Abstract
Aims
Although the frequencies of pharmacogenetic variants differ among racial groups, most pharmacogenetic algorithms for genotype-guided warfarin dosing only include two CYP2C9 alleles (*2 and *3) and a single VKORC1 allele (g.-1639G>A or g.1173C>T) commonly found among Caucasians. Therefore, this study sought to identify other CYP2C9 and VKORC1 alleles important in warfarin dose variability and to determine their frequencies in different racial and ethnic groups.
Materials & methods
The CYP2C9 and VKORC1 genes were sequenced in selected sensitive (<21 mg/week) and resistant (>49 mg/week) individuals with discrepant therapeutic and algorithm-predicted warfarin doses based on prior CYP2C9 and VKORC1 genotyping. The CYP2C9 and VKORC1 allele frequencies were determined in healthy, racially self-identified blood donors.
Results
Sequencing identified an African–American male with a lower than predicted therapeutic warfarin dose (14.4 mg/week), previously genotyped as CYP2C9*1/*1, who was homozygous for CYP2C9*8 (c.449G>A; p.R150H). Genotyping 600 African–American alleles identified CYP2C9*8 as their most frequent variant CYP2C9 allele (0.047), and the combined allele frequency of CYP2C9*2, *3, *5, *6, *8 and *11 was 0.133. Given most warfarin pharmacogenetic dosing algorithms only include CYP2C9*2 and *3, the inclusion of CYP2C9*8 alone could reclassify the predicted metabolic phenotypes of almost 10% of African–Americans, or when combined with CYP2C9*5, *6 and *11, more than 15%. In addition, the African–American VKORC1 g.-1639A allele frequency was 0.108 and three g.1331G>A (p.V66M) carriers were identified.
Conclusions
CYP2C9*8 is prevalent among African–Americans (~1 in 11 individuals). Thus, in this racial group, the incorporation of CYP2C9*8 into genotyping panels may improve dose prediction of CYP2C9-metabolized drugs, including warfarin.
Keywords: African-American, allele frequencies, CYP2C9*8, pharmacogenetics, VKORC1, warfarin
The highly polymorphic cytochrome P450 2C9 (CYP2C9) gene has over 50 known variant alleles, many of which result in decreased enzyme activity [1,101]. CYP2C9 encodes a microsomal monooxygenase involved in the oxidative metabolism of many endogenous and xenobiotic compounds including anticoagulant, anticonvulsant, nonsteroidal anti-inflammatory and antihypertensive drugs. Prominent among these is warfarin, a commonly used anticoagulant that is metabolized, in part, by CYP2C9, and variant CYP2C9 alleles are associated with reduced therapeutic warfarin doses [2–6]. In addition, common polymorphisms and rare sequence variants in the vitamin K epoxide reductase complex 1 (VKORC1) gene alter warfarin pharmacodynamics and also affect warfarin dose requirements [3–9]. Consequently, genetic variation in CYP2C9 and VKORC1 combined with clinical and environmental factors (e.g., age, gender, body weight, concomitant medications and diet), account for approximately half of interindividual warfarin dose variation [10–13].
The identification of variant CYP2C9 and VKORC1 alleles led to their inclusion in warfarin-dosing algorithms (for review, see [14]). However, a current limitation of pharmacogenetic-predicted warfarin dosing is its application to multiracial and multi-ethnic patient populations. In part, this is owing to differences in CYP2C9 and VKORC1 allele frequencies between racial groups as CYP2C9*2 (c.430C>T; p.R114C) and *3 (c.1075A>C; p.I359L) are common in Caucasians, but less frequent in African–Americans and Asians [15]. Since these variant alleles are used in the majority of pharmacogenetic-based dosing algorithms, their lower frequencies in African–Americans may account for why recent algorithms derived from multi-ethnic cohorts better explain dose variation in Caucasians than in African–Americans [16,17]. For example, the multi-ethnic algorithm recently reported by Gage et al. accounted for 57% of the dose variability in Caucasians, but only 31% in African–Americans [16].
To determine if other CYP2C9 and VKORC1 alleles are important in warfarin dose variability in African–Americans and other non-Caucasian populations, the CYP2C9 and VKORC1 genes were sequenced in selected patients from a multi-ethnic cohort with sensitive (<21 mg/week) and resistant (>49 mg/week) warfarin doses. An African–American male was identified who required a very low warfarin dose (14.4 mg/week) to achieve stable anticoagulation intensity (International Normalized Ratio [INR] 2–3) and who was homozygous for the CYP2C9*8 variant allele. Consequently, the frequencies of variant CYP2C9 (*2, *3, *4, *5, *6, *8, *11 and *13) and VKORC1 (g.-1639G>A, g.1331G>A) alleles were determined in a cohort of 300 healthy African–American individuals. Of note, the CYP2C9*8 allele had the highest allele frequency (0.047) among African–Americans and the combined allele frequency of CYP2C9*5, *6, *8 and *11 was 0.085. These studies underscore the importance of incorporating at least CYP2C9*8 into pharmacogenetic-based dosing algorithms for CYP2C9-metabolized drugs, including warfarin.
Materials & methods
Human subjects
Since July 2007, adult (>18 years) blood samples have been collected with Institutional Review Board approval and informed consent from patients with arrhythmias or venous thrombosis treated with warfarin in a large anticoagulation program at an urban academic medical center. The anticoagulation clinic serves a population of over 500 patients, of which approximately 55% are Caucasian (~30% Ashkenazi Jewish) 25% Hispanic, 15% are African–American and 5% are Asian. A total of 114 patients were enrolled in this study and 65 were either sensitive (<21 mg/week) or resistant (>49 mg/week) with a target INR of 2–3. Of these, six patients had discrepant therapeutic and algorithm-predicted warfarin doses based on prior CYP2C9 and VKORC1 genotyping; therefore, their CYP2C9 and/or VKORC1 exonic regions were sequenced. To determine the frequency of CYP2C9 and VKORC1 alleles, blood samples from healthy donors who indicated their racial background and gave informed consent for the use of their DNA for research were obtained from the New York Blood Center with Institutional Review Board approval.
Genotyping
The CYP2C9 allele designations refer to those defined by the Cytochrome P450 Allele Nomenclature Committee [1,101]. All patients enrolled in the study were initially genotyped for eight CYP2C9 alleles (*1, *2, *3, *4, *5, *6, *11 and *13) and 12 VKORC1 variants (g.-4931T>C, g.-1639G>A, g.1173C>T, g.2255C>T, g.3730G>A, g.85G>T [p.V29L], g.106G>T [p.D36Y], g.121G>T [p.A41S], g.134T>C [p.V45A], g.172A>G [p.R58G], g.1331G>A [p.V66M] and g.3487T>G [p.L128R]). The coagulation factor VII (F7) and apolipoprotein E (APOE) alleles were also genotyped in selected patients by direct sequencing, and additional CYP2C9 and VKORC1 genotyping was performed to determine allele and genotype frequencies.
Genotyping of six CYP2C9 alleles (*1, *2, *3, *4, *5 and *6), and seven VKORC1 nucleotide variants (g.-1639G>A, g.85G>T [p.V29L], g.121G>T [p.A41S], g.134T>C [p.V45A], g.172A>G [p.R58G], g.1331G>A [p.V66M] and g.3487T>G [p.L128R]) was performed using the Tag-It™ Mutation Detection Kit (Luminex Molecular Diagnostics, ON, Canada) as previously described [18]. The CYP2C9 and VKORC1 genotypes were determined using Tag-It Data Analysis Software (Luminex Molecular Diagnostics) and the wild-type CYP2C9*1 allele was assigned in the absence of other detectable variant alleles. Genotyping of the VKORC1 g.106G>T (p.D36Y), g.698C>T (6009C>T, Accession Number AY587020) and g.3730G>A (9041G>A, Accession Number AY587020) alleles was performed as previously described [18].
The CYP2C9*8, *11 and *13 alleles were amplified by PCR and interrogated using RFLP assays. CYP2C9*8 and *13 were amplified using CYP2C9 exon 2/3 primers and *11 was amplified using exon 7 primers (Table 1). Reactions were performed in 25 μl containing approximately 100 ng of DNA, 1X PCR buffer (Invitrogen, CA, USA), 2.5 mM MgCl2, 0.2 mM of each dNTP, 0.4 mM of each primer and 1.0 unit of Platinum® Taq DNA Polymerase (Invitrogen). Amplification consisted of an initial denaturation step at 94°C for 5 min followed by 35 amplification cycles (94°C for 30 s, 58°C for 30 s, and 72°C for 45 s) and a final incubation at 72°C for 5 min. Genotyping of CYP2C9*8, *11 and *13 was performed using AciI, BstNI and MspI (New England BioLabs, MA, USA), respectively. The VKORC1 g.-4931T>C, g.1173C>T, and g.2255C>T alleles were genotyped by PCR-RFLP using the primers, annealing temperatures and MgCl2 concentrations listed in Table 1 and AciI, MlyI and NcoI (New England BioLabs), respectively. Digested PCR products were visualized by agarose gel electrophoresis and positive control samples were confirmed by sequencing.
Table 1.
Primer sequences for PCR amplification.
| Primer | Sequence (5′–3′) | Ta (°C) | MgCl2 (mM) | Product (nt) |
|---|---|---|---|---|
| CYP2C9 | ||||
| Exon 1-FWD* | GCCTTCAGGAATTTTTTTTA | 53 | 2.5 | 929 |
| Exon 1-REV* | TTTTACTTTACCATTACCTCTTG | |||
| Exon 2/3-FWD‡ | TACAAATACAATGAAAATATCATG | 58 | 2.5 | 690 |
| Exon 2/3-REV‡ | CTAACAACCAGGACTCATAATG | |||
| Exon 4-FWD | TGTTAAGGGAATTTGTAGGTAAGA | 58 | 2.5 | 366 |
| Exon 4-REV | TGCACTTCAGAGCTTGATCC | |||
| Exon 5-FWD§ | CAGAGCTTGGTATATGGTATG | 58 | 2.5 | 321 |
| Exon 5-REV§ | GTAAACACAGAACTAGTCAAC | |||
| Exon 6-FWD* | GTTTGGGCAAGTTGGTCTA | 58 | 2.5 | 395 |
| Exon 6-REV* | AGAAACAGGAAGGAGGACAC | |||
| Exon 7-FWD | CTGAATTGCTACAACAAATGTG | 58 | 2.5 | 315 |
| Exon 7-REV | GATACTATGAATTTGGGGACTTC | |||
| Exon 8-FWD | ACCACTGTTTCTTCAACCTTCATGG | 58 | 2.5 | 370 |
| Exon 8-REV | TTAGGATGTATCATGAGCAGGGTG | |||
| Exon 9-FWD* | TATTGCATATTCTGTTTGTGC | 58 | 2.5 | 803 |
| Exon 9-REV* | CAAGTAACTCTAACACTCACCC | |||
| VKORC1 | ||||
| g.-4931T>C-FWD | TCCAGCTTTAGCCACCATCT | 60 | 2.5 | 560 |
| g.-4931T>C-REV | TTTTGACCATATTTTCTTCTAGCATTT | |||
| g.1173C>T-FWD | AGGGTCAGTGACATGGAATCCTGA | 60 | 1.5 | 178 |
| g.1173C>T-REV | GGGTGGAACCAGGTTAGGACTGT | |||
| g.2255C>T-FWD | ATGCCATATGCCGGAGATGAGACT | 60 | 1.5 | 222 |
| g.2255C>T-REV | ACTGGTCTCTGAAGCTCTTGCCAT | |||
| Exon 1-FWD | AAGTGCTGGGATTACAAGCGTGAG | 60 | 1.5 | 720 |
| Exon 1-REV | GTCCCTTGCCTCGCACTCTTATTT | |||
| Exon 2-FWD | AGGGTCAGTGACATGGAATCCTGA | 60 | 1.5 | 535 |
| Exon 2-REV | GGGACCTAGGATGTCTTTAAGGG | |||
| Exon 3-FWD | TTTAGAGACCCTTCCCAGCAGCTC | 60 | 1.5 | 700 |
| Exon 3-REV | CACTCTCCCTCTGACTCACCCTT | |||
| F7 | ||||
| g.-401G>T-FWD¶ | TAAGAAACCAGCCTCCCTTG | 58 | 1.5 | 229 |
| g.-401G>T-REV¶ | CGTGCAGGTGTTAAGGTGTG | |||
| APOE | ||||
| APOE-FWD | TGAAGGCCTACAAATCGGAACTGG | 62 | 1.5 | 459 |
| APOE-REV | GGCTGCCCATCTCCTCCAT | |||
The F7 g.-401G>T allele was PCR-amplified as described above using primers, annealing temperatures and MgCl2 concentrations listed in Table 1. The common APOE alleles (ε2, ε3 and ε4) were PCR-amplified in 25 μl containing approximately 100 ng of DNA, 1X GC-RICH Reaction buffer (includes 1.5 mM MgCl2), 0.2 mM of each dNTP, 0.4 mM of each primer (Table 1), 0.5 M GC-RICH Resolution Solution, and 0.4 units of GC-RICH Enzyme mix (Roche Applied Science, Basel, Switzerland). Amplification of APOE consisted of an initial denaturation step at 95°C for 5 min followed by 40 amplification cycles (95°C for 30 s, 62°C for 30 s and 72°C for 45 s) and a final incubation at 72°C for 5 min. Individual F7 and APOE PCR products were purified using QIAquick columns (Qiagen, Hilden, Germany) and bidirectionally sequenced using amplification primers.
CYP2C9 & VKORC1 exonic sequencing
Primers for CYP2C9 sequencing were aligned against other CYP2C family members to ensure specific amplification of the CYP2C9 gene and are listed with the VKORC1 exon primers in Table 1. Amplification reactions were performed as described above using the indicated MgCl2 concentrations and annealing temperatures (Table 1). Individual PCR products were purified using QIAquick columns and bidirectionally sequenced using appropriate amplification primers.
Results
Analysis of CYP2C9 & VKORC1
Of the 65 patients who were designated sensitive (<21 mg/week) or resistant (>49 mg/week), six were identified whose therapeutic dose was discrepant from their algorithm-predicted dose based on the alleles tested below. Of these, one patient was identified with a nonsynonymous variant following CYP2C9 and VKORC1 exonic sequencing. This patient was a 61.1 year old HIV-positive African–American male (height: 188 cm; weight: 86.2 kg) with a non-ischemic cardiomyopathy (New York Heart Association class III heart failure) and stage three chronic kidney disease who was treated with anticoagulation since 2002 for paroxysmal atrial fibrillation with a target INR of 2–3 and a mean therapeutic warfarin dose of 14.4 mg/week. Other medications included aspirin, amiodarone, 200 mg daily, carvedilol, 6.25 mg twice daily, lisinopril, 5 mg daily, spironolactone, 25 mg daily, furosemide, 120 mg twice daily, lamivudine, 150 mg twice daily, zidovudine, 300 mg twice daily, potassium chloride and docusate. The patient had not developed bleeding complications during anticoagulation, nor had he required a blood transfusion. His blood chemistry values including liver function tests were normal and hepatitis serologies revealed no evidence of infection.
Patients enrolled in the study were genotyped for eight CYP2C9 and 12 VKORC1 variant alleles (see Materials & methods) and the index patient described above had the following genotypes used for algorithm-based dose prediction: CYP2C9*1/*1; VKORC1 g.-4931T/C, g.-1639G/G, g.1173C/C, g.2255C/T, g.3730G/G; F7 g.-401G/G; APOE ε2/ε3. None of the seven tested warfarin resistant VKORC1 mutations were identified in this patient. The predicted doses from the 15 tested algorithms ranged from 24.9 to 52.2 mg/week and are summarized in Table 2. Based on the clinical features and identified genotypes, all algorithms predicted a significantly higher maintenance warfarin dose (24.9 to 52.2 mg/week) than this patient actually required, even when adjusted for renal clearance [19]. In addition, although amiodarone inhibits CYP2C9 and decreases warfarin requirements, the eight algorithms [6,16,17,20–22], which included an adjustment for amiodarone or concomitant drug interaction predicted doses ranging from 24.9 to 50.1 mg/week, all greater than the observed 14.4 mg/week therapeutic dose.
Table 2.
Mean therapeutic and algorithm-predicted warfarin doses in the index patient.
| Warfarin dose | mg/week | Ref. |
|---|---|---|
| Mean therapeutic dose | 14.4 | – |
| Algorithm-predicted dose‡ | ||
| Sconce et al. (2005) | 47.6 | [41] |
| Wadelius et al. (2005) | 49.7 | [21] |
| Herman et al. (2006) | 42.5 | [51] |
| Takahashi et al. (2006) | 50.3 | [5] |
| Tham et al. (2006) | 32.9 | [52] |
| Anderson et al. (2007) | 45.8 | [53] |
| Miao et al. (2007) | 49.1 | [54] |
| Zhu et al. (2007) | 52.2 | [40] |
| Gage et al. (2008) | 34.6 | [16] |
| Schelleman et al. (2008) (C) | 41.9 | [17] |
| Schelleman et al. (2008) (AA) | 28.4 | [17] |
| Schelleman et al. (2008) (C + AA) | 41.5 | [17] |
| Wu et al. (2008) | 24.9 | [20] |
| Klein et al. (2009) | 35.4 | [6] |
| Wadelius et al. (2009) | 50.1 | [22] |
Based on the following genotypes: CYP2C9: *1/*1; VKORC1 (Accession Number AY587020) g.-4931T>C (381T>C): T/C, g.-1639G>A (3673G>A): G/G, g.1173C>T (6484C>T): C/C, g.2255C>T (7566C>T): C/T, g.3730G>A (9041G>A): G/G; F7 g.-401G>T: G/G; APOE: ε2/ε3.
AA: African–American; C: Caucasian.
Sequencing identified CYP2C9*8
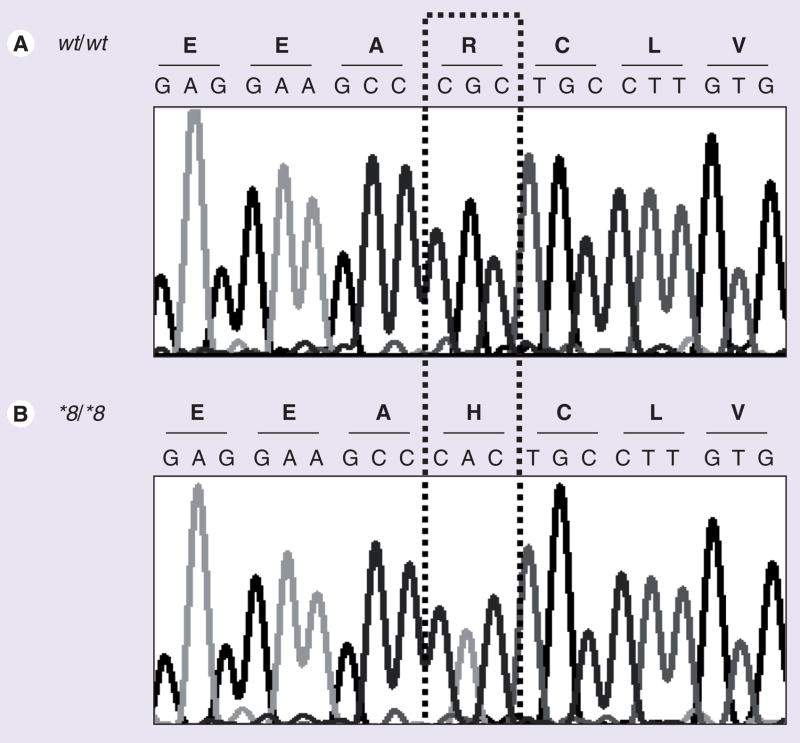
The sensitivity to warfarin and the discrepancy between the therapeutic (14.4 mg/week) and algorithm-predicted (24.9 to 52.2 mg/week) warfarin doses in this patient prompted CYP2C9 and VKORC1 sequencing. In the CYP2C9 gene, one homozygous base substitution was identified in exon 3 (rs7900194, c.449A/A) and three heterozygous nucleotide changes were identified in exon 9 (rs9332240, c.1540C/T; rs9332241, c.1561C/T; rs9332243, c.1628C/T). The exon 3 variant, a known arginine to histidine substitution (p.R150H; Figure 1), has been classified by the Cytochrome P450 Allele Nomenclature Committee as CYP2C9*8. All three exon 9 transition polymorphisms were in the 3′;-UTR and have not been reported in known CYP2C9 haplotypes according to the Cytochrome P450 Allele Nomenclature Committee. No additional VKORC1 coding region or intron–exon boundary changes were identified.
Figure 1. Sequence analysis of CYP2C9 exon 3 in the African–American index patient compared with that from a homozygous wild-type individual.
(A) Electropherogram illustrates the wild-type genomic DNA and amino acid sequences. (B) Electropherogram illustrates the CYP2C9*8/*8 genomic DNA (c.449G>A) and amino acid (p.R150H) sequences identified in the African–American index patient.
wt: Wild-type.
African–American CYP2C9 allele frequencies
The CYP2C9*2, *3, *4, *5, *6, *8, *11 and *13 allele frequencies were determined in 300 anonymous healthy African–American individuals from the greater New York, USA, metropolitan area and were compared with previous African–American CYP2C9 population data (Table 3). Representative PCR-RFLP genotyping for CYP2C9*8, *11 and *13 is illustrated in Figure 2. No CYP2C9*4 or *13 alleles were identified and *8 was the most prevalent variant allele (0.047) in this cohort with a combined heterozygote and homozygote frequency of approximately 1 in 11 individuals. The wild-type (*1) allele frequency was 0.867 and the identified variant alleles, *2, *3, *5, *6, *8 and *11, had a combined frequency of 0.133 (~1 in 4 individuals). To determine if CYP2C9*8 is specific to the African–American population, genomic DNAs (100 each) from Caucasian, Ashkenazi Jewish, Hispanic and Asian individuals were interrogated and the allele was only detected in Hispanics and Asians with frequencies of 0.015 and 0.010, respectively. The Hispanic frequency may represent an African contribution to the Caribbean Hispanic community of the New York metropolitan area [23].
Table 3.
African–American CYP2C9 allele frequencies.
| Allele | This study (n = 600) | Blaisdell (2004) [33](n = 28)‡ | Takahashi (2006) [5] (n = 128) | Momary (2007) [30] (n = 202) | Kealey (2007) [55] (n = 336) | Limdi (2008) [29] (n = 536) | |
|---|---|---|---|---|---|---|---|
| Frequency | 95% CI | ||||||
| CYP2C9*1 | 0.867 | 0.839–0.894 | 0.831‡ | 0.953 | 0.946 | 0.961 | 0.937 |
| CYP2C9*2 | 0.028 | 0.015–0.042 | 0.000 | 0.000 | 0.030 | 0.036 | 0.013 |
| CYP2C9*3 | 0.020 | 0.009–0.031 | 0.000 | 0.008 | 0.015 | 0.003 | 0.019 |
| CYP2C9*4 | 0.000 | 0.000–0.000 | 0.000 | 0.000 | ND | ND | ND |
| CYP2C9*5 | 0.015 | 0.005–0.025 | 0.000 | 0.008 | 0.010 | ND | 0.009 |
| CYP2C9*6 | 0.010 | 0.002–0.018 | 0.000 | 0.008 | ND | ND | 0.007 |
| CYP2C9*8 | 0.047 | 0.030–0.064 | 0.036 | ND | ND | ND | ND |
| CYP2C9*11 | 0.013 | 0.004–0.023 | 0.000 | 0.023 | ND | ND | 0.015 |
| CYP2C9*13 | 0.000 | 0.000–0.000 | 0.000 | ND | ND | ND | ND |
This study performed CYP2C9 sequencing on 14 or 15 African–American samples. Of these, 1 of 14 had CYP2C9*8, 4 of 15 (0.133) had CYP2C9*9 (p.H251R) and several noncoding or synonymous variants were identified. The frequency of CYP2C9*1 was therefore calculated as 1 – the sum of CYP2C9*8 and *9.
n: Number of alleles; ND: Not determined.
Figure 2. Representative PCR-RFLP genotyping of CYP2C9 *8, *11 and *13.
Digested PCR products were visualized by agarose gel electrophoresis and compared with 100 bp size markers. Genotypes of all samples are noted above appropriate gel lanes.
M: Size marker; H2O: Reagent control; wt: Wild-type.
All African–American CYP2C9 allele frequencies were in Hardy–Weinberg equilibrium and the genotype frequencies are summarized in Table 4. Based on their genotypes, the assigned metabolic phenotypes [24–27] among the African–American individuals in our cohort were distributed as extensive (75.7%), intermediate (22.7%) and poor (1.7%) metabolizers. Notably, the high CYP2C9*8 frequency (0.047) made *1/*8 the most common intermediate metabolizer genotype in this population (8.7%). In addition, the inclusion of CYP2C9*5, *6, *8 and *11 in the genotyping panel reclassified approximately 16% of African–Americans compared with the predicted phenotypes using a panel consisting of only CYP2C9*2 and *3.
Table 4.
African–American CYP2C9 genotype frequencies (n = 300).
| Predicted metabolizer phenotype/genotype | Number of subjects | Observed (expected‡) frequency (%) |
|---|---|---|
| Extensive metabolizer | ||
| *1/*1 | 227 | 75.7 (75.1) |
| Intermediate metabolizer | ||
| *1/*2 | 13 | 4.3 (4.9) |
| *1/*3 | 10 | 3.3 (3.5) |
| *1/*5 | 8 | 2.7 (2.6) |
| *1/*6 | 5 | 1.7 (1.7) |
| *1/*8 | 26 | 8.7 (8.1) |
| *1/*11 | 6 | 2.0 (2.3) |
| Total | 68 | 22.7 (23.1) |
| Poor metabolizer | ||
| *2/*2 | 1 | 0.3 (0.1) |
| *2/*3 | 1 | 0.3 (0.1) |
| *3/*11 | 1 | 0.3 (0.1) |
| *5/*6 | 1 | 0.3 (0.0) |
| *8/*11 | 1 | 0.3 (0.1) |
| Total | 5 | 1.7 (0.4) |
Predicted Hardy–Weinberg frequencies.
African–American VKORC1 allele frequencies
The VKORC1 allele frequencies were determined in the African–American cohort and were compared with previous African–American population data (Table 5). The VKORC1 g.-1639A sensitive allele, which is in strong linkage disequilibrium (LD) with g.1173C>T, g.1542G>C and g.2255C>T in Caucasians [7,8], had a frequency of 0.108, consistent with other reports of the g.-1639G>A and g.1173C>T tag-SNPs [5,28,29], but lower than the reported g.1542G>C frequency in African–Americans (0.255) [30]. No g.85G>T (p.V29L), g.121G>T (p.A41S), g.134T>C (p.V45A), g.172A>G (p.R58G), or g.3487T>G (p.L128R) warfarin resistant alleles were detected; however, three African–American g.1331G>A (p.V66M) carriers were identified. These three samples were interrogated for additional VKORC1 alleles and the genotypes are listed in Table 6. In addition, the VKORC1 g.106G>T (p.D36Y) warfarin resistant allele found at a high frequency in Ashkenazi Jewish [18,31] and Ethiopian [32] individuals was analyzed and no African–American carriers were identified.
Table 5.
African–American VKORC1 allele frequencies.
| Allele | This study (n = 600) | Takahashi (2006) [5] (n = 128)* | Momary (2007) [30] (n = 216)‡ | Schelleman (2007) [28] (n = 318) | Limdi (2008) [29] (n = 518)* | |
|---|---|---|---|---|---|---|
| Frequency | 95% CI | |||||
| VKORC1 g.-1639G | 0.892 | 0.867–0.917 | 0.914 | 0.745 | 0.896 | 0.894 |
| VKORC1 g.-1639A | 0.108 | 0.083–0.133 | 0.086 | 0.255 | 0.104 | 0.106 |
| VKORC1 g.1331G | 0.995 | 0.989–1.000 | 0.984 | ND | ND | ND |
| VKORC1 g.1331A (p.V66M) | 0.005 | 0.000–0.011 | 0.016 | ND | ND | ND |
Based on tag-SNP g.1173C>T (6484C>T).
Based on tag-SNP g.1542G>C (6853G>C).
n: Number of alleles; ND: Not determined.
Table 6.
VKORC1 Tag-SNP genotypes among g.1331G>A (p.V66M) carriers.
| Subject | g.-4931T>C (381*) | g.-1639G>A (3673*) | g.698C>T (6009*) | g.1173C>T (6484*) | g.1542G>C (6853*) | g.2255C>T (7566*) | g.3730G>A (9041*) |
|---|---|---|---|---|---|---|---|
| 1 | C/C | G/G | C/C | C/C | G/G | C/T | G/G |
| 2 | C/C | G/G | C/C | C/C | G/C | C/T | G/G |
| 3 | C/T | G/G | C/C | C/C | G/G | C/T | G/A |
Position based on Accession Number AY587020.
All African–American VKORC1 allele frequencies were in Hardy–Weinberg equilibrium and the g.-1639G>A genotype frequencies are summarized in Table 7. By contrast to the Caucasian and Asian populations, the low g.-1639A frequency (0.108) resulted in only approximately 20% of African–American individuals carrying either the G/A or A/A genotype.
Table 7.
African–American VKORC1 g.-1639G>A genotype frequencies (n = 300).
| Genotype | Number of subjects | Observed (expected*) frequency (%) |
|---|---|---|
| G/G | 241 | 80.3 (79.5) |
| G/A | 53 | 17.7 (19.3) |
| A/A | 6 | 2.0 (1.2) |
Predicted Hardy–Weinberg frequencies.
Discussion
By sequencing the CYP2C9 and VKORC1 genes in patients with discrepant therapeutic and algorithm-predicted warfarin doses, an African–American male was identified who required a very low warfarin dose (14.4 mg/week) to achieve stable anticoagulation (INR of 2–3) and who was homozygous for the CYP2C9*8 variant allele. To date, the frequency of the CYP2C9*8 allele has not been systematically determined in distinct racial and ethnic populations. Therefore, we investigated the frequency of CYP2C9*8 in healthy cohorts from various racial and ethnic groups and detected the allele only in African–American (0.047), Hispanic (0.015) and Asian (0.010) individuals, with frequencies of approximately 1 in 11, 1 in 34 and 1 in 50, respectively.
The CYP2C9*8 c.449G>A transition polymorphism results in a nonsynonymous amino acid substitution (p.R150H) and was originally identified by resequencing CYP2C9 in lymphoblastoid lines derived from African–American and African–Pygmy individuals [33]. In addition, Blaisdell et al. determined the CYP2C9*8 allele frequency to be 0.036 in a small African–American cohort (n = 14), and measured its in vitro metabolizing activity towards tolbutamide [33]. Kinetic studies revealed a slight reduction in Km and an increased intrinsic clearance compared with CYP2C9*1, suggesting that *8 may have increased, not decreased, catalytic activity towards tolbutamide. In addition, an in vivo pharmacokinetic study of CYP2C9*8 using the urinary losartan:E-3174 ratio as a marker of CYP2C9 oxidation in Beninese subjects indicated that CYP2C9*8 activity was not significantly different than wild-type; however, the number of heterozygous *1/*8 human subjects in the study was small (n = 2), which may have confounded the results [34].
By contrast, when using phenytoin as a substrate for in vivo CYP2C9 activity, heterozygous CYP2C9*8 carriers displayed significantly impaired metabolic activity [26], suggesting that variant CYP2C9 alleles may have differing catalytic activities when measured in vitro or in vivo, and/or with different substrates. Similar contradictory phenotypes have been reported for CYP2C9*11 with tolbutamide [33] or losartan [34] as substrates. However, CYP2C9*11 recently has been associated with reduced therapeutic warfarin [25] and acenocoumarol [35] dose requirements. Importantly, heterozygosity for the CYP2C9*8 allele was also recently found to significantly reduce warfarin requirements in a multi-ethnic cohort, indicating that this allele has impaired in vivo metabolism of warfarin [27].
In addition to oral anticoagulants, CYP2C9 is involved in the metabolism of hypoglycemic, anticonvulsant, nonsteroidal anti-inflammatory and antihypertensive drugs. For example, the hydroxylation and elimination of phenytoin, a commonly used anti-epileptic, is impaired by the CYP2C9*2 and *3 variants both in vitro [36] and in vivo [37]. Moreover, the CYP2C9*6 variant, originally identified in an African–American with phenytoin toxicity, results in defective phenytoin clearance [38] and similar phenotypic responses to phenytoin have been reported for CYP2C9*5 and *11 [26]. Given the combined allele frequency of 0.133 for CYP2C9*2, *3, *5, *6, *8 and *11 in the African–American population, our data suggest that approximately 1 in 4 African–Americans may be at risk for phenytoin or other important CYP2C9-mediated drug toxicity.
Although markedly more common in Caucasians and Asians [5,7], the VKORC1 g.-1639G>A promoter polymorphism allele frequency in our African–American cohort (0.108) was consistent with other studies involving African–American populations. The g.-1639A allele reduces hepatic VKORC1 expression [8,9,39] and is frequently identified in warfarin-sensitive individuals [3,5,7,8,11,21,22,28,40,41]. Importantly, this allele is in strong LD with g.1173C>T, g.1542G>C and g.2255C>T among Caucasians [7,8]. However, the haplotype structure of VKORC1 among African–Americans is much more complex [8,42,43]. This may, in part, be why the commonly tested VKORC1 alleles (g.-1639G>A and g.1173C>T) contribute less to warfarin dose variability in African–Americans than Caucasians [16,28]. Furthermore, the weaker LD between g.-1639G>A and g.1542G>C among African–Americans [29,43] could explain the discrepancy between the g.-1639G>A frequency in our study and the previously reported g.1542G>C frequency [30]. Recently, a comprehensive analysis of VKORC1 polymorphisms identified 12 common African–American haplotypes and concluded that g.-1639G>A or g.1173C>T were still the best predictors of warfarin dose variability in this racial group [42], supporting their use in multi-ethnic dosing algorithms.
Interestingly, three VKORC1 g.1331G>A (p.V66M) carriers were detected in our African–American cohort. This variant was originally identified shortly after the discovery of VKORC1 in an African–Caribbean individual who required more than 25 mg/day of warfarin for therapeutic anticoagulation [44] and subsequently was reported in other warfarin-resistant individuals requiring at least 20 mg/day [45,46]. Like other VKORC1 resistant mutations, p.V66M occurs within the cytoplasmic loop of the VKORC1 protein, and it is hypothesized that this domain may serve as an important accessory binding site for coumarin anticoagulants [46]. To investigate if p.V66M occurs on a common haplotype among the African–American p.V66M carriers, additional VKORC1 genotyping was performed. Based on these studies (Table 6), in this racial group the p.V66M allele likely occurs on a CGCCGTG haplotype background which corresponds to the H4 or BHT4 haplotype using the nomenclature of Rieder [8] or Limdi et al. [42], respectively. Given the diverse VKORC1 African–American haplotype structure [8,42,43], further testing on additional p.V66M carriers is required to confirm this hypothesis.
Although the frequency of VKORC1 g.1331G>A (p.V66M) is low among African–Americans (~1 in 100), the extreme resistance to warfarin consistently associated with the allele suggests that its inclusion into genotyping panels, if cost-effective, may also be considered. In addition, the CYP4F2*3 (p.V433M) variant recently shown to be responsible for approximately 1–2% of the variation in warfarin dosing among Caucasians [47,48] may also warrant inclusion into genotyping panels following evaluation in multi-ethnic cohorts.
Conclusion
The frequency of the CYP2C9*8 allele in the African–American population (~1 in 11 individuals) and the finding that homozygosity and heterozygosity [27] are associated with reduced warfarin-dose requirements underscores the importance of its incorporation into future pharmacogenetic-based dosing algorithms. Furthermore, as the majority of warfarin algorithms only include CYP2C9*2 and *3 (with VKORC1 g.-1639G>A or g.1173C>T), our data indicate that the inclusion of CYP2C9*8 alone could reclassify the predicted metabolic phenotypes of almost 10% of African–Americans, and when combined with CYP2C9*5, *6 and *11, more than 15%. Thus, in the African–American population, the incorporation of CYP2C9*8 into genotyping panels may improve dose prediction of CYP2C9-metabolized drugs, including warfarin.
Future perspective
Dosing algorithms that include common CYP2C9 and VKORC1 polymorphisms have been developed and currently are being prospectively evaluated to determine if they predict the initial warfarin dose more accurately than standard clinical algorithms. Given that the majority of pharmacogenetic algorithms perform better in Caucasians than other racial and ethnic groups, it is likely that other genetic variants and nongenetic factors will be identified that significantly influence warfarin dosing and drug response in specific subpopulations. The identification of novel and racial-specific CYP450 alleles and other pharmacogenetic variants that influence drug metabolism will permit improved phenotypic prediction and drug selection. Inclusion of known and novel variants into future clinical testing panels must be accompanied by careful interpretation of genotype–phenotype relationships, physician awareness and cost-effective testing.
Executive summary
Although the frequencies of pharmacogenetic variants differ among racial groups, most pharmacogenetic algorithms for genotype-guided warfarin dosing only include two CYP2C9 alleles (*2 and *3) and a single VKORC1 allele (g.-1639G>A or g.1173C>T) commonly found among Caucasians.
To identify other CYP2C9 and VKORC1 alleles important in warfarin dose variability, these genes were sequenced in selected sensitive (<21 mg/week) and resistant (>49 mg/week) individuals with a target International Normalized Ratio of 2–3.
Identification of CYP2C9*8 & African–American allele frequencies
Sequencing identified an African–American male with a lower than predicted therapeutic warfarin dose (14.4 mg/week), previously genotyped as CYP2C9*1/*1, who was homozygous for CYP2C9*8 (c.449G>A; p.R150H).
Genotyping 600 alleles from healthy African–American blood donors identified CYP2C9*8 as their most frequent variant CYP2C9 allele (0.047; ~1 in 11 individuals) and the combined allele frequency of CYP2C9*2, *3, *5, *6, *8 and *11 was 0.133 (~1 in 4 individuals).
VKORC1 African–American allele frequencies
The VKORC1 g.-1639A allele frequency was 0.108 and three warfarin resistant g.1331G>A (p.V66M) carriers were identified (1 in ~100 individuals).
Conclusion
Inclusion of CYP2C9*8 alone reclassified the predicted metabolic phenotypes of almost 10% of African–Americans, and when combined with CYP2C9*5, *6 and *11, more than 15%.
For the African–American population, the incorporation of CYP2C9*8 into genotyping panels may improve dose prediction of CYP2C9-metabolized drugs, including warfarin.
Supplementary Material
Acknowledgments
Financial & competing interests disclosure
This research was supported in part by a NIH Cooperative Study grant (N268200800003C) and a NIH grant (5 M01 RR00071) from the Division of Research Resources for the Mount Sinai General Clinical Research Center. SAS and MJ are the recipients of Biochemical/Molecular Genetics Fellowships from the Genzyme Corporation. The authors would like to thank Elizabeth Rothlauf and Arun Agarwal of the Mount Sinai Medical Center for assistance with patient enrollment, and the staffs of the Nucleic Acid Testing Laboratory and Department of Special Patient and Community Health Services of New York Blood Center for assistance with sample collection. Heterozygous CYP2C9*13 positive control DNA samples were generously provided by Professor Seok-Yong Lee, Sungkyunkwan University, Suwon, Korea. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Footnotes
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Sim SC, Ingelman-Sundberg M. The human cytochrome P450 Allele Nomenclature Committee Web site: submission criteria, procedures, and objectives. Methods Mol Biol. 2006;320:183–191. doi: 10.1385/1-59259-998-2:183. [DOI] [PubMed] [Google Scholar]
- 2.Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 3.Aquilante CL, Langaee TY, Lopez LM, et al. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther. 2006;79:291–302. doi: 10.1016/j.clpt.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Carlquist JF, Horne BD, Muhlestein JB, et al. Genotypes of the cytochrome p450 isoform, CYP2C9, and the vitamin K epoxide reductase complex subunit 1 conjointly determine stable warfarin dose: a prospective study. J Thromb Thrombolysis. 2006;22:191–197. doi: 10.1007/s11239-006-9030-7. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi H, Wilkinson GR, Nutescu EA, et al. Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African–Americans. Pharmacogenet Genomics. 2006;16:101–110. doi: 10.1097/01.fpc.0000184955.08453.a8. [DOI] [PubMed] [Google Scholar]
- 6 ▪▪.Klein TE, Altman RB, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. Recent large study underscoring the benefit of pharmacogenetic-guided warfarin dosing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geisen C, Watzka M, Sittinger K, et al. VKORC1 haplotypes and their impact on the inter-individual and inter-ethnical variability of oral anticoagulation. Thromb Haemost. 2005;94:773–779. doi: 10.1160/TH05-04-0290. [DOI] [PubMed] [Google Scholar]
- 8.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 9.Yuan HY, Chen JJ, Lee MT, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet. 2005;14:1745–1751. doi: 10.1093/hmg/ddi180. [DOI] [PubMed] [Google Scholar]
- 10.Hirsh J, Dalen J, Anderson DR, et al. Oral anticoagulants: mechanism of action, clinical effectiveness, and optimal therapeutic range. Chest. 2001;119:8S–21S. doi: 10.1378/chest.119.1_suppl.8s. [DOI] [PubMed] [Google Scholar]
- 11.Wadelius M, Chen LY, Eriksson N, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007;121:23–34. doi: 10.1007/s00439-006-0260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimmel SE. Warfarin therapy: in need of improvement after all these years. Expert Opin Pharmacother. 2008;9:677–686. doi: 10.1517/14656566.9.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13 ▪.Limdi NA, Veenstra DL. Warfarin pharmacogenetics. Pharmacotherapy. 2008;28:1084–1097. doi: 10.1592/phco.28.9.1084. Comprehensive overview of warfarin pharmacogenetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14 ▪.Schelleman H, Limdi NA, Kimmel SE. Ethnic differences in warfarin maintenance dose requirement and its relationship with genetics. Pharmacogenomics. 2008;9:1331–1346. doi: 10.2217/14622416.9.9.1331. Excellent review on pharmacogenetic warfarin-dosing algorithms. [DOI] [PubMed] [Google Scholar]
- 15.Xie HG, Prasad HC, Kim RB, Stein CM. CYP2C9 allelic variants: ethnic distribution and functional significance. Adv Drug Deliv Rev. 2002;54:1257–1270. doi: 10.1016/s0169-409x(02)00076-5. [DOI] [PubMed] [Google Scholar]
- 16.Gage B, Eby C, Johnson J, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84(3):326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schelleman H, Chen J, Chen Z, et al. Dosing algorithms to predict warfarin maintenance dose in Caucasians and African Americans. Clin Pharmacol Ther. 2008;84:332–339. doi: 10.1038/clpt.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott SA, Edelmann L, Kornreich R, Desnick RJ. Warfarin pharmacogenetics: CYP2C9 and VKORC1 genotypes predict different sensitivity and resistance frequencies in the Ashkenazi and Sephardi Jewish populations. Am J Hum Genet. 2008;82:495–500. doi: 10.1016/j.ajhg.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Limdi NA, Beasley TM, Baird MF, et al. Kidney function influences warfarin responsiveness and hemorrhagic complications. J Am Soc Nephrol. 2009;20(4):912–921. doi: 10.1681/ASN.2008070802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu AH, Wang P, Smith A, et al. Dosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multi-ethnic population: comparison with other equations. Pharmacogenomics. 2008;9:169–178. doi: 10.2217/14622416.9.2.169. [DOI] [PubMed] [Google Scholar]
- 21.Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 22.Wadelius M, Chen LY, Lindh JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2008;113(4):784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benn-Torres J, Bonilla C, Robbins CM, et al. Admixture and population stratification in African Caribbean populations. Ann Hum Genet. 2008;72:90–98. doi: 10.1111/j.1469-1809.2007.00398.x. [DOI] [PubMed] [Google Scholar]
- 24 ▪.Kirchheiner J, Brockmoller J. Clinical consequences of cytochrome P450 2C9 polymorphisms. Clin Pharmacol Ther. 2005;77:1–16. doi: 10.1016/j.clpt.2004.08.009. Important discussion of the clinical relevance of CYP2C9 metabolizer phenotypes. [DOI] [PubMed] [Google Scholar]
- 25.Tai G, Farin F, Rieder MJ, et al. In-vitro and in-vivo effects of the CYP2C9*11 polymorphism on warfarin metabolism and dose. Pharmacogenet Genomics. 2005;15:475–481. doi: 10.1097/01.fpc.0000162005.80857.98. [DOI] [PubMed] [Google Scholar]
- 26 ▪.Allabi AC, Gala JL, Horsmans Y. CYP2C9, CYP2C19, ABCB1 (MDR1) genetic polymorphisms and phenytoin metabolism in a Black Beninese population. Pharmacogenet Genomics. 2005;15:779–786. doi: 10.1097/01.fpc.0000174787.92861.91. Study demonstrating decreased phenytoin metabolism in CYP2C9*5, *6, *8 and *11 carriers. [DOI] [PubMed] [Google Scholar]
- 27.Hujsak P, Wang P, Smith A, Wu AH. Addition of CYP450 2C9*8 and CYP450 4F2 to a dosing algorithm for warfarin dosing [abstract] Am Soc of Hum Genet. 2008;371 Abstract 1924. [Google Scholar]
- 28.Schelleman H, Chen Z, Kealey C, et al. Warfarin response and vitamin K epoxide reductase complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007;81:742–747. doi: 10.1038/sj.clpt.6100144. [DOI] [PubMed] [Google Scholar]
- 29.Limdi NA, Arnett DK, Goldstein JA, et al. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European–Americans and African–Americans. Pharmacogenomics. 2008;9:511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Momary KM, Shapiro NL, Viana MA, Nutescu EA, Helgason CM, Cavallari LH. Factors influencing warfarin dose requirements in African–Americans. Pharmacogenomics. 2007;8:1535–1544. doi: 10.2217/14622416.8.11.1535. [DOI] [PubMed] [Google Scholar]
- 31.Loebstein R, Dvoskin I, Halkin H, et al. A coding VKORC1 Asp36Tyr polymorphism predisposes to warfarin resistance. Blood. 2007;109:2477–2480. doi: 10.1182/blood-2006-08-038984. [DOI] [PubMed] [Google Scholar]
- 32.Aklillu E, Leong C, Loebstein R, Halkin H, Gak E. VKORC1 Asp36Tyr warfarin resistance marker is common in Ethiopian individuals. Blood. 2008;111:3903–3904. doi: 10.1182/blood-2008-01-135863. [DOI] [PubMed] [Google Scholar]
- 33 ▪▪.Blaisdell J, Jorge-Nebert LF, Coulter S, et al. Discovery of new potentially defective alleles of human CYP2C9. Pharmacogenetics. 2004;14:527–537. doi: 10.1097/01.fpc.0000114759.08559.51. Initial identification of the CYP2C9*8 allele. [DOI] [PubMed] [Google Scholar]
- 34.Allabi AC, Gala JL, Horsmans Y, et al. Functional impact of CYP2C95, CYP2C96, CYP2C98, and CYP2C911 in vivo among black Africans. Clin Pharmacol Ther. 2004;76:113–118. doi: 10.1016/j.clpt.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Rettie AE, Farin FM, Beri NG, Srinouanprachanh SL, Rieder MJ, Thijssen HH. A case study of acenocoumarol sensitivity and genotype–phenotype discordancy explained by combinations of polymorphisms in VKORC1 and CYP2C9. Br J Clin Pharmacol. 2006;62:617–620. doi: 10.1111/j.1365-2125.2006.02688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veronese ME, Doecke CJ, Mackenzie PI, et al. Site-directed mutation studies of human liver cytochrome P-450 isoenzymes in the CYP2C subfamily. Biochem J. 1993;289(Pt 2):533–538. doi: 10.1042/bj2890533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odani A, Hashimoto Y, Otsuki Y, et al. Genetic polymorphism of the CYP2C subfamily and its effect on the pharmacokinetics of phenytoin in Japanese patients with epilepsy. Clin Pharmacol Ther. 1997;62:287–292. doi: 10.1016/S0009-9236(97)90031-X. [DOI] [PubMed] [Google Scholar]
- 38.Kidd RS, Curry TB, Gallagher S, Edeki T, Blaisdell J, Goldstein JA. Identification of a null allele of CYP2C9 in an African–American exhibiting toxicity to phenytoin. Pharmacogenetics. 2001;11:803–808. doi: 10.1097/00008571-200112000-00008. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadee W. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood. 2008;112:1013–1021. doi: 10.1182/blood-2008-03-144899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y, Shennan M, Reynolds KK, et al. Estimation of warfarin maintenance dose based on VKORC1 (−1639 G>A) and CYP2C9 genotypes. Clin Chem. 2007;53:1199–1205. doi: 10.1373/clinchem.2006.078139. [DOI] [PubMed] [Google Scholar]
- 41.Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 42.Limdi NA, Beasley TM, Crowley MR, et al. VKORC1 polymorphisms, haplotypes and haplotype groups on warfarin dose among African–Americans and European–Americans. Pharmacogenomics. 2008;9:1445–1458. doi: 10.2217/14622416.9.10.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crawford DC, Ritchie MD, Rieder MJ. Identifying the genotype behind the phenotype: a role model found in VKORC1 and its association with warfarin dosing. Pharmacogenomics. 2007;8:487–496. doi: 10.2217/14622416.8.5.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrington DJ, Underwood S, Morse C, Shearer MJ, Tuddenham EG, Mumford AD. Pharmacodynamic resistance to warfarin associated with a Val66Met substitution in vitamin K epoxide reductase complex subunit 1. Thromb Haemost. 2005;93:23–26. doi: 10.1160/TH04-08-0540. [DOI] [PubMed] [Google Scholar]
- 45.Bodin L, Perdu J, Diry M, Horellou MH, Loriot MA. Multiple genetic alterations in vitamin K epoxide reductase complex subunit 1 gene (VKORC1) can explain the high dose requirement during oral anticoagulation in humans. J Thromb Haemost. 2008;6:1436–1439. doi: 10.1111/j.1538-7836.2008.03049.x. [DOI] [PubMed] [Google Scholar]
- 46.Harrington DJ, Gorska R, Wheeler R, et al. Pharmacodynamic resistance to warfarin is associated with nucleotide substitutions in VKORC1. J Thromb Haemost. 2008;6:1663–1670. doi: 10.1111/j.1538-7836.2008.03116.x. [DOI] [PubMed] [Google Scholar]
- 47.Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeuchi F, McGinnis R, Bourgeois S, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:E1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Si D, Guo Y, Zhang Y, Yang L, Zhou H, Zhong D. Identification of a novel variant CYP2C9 allele in Chinese. Pharmacogenetics. 2004;14:465–469. doi: 10.1097/01.fpc.0000114749.08559.e4. [DOI] [PubMed] [Google Scholar]
- 50.Dickmann LJ, Rettie AE, Kneller MB, et al. Identification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Mol Pharmacol. 2001;60:382–387. doi: 10.1124/mol.60.2.382. [DOI] [PubMed] [Google Scholar]
- 51.Herman D, Peternel P, Stegnar M, Breskvar K, Dolzan V. The influence of sequence variations in factor VII, γ-glutamyl carboxylase and vitamin K epoxide reductase complex genes on warfarin dose requirement. Thromb Haemost. 2006;95:782–787. [PubMed] [Google Scholar]
- 52.Tham LS, Goh BC, Nafziger A, et al. A warfarin-dosing model in Asians that uses single-nucleotide polymorphisms in vitamin K epoxide reductase complex and cytochrome P450 2C9. Clin Pharmacol Ther. 2006;80:346–355. doi: 10.1016/j.clpt.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 53.Anderson JL, Horne BD, Stevens SM, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116:2563–2570. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 54.Miao L, Yang J, Huang C, Shen Z. Contribution of age, body weight, and CYP2C9 and VKORC1 genotype to the anticoagulant response to warfarin: proposal for a new dosing regimen in Chinese patients. Eur J Clin Pharmacol. 2007;63:1135–1141. doi: 10.1007/s00228-007-0381-6. [DOI] [PubMed] [Google Scholar]
- 55.Kealey C, Chen Z, Christie J, et al. Warfarin and cytochrome P450 2C9 genotype: possible ethnic variation in warfarin sensitivity. Pharmacogenomics. 2007;8:217–225. doi: 10.2217/14622416.8.3.217. [DOI] [PubMed] [Google Scholar]
Website
- 101.Home Page of the Human Cytochrome P450 (CYP) Allele Nomenclature Committee . www.cypalleles.ki.se/cyp2c9.htm.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.