Abstract
Eukaryotic translation initiation factor 5A (eIF5A) is a highly conserved protein essential for eukaryotic cell proliferation and is the only protein containing hypusine, [Nε-(4-amino-2-hydroxybutyl)lysine]. eIF5A is activated by the posttranslational synthesis of hypusine. eIF5A also undergoes an acetylation at specific Lys residue(s). In this study, we have investigated the effect of hypusine modification and acetylation on the subcellular localization of eIF5A. Immunocytochemical analyses showed differences in the distribution of non-hypusinated eIF5A precursor and the hypusine-containing mature eIF5A. While the precursor is found in both cytoplasm and nucleus, the hypusinated eIF5A is primarily localized in cytoplasm. eIF5A mutant proteins, defective in hypusine modification (K50A, K50R) were localized in a similar manner to the eIF5A precursor, whereas hypusine-modified mutant proteins (K47A, K47R, K68A) were localized mainly in the cytoplasm. These findings provide strong evidence that the hypusine modification of eIF5A dictates its localization in the cytoplasmic compartment where it is required for protein synthesis.
Keywords: hypusine, eIF5A, acetylation, deoxyhypusine synthase, deoxyhypusine hydroxylase, polyamine, nuclear export
Introduction
eIF5A is a small, essential eukaryotic protein that contains a modified lysine residue, hypusine [Nε-(4-amino-2-hydroxybutyl)lysine] (see reviews [1–3]) Hypusine is formed post-translationally in only one cellular protein, the eIF5A precursor, eIF5A(Lys), by two consecutive enzymatic steps. In the first step, deoxyhypusine synthase (DHS) catalyzes the transfer of the aminobutyl moiety from the polyamine spermidine to a specific lysine residue (Lys50 for the human eIF5A) to form deoxyhypusine [Nε-(4-aminobutyl)lysine] residue. This intermediate is subsequently hydroxylated by deoxyhypusine hydroxylase (DOHH) [4] to produce hypusine and thereby the biologically active eIF5A. Evidence in support of the essential role of hypusine and eIF5A in eukaryotic and mammalian cell proliferation has been reported from several laboratories ([1–3, 5]).
Although the essential nature of eIF5A in eukaryotic cell proliferation is firmly established, the precise cellular function of this putative initiation factor has remained obscure for decades. eIF5A was initially isolated from the high salt washes of reticulocyte lysate ribosomes, along with other initiation factors [6]. eIF5A exists mainly as the fully modified hypusine form in mammalian tissues and cells [7]. eIF5A stimulates methionyl-puromycin synthesis in vitro in a hypusine-dependent manner [8, 9]. Recent evidence for the hypusine-dependent association of eIF5A with actively translating ribosomes [10, 11] and the increase in the polysome/monosome ratio in certain eIF5A mutant strains suggest a role for eIF5A in the elongation rather than the initiation step of translation [3, 12]. S. cerevisiae strains harboring temperature-sensitive eIF5A mutants exhibit diverse cellular changes, suggesting a direct or indirect role of eIF5A in cell wall integrity, mRNA decay, actin polarization, apoptosis and cell cycle progression [3]. It is not yet clear whether eIF5A has an independent function in addition to that as a translation factor and how depletion or dysfunction of eIF5A leads to the pleiotropic phenotypes of the temperature-sensitive eIF5A mutant strains.
Subcellular localization of eIF5A in conjunction with its potential function in nuclear export has been investigated by several laboratories and the results reported are inconsistent. Ruhl et al. described eIF5A as a cellular cofactor for the HIV-1 Rev protein that is involved in nuclear export of unspliced viral mRNAs [13] and its localization in the nucleus [13]. However, a contradictory report from another laboratory showed localization of eIF5A primarily in the cytoplasm in various mammalian cells and a lack of effect of Rev overexpression on eIF5A distribution [14]. Subsequently, Jao and Chen presented evidence [15] that different fixation methods (HCHO or methanol) result in different patterns of eIF5A subcellular distribution. By direct visualization of exogenously expressed GFP-tagged eIF5A and indirect immunofluorescence detection of endogenous eIF5A, they concluded that eIF5A is present in both cytoplasm and nucleus and that eIF5A gains nuclear entry by passive diffusion. They were able to demonstrate nuclear export of GFP-Rev by an interspecies heterokaryon fusion assay, but not of GFP-eIF5A [15]. More recently, using truncated eIF5A with C-terminal GFP-tags, Parreiras et al. reported that the N-terminal 19 amino acids of eIF5A serves as a signal for nuclear localization of eIF5A [16]. eIF5A or its non-hypusinated precursor has also been implicated in the regulation of apoptosis [17–19] as a modulator of p53 in human lung cancer cell lines [17]. Furthermore, in colon carcinoma cells, induction of apoptosis by cytokines was reported to be associated with nuclear import of eIF5A [18]. Thus, an intriguing controversy remains as to whether there exists a regulated mechanism of nuclear import or export of eIF5A and whether nuclear eIF5A has a significant cellular function.
One of the causes of discrepancies in reported eIF5A subcellular localization may stem from the use of exogenously expressed, epitope-tagged (GFP or GST) eIF5A as a marker for endogenous eIF5A, without knowing that endogenous eIF5A and exogenous eIF5A differ in modification status with respect to hypusination and acetylation [13, 15–17]. Unlike endogenous eIF5A, which is efficiently modified to the hypusine form and exists predominantly as the hypusine form, exogenously produced FLAG-eIF5A protein remains largely as the non-hypusinated precursors. Furthermore, exogenously produced non-hypusinated eIF5A precursor is readily acetylated at Lys47. Our present data strongly suggest that its hypusine modification status dictates the cytoplasmic localization of eIF5A, where it is mobilized for protein synthesis. The nuclear export of eIF5A may be mediated by the nuclear exportor exportin 4 in a hypusine-dependent manner.
Materials and Methods
Construction of pCEFL/GFP-eIF5A plasmids
The ORF of human eIF5A-1 was PCR-amplified and using a primer set, 5′-atagaattcATGGCAGATGACTTGGACTTC′-3′ and 5′-atagcggccgcTTATTTTGCCATGGCCTTGATTG-3′ and subcloned into EcoR1 and Not1 sites at the C-terminus of GFP of the pCEFL/GFP vector.
Site-directed mutagenesis
Vectors encoding GFP-eIF5A mutant proteins (K47A, K47R, K50A, K50R and K68A) and FLAG-eIF5A mutant proteins (K47A, K47R, G49A, K50A, K50R, H51A, G52A, H53A, K55A and K68A) were generated using the Quick Change Site-Directed Mutagenesis Kit (Stratagene) with primer sets designed for the intended mutations.
Radiolabeling of eIF5A by culturing cells in the presence of [3H]spermidine
[3H]Spermidine (5 μCi/ml) (Perkin-Elmer/NEN) was added to the medium after transfection (using Lipofectamine 2000 (Invitrogen)). Cells were harvested after 48h and precipitated with 10% trichloraoacetic acid (TCA) containing 1mM each of unlabeled polyamines. After thorough washing, half of the TCA precipitate was hydrolyzed in 6 N HCl for the analysis of radiolabeled hypusine or deoxyhypusine, and other half of the TCA precipitate was used for SDS/PAGE for detection of the radiolabeled eIF5A proteins.
Immunostaining and Fluorescence Microscopy
Transfected cells grown on cover slips were fixed in 2% formaldehyde for 15 min. Fixed cells were blocked with 10% fetal bovine serum for 15 min and incubated with primary antibody against eIF5A (1:100 dilution in blocking solution containing 0.2% saponin) or anti-FLAG antibody for 1hr followed by the Alexa Fluor 594 or Alexa Fluor 488 secondary antibodies (Invitrogen) for 1 hr. The slides were mounted with mounting medium containing DAPI (Vector Laboratories). The images were captured using an Axiophot microscope (Zeiss). For confocal analysis, the images were analyzed by using an 1X81 inverted confocal microscope (Olympus).
Results
Exogenously produced eIF5A remains largely as the non-hypusinated precursor in mammalian cells
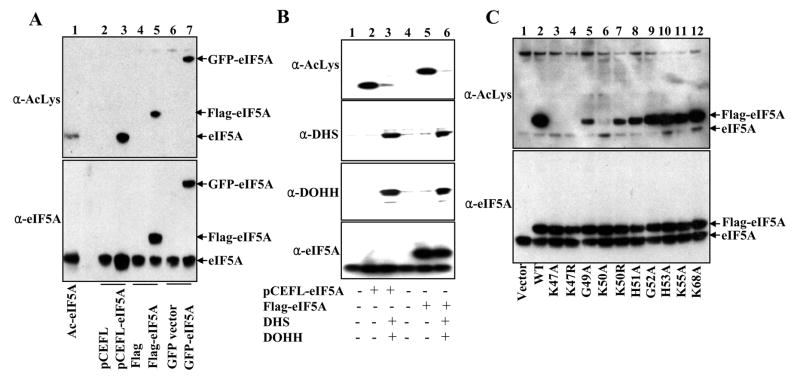
In various mammalian cells, the function and subcellular localization of eIF5A have been studied by transfection with eIF5A vector alone [13, 15–17]. However, we have found that exogenously produced eIF5A is poorly modified by endogenous DHS and/or DOHH in mammalian cells and remains largely as the unmodified precursor, eIF5A(Lys). As shown in Fig. 1, in HeLa cells transfected with p3XFLAG-CMV-7.1/eIF5A vector alone, there was little labeling of FLAG-eIF5A, even when FLAG-eIF5A protein was highly expressed as shown in the western blot (Fig. 1A, lanes 2 and 5). The lack of labeling of FLAG-eIF5A upon incubation with [3H]spermidine indicates that FLAG-eIF5A was not being modified by endogenous DHS and DOHH. Only upon cotransfection with the modification enzymes, was the FLAG-eIF5A precursor fully modified to the hypusine form (Fig. 1A, lanes 3 and 6, hypusine analysis data not shown). Similar results were obtained with GFP-eIF5A (Fig. 1B). These findings support a premise that FLAG-eIF5A or GFP-eIF5A, produced upon transfection with an eIF5A vector alone, represents largely the non-hypusinated eIF5A precursor, whereas that produced by cotransfection with all three vectors (eIF5A, DHS and DOHH) represents the hypusine form.
Fig. 1. Exogenous expression of (A) FLAG-eIF5A and (B) GFP-eIF5A precursors or their hypusine-modified forms.
HeLa cells were transfected with 3XFLAG-CMV-7.1, 3XFLAG-CMV-7.1/heIF5A-1, pCEFL, or pCEFL/GFP-heIF5A-1 with or without pCEFL/hDHS and pCEFL/hDOHH as indicated. Transfection was performed in parallel in two sets, one set of cells (6 well dishes) were used for western blotting using an eIF5A antibody (BD Bioscience) that detects eIF5A(Lys), eIF5A(Dhp) and eIF5A(Hpu) equally well. The other set (60 mm dishes) was used for labeling with [3H]spermidine (5 μCi/ml). Cells were harvested after 42 h of transfection.
Exogenously produced eIF5A is acetylated at Lys47
Previously, several lines of evidence were reported on the acetylation of eIF5A at specific lysine residues [7]. We used an AcLys antibody to detect eIF5A acetylation in vivo in HeLa cells transfected with three eIF5A vectors, non-tagged, FLAG-tagged and GFP-tagged. In each case, exogenously expressed eIF5A precursor was by far the major acetylated protein in these cells (lanes 3, 5, and 7 in Fig. 2A). However, acetylation was hardly detectable in the endogenous hypusine-containing eIF5A. The exogenous hypusine-containing eIF5A, produced upon cotransfection with three vectors encoding eIF5A, DHS and DOHH, showed much weaker AcLys signals than the non-hypusinated eIF5A precursors did (compare lanes 3 with 2, and lanes 6 with 5 in Fig 2B), suggesting that non-hypusinated eIF5A precursors are better substrates for cellular acetyltransferase than their hypusinated counterparts. In order to determine the eIF5A acetylation site in vivo, we tested a number of mutant proteins of FLAG-eIF5A for their acetylation status. While all the mutant proteins were expressed at similar levels (Fig. 2C lower panel), the degree of acetylation (Fig. 2C upper panel) varied widely. There was no acetylation with two mutants, K47A and K47R, confirming that Lys47 is indeed the major site of eIF5A acetylation. K50R was acetylated, whereas acetylation in K50A was significantly diminished, suggesting that a basic charge at residue 50 is important for acetylation at Lys47 (Fig. 2C).
Fig. 2. Identification of the major acetylation site of eIF5A as Lys47.
HeLa cells were transfected as indicated and harvested after 48 h of transfection. 10 μg of proteins was used for western blotting using either AcLys antibody (Santa Cruze Biotechnology), eIF5A antibody (BD Bioscience), or DHS and DOHH rabbit polyclonal antibodies.
Differences between the subcellular distribution patterns of non-hypusinated eIF5A precursor and hypusine-modified eIF5A
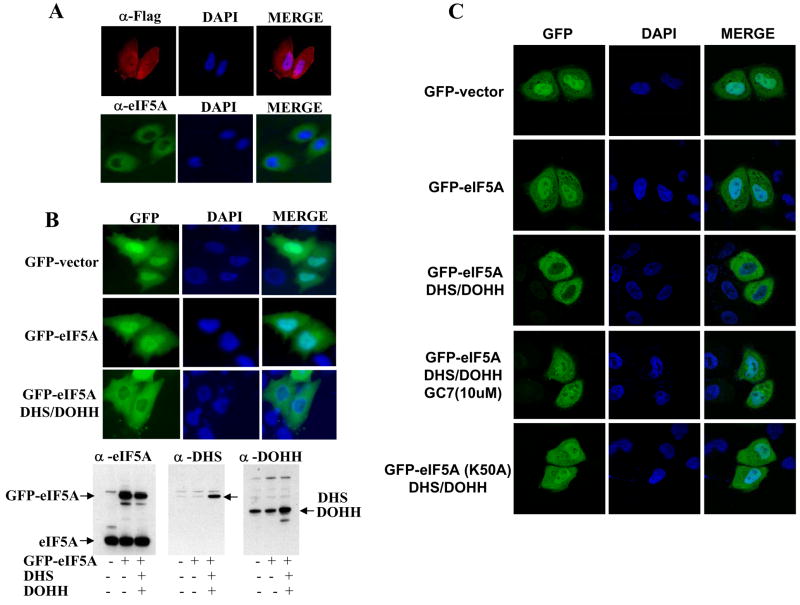
First, we compared the cellular localization of endogenous eIF5A (hypusine form) with the exogenously produced eIF5A precursor, FLAG-eIF5A(Lys). Upon indirect immuno-fluorescence detection with eIF5A antibody, endogenous eIF5A was primarily localized in the cytoplasm (Fig 3A, lower panels). In contrast, the exogenously produced FLAG-eIF5A (precursor form) detected with the FLAG antibody showed whole cell distribution i.e. in both cytoplasm and nucleus (Fig 3A, upper panels). When the fluorescence of GFP-eIF5A was directly visualized, a consistent difference was observed between non-hypusinated and hypusinated eIF5A (compare middle and lower panels in Fig. 3B). Whereas the GFP-eIF5A precursor exhibited a whole cell distribution, GFP-eIF5A(Hpu) produced from cells cotransfected with vectors encoding DHS and DOHH was excluded from the nucleus, showing primarily cytoplasmic localization (Fig 3B, the lower left panel), similar to that of endogenous eIF5A. Cotransfection of GFP with DHS and DOHH did not alter GFP distribution (not shown). This finding suggests that hypusine modification promotes nuclear export of eIF5A. A clearer difference in subcellular localization was observed by imaging with a confocal microscope (Fig. 3C, second and third row of panels). Unlike the GFP-eIF5A (Hpu), the GFP-eIF5A mutant (K50A) which cannot be modified to the hypusine form, was not excluded from nucleus when cotransfected with DHS and DOHH. The loss of nuclear exclusion was also observed when hypusine modification was inhibited by GC7, an inhibitor of DHS.
Fig. 3. Comparison of subcellular distribution of endogenous eIF5A(Hpu), exogenous eIF5A(Lys) and exogenous eIF5A(Hpu).
(A) HeLa cells were fixed after 48 h of transfection, incubated with primary antibodies, eIF5A antibody (NIH353, rabbit polyclonal) or anti FLAG antibody (mouse monoclonal from Sigma) followed by the secondary antibodies, Alexa Fluor 488-conjugated secondary anti-rabbit antibodies (green, Invitrogen) or Alexa Fluor 594-conjugated secondary anti-mouse antibodies (red, Invitrogen) for detection of endogenous eIF5A and exogenous FLAG-eIF5A, respectively. (B) HeLa cells, transfected with pCEFL/GFP-eIF5A without or with vectors encoding DHS and DOHH, were fixed and GFP-eIF5A precursor and GFP-eIF5A(Hpu) were directly visualized. (C) Fluorescence of GFP or GFP-eIF5A formed in HeLa cells transfected as in (B) was visualized by a confocal microscope.
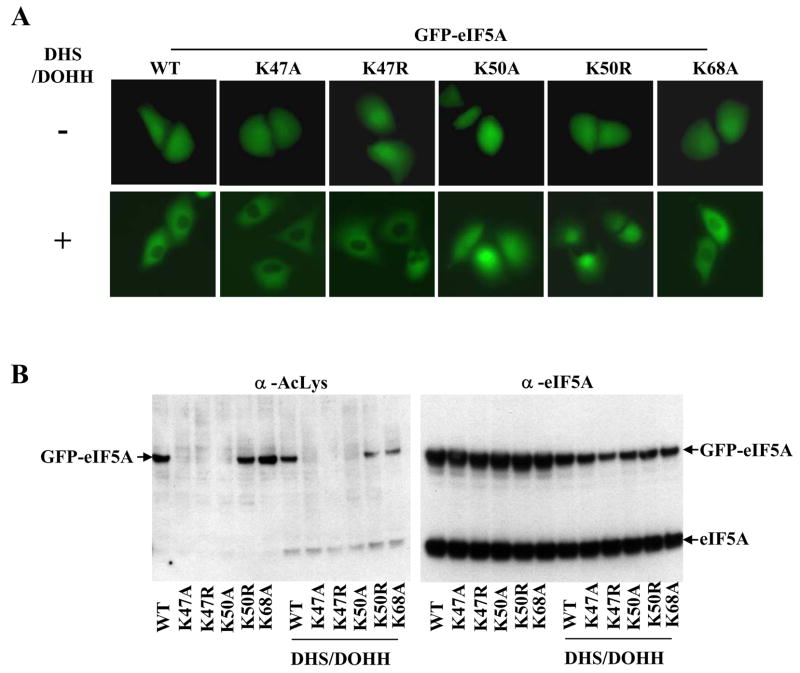
In order to confirm this finding, we compared the subcellular distribution GPF-eIF5A mutants with individual amino acid substitutions around Lys50, including K47A, K47R, K50A, K50R, and K68A. All of these mutants displayed similar distribution patterns upon transfection with GFP-eIF5A vector alone (Fig. 4A, upper panels). On the other hand, differences were observed when these GFP-eIF5A mutant proteins were coexpressed with DHS and DOHH (Fig. 4A, lower panels). Those that can undergo hypusine modification (namely, GPF-eIF5A wt, K47A, K47R and K68A) appeared to be excluded from nucleus upon cotransfection with DHS and DOHH. On the other hand, the subcellular distribution of two mutants, K50A and K50R which cannot be modified to the hypusine form, did not change when DHS and DOHH were coexpressed. These findings demonstrate that the overexpression of DHS and DOHH per se does not affect the eIF5A subcellular localization, but its hypusine modification status does, and further support the notion that eIF5A is exported from nucleus to cytoplasm in a hypusine-dependent manner.
Fig. 4. Comparison of the cellular distribution of GFP-eIF5A mutant proteins.
(A) direct visualization of GFP fluorescence after 48h of transfection. HeLa cells were transfected with vectors encoding GFP-eIF5A wt and mutants, without (upper panels) or with (lower panels) vectors encoding DHS and DOHH. (B) western blots of the above samples using AcLys and eIF5A antibodies (BD Bioscience).
Acetylation of cellular proteins has various effects on the activity of the proteins, by affecting protein-protein or protein-nucleic acid interactions, or by altering their subcellular localization. If eIF5A acetylation were to affect eIF5A localization, one might observe different cellular distribution among the acetylatable and nonacetylatable GFP-eIF5A precursor forms. However, upon direct visualization of the subcellular localization of GFP-eIF5A, little difference was observed between those acetylatable forms (wt, K50R, K68A) and those non-acetylatable forms (K47A, K47R and K50A) (Fig. 4A, upper panels, Fig 4B left panel). Therefore, Lys47 acetylation does not seem to be a major factor regulating eIF5A subcellular localization. However, effects of eIF5A acetylation cannot be entirely excluded, as one may not see a visible difference if the acetylated eIF5A represents a small fraction of the total GFP-eIF5A precursor.
Discussion
We have consistently observed differences in subcellular distribution between the endogenous eIF5A (hypusine form) and exogenously expressed eIF5A (non-hypusinated precursor) (Fig 3). Our data demonstrate that the hypusine modification of eIF5A promotes its localization in the cytoplasmic compartment where it is required for protein synthesis. Thus, hypusine modification not only converts an inactive precursor to an active eIF5A, but also directs its subcellular localization, probably by influencing its nuclear export by exportin 4.
Previous studies on eIF5A function or eIF5A localization in mammalian cells have been performed by overexpression of eIF5A (tagged or nontagged) alone [13, 15–17]. However, these studies did not address the locus or function of the active hypusinated eIF5A, since it was not known that the overexpressed protein is largely in the form of non-hypusinated eIF5A precursor (Fig. 1). Most of the endogenous eIF5A exists as the mature hypusine protein in mammalian cells, suggesting efficient modification by endogenous DHS and DOHH. On the other hand, the exogenous eIF5A produced by transfection using mammalian cDNA vectors is not effectively modified by the endogenous enzymes. Only after coexpression with DHS and DOHH, are the exogenous eIF5A precursors completely modified to the hypusine form (Fig. 1 and Ref 6). Poor modification of exogenous eIF5A is not likely to be due to reduced affinity of tagged-eIF5A for the modification enzymes, since even the non-tagged exogenous eIF5A precursor was poorly modified. Thus, differential modification of endogenous vs exogenous eIF5A cannot be simply explained by insufficient level of the endogenous enzymes. It may be due to differences between the expression pathways (transcription, translation and modification) from genomic DNA and cDNA vectors or differences in compartmentalization.
Exogenously produced eIF5A precursor exhibits a strong signal against AcLys antibody and is the major acetylated protein in HeLa and other mammalian cells. The primary acetylation site on eIF5A has been identified as Lys47. In yeast S. cerevisiae, a human eIF5A mutant K47R supported growth as well as the wild type, whereas K47A displayed a slow growth phenotype. K47D was totally inactive, suggesting that a basic charge at this position is critical for eIF5A activity in vivo [20]. Thus, eIF5A activity may be regulated in cells by the acetylation status at Lys 47 in a reversible manner. Although the steady state level of Lys47-acetylated eIF5A(Hpu) seems to be normally low, it may increase upon treatment with specific stimuli that cause activation of eIF5A acetyltransferase or inhibition of eIF5A deacetylase. It is not clear if eIF5A acetylation directly influences its subcellular localization, as little difference was observed between the distribution patterns of acetylatable and non-acetylatable GFP-eIF5A mutant proteins.
eIF5A localization has been studied in several laboratories, but somewhat inconsistent results have been reported as to its nuclear localization [13, 14]. Jao and Chen [15] partially resolved the problem, by demonstrating that different fixation methods (using MeOH or HCHO) yielded different images of eIF5A subcellular distribution and concluded that a more gentle formaldehyde fixation method preserves nuclear eIF5A from leakage. However, no distinction was made between endogenous eIF5A (hypusine form) and exogenous eIF5A (non-hypusinated precursor) in the interpretation. Our finding that exogenous eIF5A produced from transfection of mammalian cells with an eIF5A vector alone is largely the non-hypusinated eIF5A precursor permitted us to distinguish eIF5A precursor from hypusinated eIF5A. We demonstrate that hypusinated eIF5A, both the endogenous eIF5A(Hpu) and exogenous eIF5A(Hpu) (produced from cotransfection with eIF5A, DHS and DOHH vectors) is localized predominantly in the cytoplasm whereas non-hypusinated eIF5A(Lys) precursor was found in the nucleus as well as in the cytoplasm. Jao and Chen [15] concluded that nuclear entry of eIF5A is by passive diffusion through the nuclear pores, since eIF5A is a small protein and that there was no active nuclear-cytoplasmic shuttling of eIF5A from a heterokaryon fusion assay. However, their failure to detect nuclear-cytoplasmic shuttling might have been due to the fact that the GFP-eIF5A used for imaging was largely in the non-hypusinated precursor form, not the biologically active hypusine form.
eIF5A was identified as a substrate for exportin 4 [21] in HeLa cell lysates, using an exportin 4 affinity resin. Exportin 4 is a member of importin β-related receptors mediating many nucleocytoplasmic transport events. Furthermore, eIF5A binding to exportin 4 appeared to depend on the hypusine modification, since the non-hypusinated eIF5A precursor protein failed to bind to the exportin 4 resin, whereas the deoxyhypusine form elicited low exportin 4 binding in vitro. Recently, in vivo evidence for the interaction of eIF5A and exportin 4 (by indirect-immunofluorescence detection) was reported in mouse hepatocarcinoma cells [22]. Endogenous eIF5A(Hpu) was primarily localized in the cytoplasm, similar to our patterns. Depletion of exportin 4 by SiRNA altered the intracellular distribution of eIF5A(Hpu), resulting in whole cell distribution. Based on these and our findings, it is tempting to speculate that the hypusine-modification of eIF5A promotes its binding to exportin 4 and thereby its nuclear export. Another factor contributing to the cytoplasmic localization of eIF5A(Hpu) may be hypusine-dependent binding of eIF5A to its binding partners in the cytoplasm, including ribosomes. Although it is not known whether eIF5A has any nuclear function, depletion of exportin 4 by its SiRNA caused an increase in nuclear eIF5A, stimulation of growth and tumorigenicity of murine hepatocellular carcinoma cells [22]. The significance of nuclear accumulation of eIF5A precursor or hypusinated eIF5A in mammalian cell proliferation, tumorigenesis and apoptosis warrants further investigation.
Acknowledgments
We thank J. Silvio Gutkind (NIDCR/NIH) for the pCEFL/GFP vector and Edith C. Wolff (NIDCR, NIH) for critical reading of the manuscript.
Abbreviations
- eIF5A
eukaryotic translation initiation factor 5A
- eIF5A-1
primary isoform of eIF5A
- eIF5A(Lys)
non-hypusinated eIF5A precursor
- eIF5A(Dhp)
eIF5A intermediate containing deoxyhypusine
- eIF5A(Hpu)
eIF5A active form containing hypusine
- DHS
deoxyhypusine synthase
- DOHH
deoxyhypusine hydroxylase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J Biochem (Tokyo) 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen KY, Liu AY. Biochemistry and function of hypusine formation on eukaryotic initiation factor 5A. Biol Signals. 1997;6:105–109. doi: 10.1159/000109115. [DOI] [PubMed] [Google Scholar]
- 3.Zanelli CF, Valentini SR. Is there a role for eIF5A in translation? Amino Acids. 2007;33:351–358. doi: 10.1007/s00726-007-0533-0. [DOI] [PubMed] [Google Scholar]
- 4.Abbruzzese A, Park MH, Folk JE. Deoxyhypusine hydroxylase from rat testis. Partial purification and characterization. J Biol Chem. 1986;261:3085–3089. [PubMed] [Google Scholar]
- 5.Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benne R, Hershey JWB. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 7.Klier H, Csonga R, Joao HC, Eckerskorn C, Auer M, Lottspeich F, Eder J. Isolation and structural characterization of different isoforms of the hypusine-containing protein eIF-5A from HeLa cells. Biochemistry. 1995;34:14693–14702. doi: 10.1021/bi00045a010. [DOI] [PubMed] [Google Scholar]
- 8.Park MH. The essential role of hypusine in eukaryotic translation initiation factor 4D (eIF-4D). Purification of eIF-4D and its precursors and comparison of their activities. J Biol Chem. 1989;264:18531–18535. [PubMed] [Google Scholar]
- 9.Smit-McBride Z, Schnier J, Kaufman RJ, Hershey JW. Protein synthesis initiation factor eIF-4D. Functional comparison of native and unhypusinated forms of the protein. J Biol Chem. 1989;264:18527–18530. [PubMed] [Google Scholar]
- 10.Jao DL, Chen KY. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J Cell Biochem. 2006;97:583–598. doi: 10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- 11.Zanelli CF, Maragno AL, Gregio AP, Komili S, Pandolfi JR, Mestriner CA, Lustri WR, Valentini SR. eIF5A binds to translational machinery components and affects translation in yeast. Biochem Biophys Res Commun. 2006;348:1358–1366. doi: 10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]
- 12.Gregio APB, Cano VPS, Avaca JS, Valentini SR, Zanelli CF. eIF5A has a function in the elongation step of translation in yeast. BBRC. 2009;380:785–790. doi: 10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- 13.Ruhl M, Himmelspach M, Bahr GM, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington GK, Probst H, Bevec D, et al. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans- activation. J Cell Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi XP, Yin KC, Zimolo ZA, Stern AM, Waxman L. The subcellular distribution of eukaryotic translation initiation factor, eIF-5A, in cultured cells. Experimental Cell Research. 1996;225:348–356. doi: 10.1006/excr.1996.0185. [DOI] [PubMed] [Google Scholar]
- 15.Jao LED, Chen KY. Subcellular localization of the hypusine-containing eukaryotic initiation factor 5A by immunofluorescent staining and green fluorescent protein tagging. Journal of Cellular Biochemistry. 2002;86:590–600. doi: 10.1002/jcb.10235. [DOI] [PubMed] [Google Scholar]
- 16.Parreiras ESLT, Gomes MD, Oliveira EB, Costa-Neto CM. The N-terminal region of eukaryotic translation initiation factor 5A signals to nuclear localization of the protein. Biochem Biophys Res Commun. 2007;362:393–398. doi: 10.1016/j.bbrc.2007.07.185. [DOI] [PubMed] [Google Scholar]
- 17.Li AL, Li HY, Jin BF, Ye QN, Zhou T, Yu XD, Pan X, Man JH, He K, Yu M, Hu MR, Wang J, Yang SC, Shen BF, Zhang XM. A novel eIF5A complex functions as a regulator of p53 and p53-dependent apoptosis. J Biol Chem. 2004;279:49251–49258. doi: 10.1074/jbc.M407165200. [DOI] [PubMed] [Google Scholar]
- 18.Taylor CA, Sun Z, Cliche DO, Ming H, Eshaque B, Jin S, Hopkins MT, Thai B, Thompson JE. Eukaryotic translation initiation factor 5A induces apoptosis in colon cancer cells and associates with the nucleus in response to tumour necrosis factor alpha signalling. Exp Cell Res. 2007;313:437–449. doi: 10.1016/j.yexcr.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 19.Caraglia M, Marra M, Giuberti G, d’Alessandro AM, Budillon A, del Prete S, Lentini A, Beninati S, Abbruzzese A. The role of eukaryotic initiation factor 5A in the control of cell proliferation and apoptosis. Amino Acids. 2001;20:91–104. doi: 10.1007/s007260170050. [DOI] [PubMed] [Google Scholar]
- 20.Cano VS, Jeon GA, Johansson HE, Henderson CA, Park JH, Valentini SR, Hershey JW, Park MH. Mutational analyses of human eIF5A-1--identification of amino acid residues critical for eIF5A activity and hypusine modification. Febs J. 2008;275:44–58. doi: 10.1111/j.1742-4658.2007.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipowsky G, Bischoff FR, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay U, GD Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. The EMBO Journal. 2000;19:4362–4371. doi: 10.1093/emboj/19.16.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, McCombie WR, Wigler M, Hicks J, Hannon GJ, Powers S, Lowe SW. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]