caused by nerve injury. Eur J Neurosci. 2007;25:2053–64. doi: 10.1111/j.1460-9568.2007.05458.x. [DOI] [PubMed] [Google Scholar]
- Barrera P, Blom A, van Lent PL, van Bloois L, Beijnen JH, van Rooijen N, Waal Malefijt MC, van de Putte LB, Storm G, van den Berg WB. Synovial macrophage depletion with clodronate-containing liposomes in rheumatoid arthritis. Arthritis Rheum. 2000;43:1951–9. doi: 10.1002/1529-0131(200009)43:9<1951::AID-ANR5>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Beggs S, Salter MW. Stereological and somatotopic analysis of the spinal microglial response to peripheral nerve injury. Brain Behav Immun. 2007;21:624–33. doi: 10.1016/j.bbi.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackbeard J, O’Dea KP, Wallace VC, Segerdahl A, Pheby T, Takata M, Field MJ, Rice AS. Quantification of the rat spinal microglial response to peripheral nerve injury as revealed by immunohistochemical image analysis and flow cytometry. J Neurosci Methods. 2007;164:207–17. doi: 10.1016/j.jneumeth.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight AR. Effects of silica on the outcome from experimental spinal cord injury: implication of macrophages in secondary tissue damage. Neuroscience. 1994;60:263–73. doi: 10.1016/0306-4522(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Carson MJ. Microglia as liaisons between the immune and central nervous systems: functional implications for multiple sclerosis. GLIA. 2002;40:218–31. doi: 10.1002/glia.10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colburn RW, DeLeo JA, Rickman AJ, Yeager MP, Kwon P, Hickey WF. Dissociation of microglial activation and neuropathic pain behaviors following peripheral nerve injury in the rat. J Neuroimmunol. 1997;79:163–75. doi: 10.1016/s0165-5728(97)00119-7. [DOI] [PubMed] [Google Scholar]
- Contreras PC, Ragan DM, Bremer ME, Lanthorn TH, Gray NM, Iyengar S, Jacobson AE, Rice KC, De Costa BR. Evaluation of U-50,488H analogs for neuroprotective activity in the gerbil. Brain Res. 1991;546:79–82. doi: 10.1016/0006-8993(91)91161-s. [DOI] [PubMed] [Google Scholar]
- Costa WS, Mandarim-de-Lacerda CA, Bauer JA. Stereological analysis of the otic ganglia in adult rat: light microscopic study. Anat Anz. 1991;172:203–7. [PubMed] [Google Scholar]
- Deininger MH, Schluesener HJ. Cyclooxygenases-1 and -2 are differentially localized to microglia and endothelium in rat EAE and glioma. J Neuroimmunol. 1999;95:202–8. doi: 10.1016/s0165-5728(98)00257-4. [DOI] [PubMed] [Google Scholar]
- Donnelly DJ, Popovich PG. Inflammation and its role in neuroprotection, axonal regeneration and functional recovery after spinal cord injury. Exp Neurol. 2008;209:378–88. doi: 10.1016/j.expneurol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felts PA, Woolston AM, Fernando HB, Asquith S, Gregson NA, Mizzi OJ, Smith KJ. Inflammation and primary demyelination induced by the intraspinal injection of lipopolysaccharide. Brain. 2005;128:1649–66. doi: 10.1093/brain/awh516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness PN, Rogers-Wheatley L, Harris KP. Semiautomatic quantitation of macrophages in human renal biopsy specimens in proteinuric states. J Clin Pathol. 1997;50:118–22. doi: 10.1136/jcp.50.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensel JC, Tovar CA, Hamers FP, Deibert RJ, Beattie MS, Bresnahan JC. Behavioral and histological characterization of unilateral cervical spinal cord contusion injury in rats. J Neurotrauma. 2006;23:36–54. doi: 10.1089/neu.2006.23.36. [DOI] [PubMed] [Google Scholar]
- Gundersen HJ, Jensen EB. The efficiency of systematic sampling in stereology and its prediction. J Microsc. 1987;147 (Pt 3):229–63. doi: 10.1111/j.1365-2818.1987.tb02837.x. [DOI] [PubMed] [Google Scholar]
- Imai Y, Ibata I, Ito D, Ohsawa K, Kohsaka S. A novel gene iba1 in the major histocompatibility complex class III region encoding an EF hand protein expressed in a monocytic lineage. Biochem Biophys Res Commun. 1996;224:855–62. doi: 10.1006/bbrc.1996.1112. [DOI] [PubMed] [Google Scholar]
- Ito D, Tanaka K, Suzuki S, Dembo T, Fukuuchi Y. Enhanced expression of Iba1, ionized calcium-binding adapter molecule 1, after transient focal cerebral ischemia in rat brain. Stroke. 2001;32:1208–15. doi: 10.1161/01.str.32.5.1208. [DOI] [PubMed] [Google Scholar]
- Jakeman LB, Guan Z, Wei P, Ponnappan R, Dzwonczyk R, Popovich PG, Stokes BT. Traumatic spinal cord injury produced by controlled contusion in mouse. J Neurotrauma. 2000;17:299–319. doi: 10.1089/neu.2000.17.299. [DOI] [PubMed] [Google Scholar]
- Jander S, Sitzer M, Schumann R, Schroeter M, Siebler M, Steinmetz H, Stoll G. Inflammation in high-grade carotid stenosis: a possible role for macrophages and T cells in plaque destabilization. Stroke. 1998;29:1625–30. doi: 10.1161/01.str.29.8.1625. [DOI] [PubMed] [Google Scholar]
- Jones TB, Basso DM, Sodhi A, Pan JZ, Hart RP, MacCallum RC, Lee S, Whitacre CC, Popovich PG. Pathological CNS autoimmune disease triggered by traumatic spinal cord injury: implications for autoimmune vaccine therapy. J Neurosci. 2002;22:2690–700. doi: 10.1523/JNEUROSCI.22-07-02690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigerl KA, Lai W, Rivest S, Hart RP, Satoskar AR, Popovich PG. Toll-like receptor (TLR)-2 and TLR-4 regulate inflammation, gliosis, and myelin sparing after spinal cord injury. J Neurochem. 2007;102:37–50. doi: 10.1111/j.1471-4159.2007.04524.x. [DOI] [PubMed] [Google Scholar]
- Kigerl KA, McGaughy VM, Popovich PG. Comparative analysis of lesion development and intraspinal inflammation in four strains of mice following spinal contusion injury. J Comp Neurol. 2006;494:578–94. doi: 10.1002/cne.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraan MC, Haringman JJ, Ahern MJ, Breedveld FC, Smith MD, Tak PP. Quantification of the cell infiltrate in synovial tissue by digital image analysis. Rheumatology (Oxford) 2000;39:43–9. doi: 10.1093/rheumatology/39.1.43. [DOI] [PubMed] [Google Scholar]
- Mayhew TM, Olsen DR. Magnetic resonance imaging (MRI) and model-free estimates of brain volume determined using the Cavalieri principle. J Anat. 1991;178:133–44. [PMC free article] [PubMed] [Google Scholar]
- McCann MJ, O’Callaghan JP, Martin PM, Bertram T, Streit WJ. Differential activation of microglia and astrocytes following trimethyl tin-induced neurodegeneration. Neuroscience. 1996;72:273–81. doi: 10.1016/0306-4522(95)00526-9. [DOI] [PubMed] [Google Scholar]
- McPherson CA, Kubik J, Wine RN, D’Hellencourt CL, Harry GJ. Alterations in cyclin A, B, and D1 in mouse dentate gyrus following TMT-induced hippocampal damage. Neurotox Res. 2003;5:339–54. doi: 10.1007/BF03033154. [DOI] [PubMed] [Google Scholar]
- Olson EE, McKeon RJ. Characterization of cellular and neurological damage following unilateral hypoxia/ischemia. J Neurol Sci. 2004;227:7–19. doi: 10.1016/j.jns.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Papenfuss TL, Rogers CJ, Gienapp I, Yurrita M, McClain M, Damico N, Valo J, Song F, Whitacre CC. Sex differences in experimental autoimmune encephalomyelitis in multiple murine strains. J Neuroimmunol. 2004;150:59–69. doi: 10.1016/j.jneuroim.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Popovich PG, Guan Z, McGaughy V, Fisher L, Hickey WF, Basso DM. The neuropathological and behavioral consequences of intraspinal microglial/macrophage activation. J Neuropathol Exp Neurol. 2002;61:623–33. doi: 10.1093/jnen/61.7.623. [DOI] [PubMed] [Google Scholar]
- Popovich PG, Guan Z, Wei P, Huitinga I, van Rooijen N, Stokes BT. Depletion of hematogenous macrophages promotes partial hindlimb recovery and neuroanatomical repair after experimental spinal cord injury. Exp Neurol. 1999;158:351–65. doi: 10.1006/exnr.1999.7118. [DOI] [PubMed] [Google Scholar]
- Popovich PG, Hickey WF. Bone marrow chimeric rats reveal the unique distribution of resident and recruited macrophages in the contused rat spinal cord. J Neuropathol Exp Neurol. 2001;60:676–85. doi: 10.1093/jnen/60.7.676. [DOI] [PubMed] [Google Scholar]
- Popovich PG, Longbrake EE. Can the immune system be harnessed to repair the CNS? Nat. Rev Neurosci. 2008;9:481–93. doi: 10.1038/nrn2398. [DOI] [PubMed] [Google Scholar]
- Popovich PG, Wei P, Stokes BT. The cellular inflammatory response after spinal cord injury in Sprague-Dawley and Lewis rats. J Comp Neurol. 1997;377:443–64. doi: 10.1002/(sici)1096-9861(19970120)377:3<443::aid-cne10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Popovich PG, Yu JY, Whitacre CC. Spinal cord neuropathology in rat experimental autoimmune encephalomyelitis: modulation by oral administration of myelin basic protein. J Neuropathol Exp Neurol. 1997;56:1323–38. doi: 10.1097/00005072-199712000-00007. [DOI] [PubMed] [Google Scholar]
- Robinson AP, White TM, Mason DW. Macrophage heterogeneity in the rat as delineated by two monoclonal antibodies MRC OX-41 and MRC OX-42, the latter recognizing complement receptor type 3. Immunology. 1986;57:239–47. [PMC free article] [PubMed] [Google Scholar]
- Roggendorf W, Strupp S, Paulus W. Distribution and characterization of microglia/macrophages in human brain tumors. Acta Neuropathol (Berlin) 1996;92:288–93. doi: 10.1007/s004010050520. [DOI] [PubMed] [Google Scholar]
- Smith JD, Trogan E, Ginsberg M, Grigaux C, Tian J, Miyata M. Decreased atherosclerosis in mice deficient in both macrophage colony-stimulating factor (op) and apolipoprotein E. Proc Natl Acad Sci U S A. 1995;92:8264–8. doi: 10.1073/pnas.92.18.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltys Z, Orzylowska-Sliwinska O, Zaremba M, Orlowski D, Piechota M, Fiedorowicz A, Janeczko K, Oderfeld-Nowak B. Quantitative morphological study of microglial cells in the ischemic rat brain using principal component analysis. J Neurosci Methods. 2005;146:50–60. doi: 10.1016/j.jneumeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Soltys Z, Ziaja M, Pawlinski R, Setkowicz Z, Janeczko K. Morphology of reactive microglia in the injured cerebral cortex. Fractal analysis and complementary quantitative methods. J Neurosci Res. 2001;63:90–7. doi: 10.1002/1097-4547(20010101)63:1<90::AID-JNR11>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Springer T, Galfre G, Secher DS, Milstein C. Mac-1: a macrophage differentiation antigen identified by monoclonal antibody. Eur J Immunol. 1979;9:301–6. doi: 10.1002/eji.1830090410. [DOI] [PubMed] [Google Scholar]
- Sroga JM, Jones TB, Kigerl KA, McGaughy VM, Popovich PG. Rats and mice exhibit distinct inflammatory reactions after spinal cord injury. J Comp Neurol. 2003;462:223–40. doi: 10.1002/cne.10736. [DOI] [PubMed] [Google Scholar]
- Stoll G, Bendszus M. Inflammation and atherosclerosis: novel insights into plaque formation and destabilization. Stroke. 2006;37:1923–32. doi: 10.1161/01.STR.0000226901.34927.10. [DOI] [PubMed] [Google Scholar]
- Szekanecz Z, Koch AE. Macrophages and their products in rheumatoid arthritis. Curr Opin Rheumatol. 2007;19:289–95. doi: 10.1097/BOR.0b013e32805e87ae. [DOI] [PubMed] [Google Scholar]
- Tak PP, Thurkow EW, Daha MR, Kluin PM, Smeets TJ, Meinders AE, Breedveld FC. Expression of adhesion molecules in early rheumatoid synovial tissue. Clin Immunol Immunopathol. 1995;77:236–42. doi: 10.1006/clin.1995.1149. [DOI] [PubMed] [Google Scholar]
- Tsutsui S, Noorbakhsh F, Sullivan A, Henderson AJ, Warren K, Toney-Earley K, Waltz SE, Power C. RON-regulated innate immunity is protective in an animal model of multiple sclerosis. Ann Neurol. 2005;57:883–95. doi: 10.1002/ana.20502. [DOI] [PubMed] [Google Scholar]
- van Rooijen N, Sanders A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J Immunol Meth. 1994;174:83–93. doi: 10.1016/0022-1759(94)90012-4. [DOI] [PubMed] [Google Scholar]
- West MJ, Slomianka L, Gundersen HJ. Unbiased stereological estimation of the total number of neurons in thesubdivisions of the rat hippocampus using the optical fractionator. Anat Rec. 1991;231:482–97. doi: 10.1002/ar.1092310411. [DOI] [PubMed] [Google Scholar]
- Yasuda Y, Shinagawa R, Yamada M, Mori T, Tateishi N, Fujita S. Long-lasting reactive changes observed in microglia in the striatal and substantia nigral of mice after 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Brain Res. 2007;1138:196–202. doi: 10.1016/j.brainres.2006.12.054. [DOI] [PubMed] [Google Scholar]
- Youssef PP, Smeets TJ, Bresnihan B, Cunnane G, Fitzgerald O, Breedveld F, Tak PP. Microscopic measurement of cellular infiltration in the rheumatoid arthritis synovial membrane: a comparison of semiquantitative and quantitative analysis. Br J Rheumatol. 1998;37:1003–7. doi: 10.1093/rheumatology/37.9.1003. [DOI] [PubMed] [Google Scholar]