Abstract
The spliceosome uses numerous strategies to regulate its function in mRNA maturation. Ubiquitin regulates many cellular processes, but its potential roles during splicing are unknown. We have developed a new strategy that reveals a direct role for ubiquitin in the dynamics of splicing complexes. A ubiquitin mutant (I44A) that can enter the conjugation pathway but is compromised in downstream functions diminishes splicing activity by reducing the levels of the U4/U6-U5 small nuclear ribonucleoprotein (snRNP). Similarly, an inhibitor of ubiquitin’s protein-protein interactions, ubistatin A, reduces U4/U6-U5 triple snRNP levels in vitro. When ubiquitin interactions are blocked, ATP-dependent disassembly of purified U4/U6-U5 particles is accelerated, indicating a direct role for ubiquitin in repressing U4/U6 unwinding. Finally, we show that the conserved splicing factor Prp8 is ubiquitinated within purified triple snRNPs. These results reveal a previously unknown ubiquitin-dependent mechanism for controlling the pre-mRNA splicing pathway.
Noncoding introns are removed from pre-mRNAs by the spliceosome through two sequential transesterifications (reviewed in ref. 1). The spliceosome is composed of five small nuclear RNAs (snRNAs) and more than 100 proteins, some of which associate with individual snRNAs to form snRNP particles1. During each round of splicing, the spliceosome recognizes splice-site sequences within the pre-mRNA and assembles on the substrate through extensive rearrangements of its components. To ensure successful splicing, these rearrangements are highly coordinated and seem to be driven by DExD/H-box ATPases2.
According to the classical model of spliceosome assembly1, binding of the U1 snRNP to the conserved 5′ splice site (5′ ss) commits a pre-mRNA to splicing, and the U2 snRNP then binds to the intronic branch site to form the pre-spliceosome. The U4/U6-U5 triple snRNP (in which the U4 and U6 snRNAs are extensively base paired) is then added, forming a catalytically inert complex. For activation, both the U4/U6 duplex and the U1–5′ ss interactions are disrupted in favor of alternative interactions to generate the active spliceosome. The ensuing catalytic steps of splicing are then followed by mRNA release and spliceosome disassembly. Sequential rearrangements of components occur on the pre-mRNA and seem to be driven by specific ATPases that are modulated by other factors in the spliceosome, such as Prp8 (ref. 3) and Snu114 (ref. 4). A prevailing theme in spliceosome dynamics is the fluidity of protein-protein interactions, which can be regulated by phosphorylation5 and potentially by other post-translational modifications such as ubiquitination.
Ubiquitination of proteins is brought about by the sequential action of three classes of enzymes: an ATP-dependent ubiquitin-activating enzyme (E1 or Uba), a ubiquitin-conjugating enzyme (E2 or Ubc) and a ubiquitin ligase (E3; reviewed in ref. 6). E3 enzymes attach ubiquitin to target proteins, nearly always via an isopeptide bond between the C terminus of ubiquitin and the side chain amino group of a lysine residue6. Ubiquitin functions in many diverse cellular processes7 in addition to its canonical role in targeting proteins to the proteasome8. Similarly to phosphorylation, ubiquitination serves as an efficient regulatory signal because the extent and timing of ubiquitination can be modulated by the balanced action of E3 ubiquitin ligases and deubiquitinating enzymes9. In addition, by virtue of variable poly-ubiquitin chain linkages and sites of target modification, ubiquitination can confer versatile topological surfaces on the modified protein, thus affecting its function. The regulatory roles of ubiquitin generally require the recognition of ubiquitin–target protein conjugates by ubiquitin binding domains (UBDs)10.
Indirect evidence has led to speculation that ubiquitin may regulate splicing. For example, ubiquitin and ubiquitin-like proteins have been shown to copurify with splicing complexes11,12; conversely, ubiquitinated splicing factors have been identified in a proteomic screen13. The essential splicing factor Prp19 and its human ortholog contain a U-box domain with E3 ubiquitin ligase activity in vitro14,15, although specific target proteins have not been reported. Several other domains related to the ubiquitin pathway have been identified in important spliceosome proteins16–19, and one such domain (the Jab1/MPN domain of the essential U5 snRNP component Prp8)20,21 shows ubiquitin binding activity16. Finally, in fission yeast, deletion of the hub1 gene (which encodes a ubiquitin-like protein) leads to splicing defects22. Although these observations suggest that ubiquitin and related proteins function in splicing, the specific mechanisms by which ubiquitin regulates the spliceosome are undefined. This has been challenging to study, owing in part to the dynamic nature of the splicing machinery and the pleiotropic influence of ubiquitin on numerous cellular processes.
To study the function of ubiquitin in pre-mRNA splicing, we have used an in vitro strategy in budding yeast that uncouples splicing from other cellular pathways. We have used two independent means to disrupt ubiquitin recognition by UBDs in splicing extracts, and in both cases spliceosome function is blocked. The in vitro splicing defects can be traced to a common, essential step in spliceosome assembly, namely the maintenance of U4/U6-U5 triple snRNP levels through regulation of U4/U6 duplex unwinding. Purified U4/U6-U5 particles show accelerated U4/U6 unwinding activity when ubiquitin recognition is blocked or when ubiquitin conjugates are removed, indicating that ubiquitin has a direct role in maintaining the U4/U6-U5 triple snRNP by repressing U4/U6 unwinding. Finally, we show that Prp8 is present as a ubiquitin conjugate in affinity-purified particles enriched for triple snRNPs, suggesting that ubiquitin modifies the ability of Prp8 to regulate the U4/U6 unwinding activity of Brr2 (ref. 3). Because U4/U6 unwinding events in the triple snRNP and in the assembled spliceosome are thought to share a common mechanism4,23,24, our results suggest a similar function for ubiquitin in governing U4/U6 unwinding during spliceosome activation.
RESULTS
A ubiquitin mutant inhibits splicing in vitro
To investigate the role of ubiquitin in pre-mRNA splicing, we devised a new approach that tests the requirement for ubiquitin-UBD interactions in vitro. Most known ubiquitin-UBD interactions involve a specific hydrophobic patch on the surface of ubiquitin (reviewed in ref. 10). Ile44 is a crucial residue within this patch, because I44A mutant ubiquitin is severely impaired in most ubiquitin-UBD interactions tested to date10,16. The ubiquitin-conjugation machinery is capable of attaching I44A ubiquitin to protein targets in two reported cases25, indicating that at least a subset of ubiquitination factors can use I44A ubiquitin as a substrate. We reasoned that the conjugation of I44A mutant ubiquitin to potential target proteins could disrupt spliceosome function at any point that requires the recognition of a ubiquitin conjugate by a UBD.
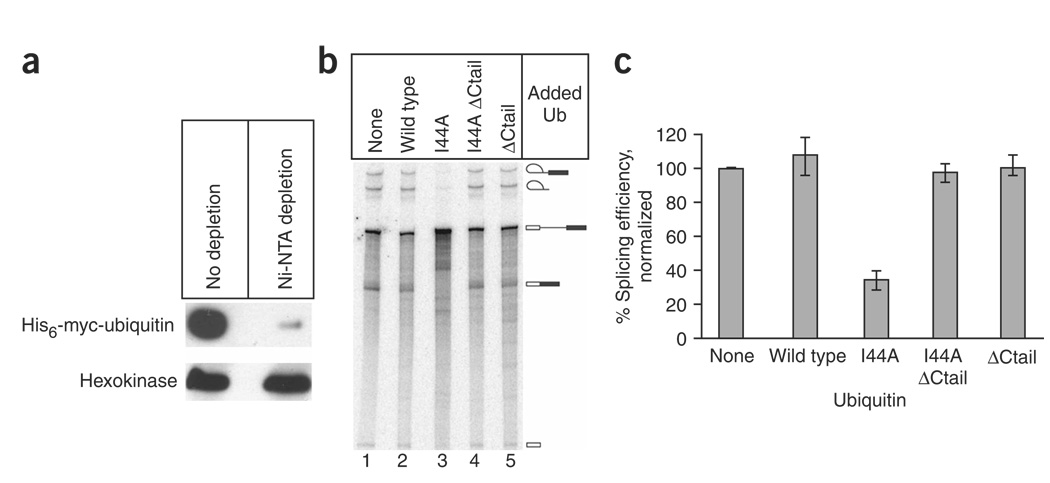
In budding yeast, the I44A ubiquitin mutation is lethal26, so to deplete and reconstitute ubiquitin in extract we used a strain in which all ubiquitin molecules carry N-terminal hexahistidine (His6) and myc-epitope tags27. We made splicing extract from this strain and then depleted ubiquitin from the extract using nickel–nitrilotriacetic acid (Ni2+-NTA) chromatography. This technique succeeded in removing most ubiquitin from the extract, although small amounts remained (Fig. 1a). The partially ubiquitin-depleted extract retained splicing activity, and the addition of purified recombinant wild-type ubiquitin had no appreciable effect (Fig. 1b, lanes 1 and 2, and Fig. 1c). In contrast, the addition of equivalent amounts of I44A ubiquitin resulted in a substantial decrease in splicing activity in vitro (Fig. 1b, lane 3, and Fig. 1c). This inhibition was observed at I44A ubiquitin concentrations as low as 0.5 mM (data not shown). Splicing was also substantially inhibited by I44A ubiquitin in extracts that were not depleted of endogenous ubiquitin (Supplementary Fig. 1 online). The I44A ubiquitin was conjugated to proteins in a crude splicing extract, although at a slightly lower efficiency than for wild-type ubiquitin (Supplementary Fig. 2 online). The ability of the ubiquitin-depleted extract to splice (Fig. 1b, lane 1) could be due to the presence of the residual endogenous ubiquitin (Fig. 1a), given that ubiquitin is present in an approximately 100-fold excess over most splicing factors in yeast cells28.
Figure 1.
A mutant form of ubiquitin (Ub) inhibits pre-mRNA splicing activity in vitro. (a) Splicing extract from a Saccharomyces cerevisiae strain expressing His6-myc–tagged ubiquitin was depleted of ubiquitin by Ni2+ affinity chromatography. Ubiquitin levels before and after depletion were assayed by probing a western blot with anti-myc antibodies. Hexokinase (below) was used as a control. (b) Ubiquitin-depleted splicing extract was used in 30-min in vitro splicing assays following the addition of wild-type and mutant forms of ubiquitin, as specified above each lane. Each ‘ΔCtail’ mutant had a four-amino-acid truncation at its C terminus to prevent entry into the target conjugation pathway. The mobilities of pre-mRNA, splicing intermediates and splicing products are indicated on the right. (c) Data from b and two equivalent splicing experiments were quantitatively analyzed with a PhosphorImager, and splicing efficiency (defined as (spliced products + splicing intermediates)/total radiolabeled RNA, normalized to the nonsupplemented extract) was plotted. The identities of the added ubiquitin derivatives are given at the bottom. Error bars indicate the range of the three experiments.
To determine whether the inhibitory activity of I44A ubiquitin requires entry into the target conjugation pathway, we expressed and purified recombinant I44A ubiquitin that lacked the C-terminal tail. Mutations in these tail residues do not affect the folding of ubiquitin’s globular domain29, but the ‘tailless’ deletion nonetheless abolishes conjugation to target proteins26,30. The splicing inhibition caused by the I44A mutation was completely alleviated by the removal of the mutant protein’s C-terminal tail (ΔCtail; Fig. 1b, lane 4, and Fig. 1c). Thus, the simple presence of high levels of the I44A mutant ubiquitin globular domain is not sufficient to inhibit splicing. These results strongly indicate that I44A ubiquitin entry into the target conjugation pathway is required for its effect on spliceosome function. This could be due to dominant interference of the I44A ubiquitin with the target conjugation pathway itself, or to a failed interaction between an I44A ubiquitin–target protein conjugate and a spliceosomal UBD. Either way, the results suggest a functional role for ubiquitination in the pre-mRNA splicing pathway. The proteasome inhibitor MG132 (ref. 31) had no effect on pre-mRNA splicing activity in vitro (Supplementary Results and Supplementary Fig. 3 online), suggesting that ubiquitin’s role in splicing is proteasome independent.
I44A ubiquitin blocks pre-spliceosome maturation
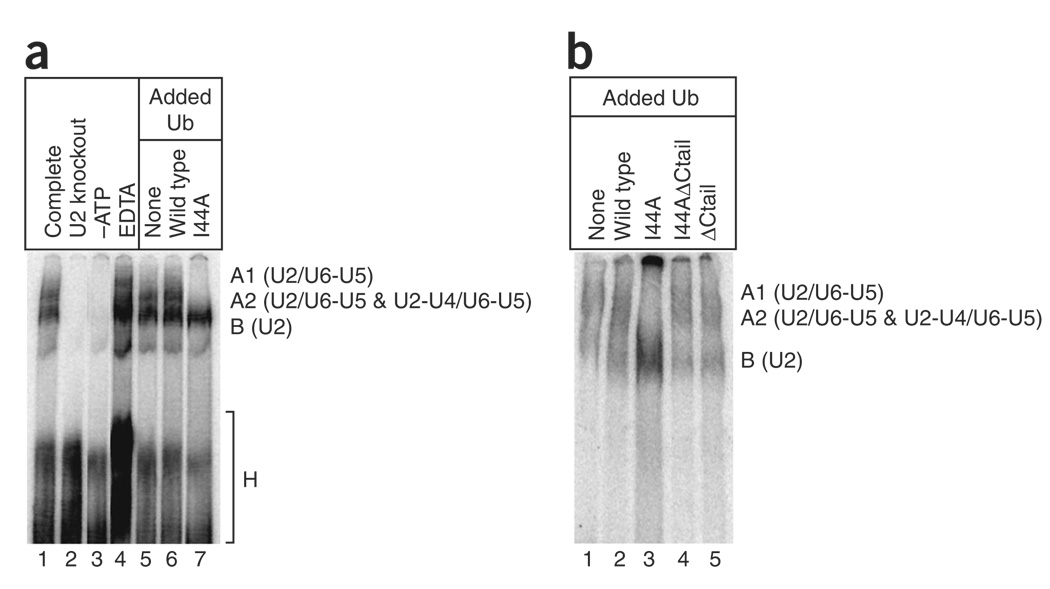
Previous native gel analyses have delineated a spliceosome assembly pathway in yeast (reviewed in ref. 1). The B complex (also known as the pre-spliceosome) contains the U2 snRNP bound to the intron branch point and progresses to the A2-1 complex upon the addition of the U4/U6-U5 triple snRNP. Subsequent RNA rearrangements, including the unwinding of U4 and U6, lead to the A1 and catalytically active A2-2 complexes. To determine whether the I44A ubiquitin blocks any of these stages of spliceosome assembly, we performed a native gel analysis. When ubiquitin-depleted splicing extract was supplemented with buffer or wild-type ubiquitin, the profile was indistinguishable from that of a control reaction (Fig. 2a, lanes 1, 5 and 6). In contrast, the addition of I44A ubiquitin led to a strong block in spliceosome assembly at the B complex stage (Fig. 2a, lane 7). Northern analyses of native gels (Supplementary Results and Supplementary Fig. 4 online) confirmed the identities of the complexes observed in Figure 2a. Removal of the C-terminal tail from I44A ubiquitin relieved the block (Fig. 2b, lanes 3 and 4, and Supplementary Fig. 4), suggesting that I44A ubiquitin entry into the target conjugation pathway stalls assembly at the B complex. The I44A ubiquitin could inhibit pre-spliceosome maturation by preventing the formation of functional U4/U6-U5 triple snRNP or by blocking the stable addition of correctly preformed triple snRNP to the B complex.
Figure 2.
The U2 snRNP-containing pre-spliceosome accumulates in the presence of I44A ubiquitin. (a) Radiolabeled pre-mRNA was incubated in splicing extract for 30 min, and the assembled complexes were then analyzed by native gel electrophoresis. The mobilities of H (nonspecific), B (U2), A2 (U2-U4/U6-U5 and U2/U6-U5), and A1 (U2/U6-U5) complexes are indicated on the right. The kinetically distinct A2-1 and A2-2 complexes47 were not resolved in this experiment. Lanes 1–4 were from control reactions that served to guide the identification of the complexes (lane 1, standard splicing reaction; lane 2, extract depleted of U2 snRNA by oligonucleotide-directed RNase H cleavage; lane 3, extract depleted of ATP by incubation with glucose; lane 4, extract supplemented with 5 mM EDTA, which leads to A1 complex accumulation47). Lanes 5–7 are from reactions with ubiquitin-depleted extract supplemented with buffer (lane 5), 1 mM wild-type ubiquitin (lane 6) or 1 mM I44A ubiquitin (lane 7). (b) As in a, except that the complete, U2 knockout, –ATP and EDTA reactions were omitted, and reactions supplemented with I44A ΔCtail ubiquitin (lane 4) and ΔCtail ubiquitin (lane 5) were included.
I44A ubiquitin interferes with U4/U6-U5 accumulation
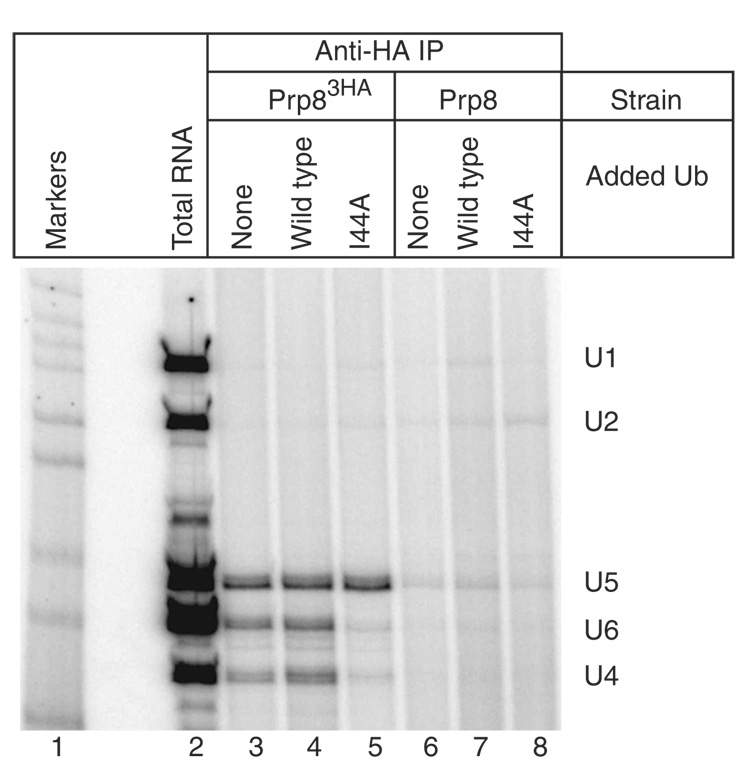
To determine whether the spliceosome assembly defect induced by I44A ubiquitin is due to a defect in the U4/U6-U5 triple snRNP, we assayed U4/U6-U5 levels in the presence of I44A ubiquitin in vitro. Splicing extract prepared from a strain expressing triple-HA–tagged Prp8 (Prp83HA, a stable component of the U5 snRNP and the U4/U6-U5 triple snRNP32) was incubated with wild-type or I44A ubiquitin in the presence of ATP (to allow ubiquitin activation) and then immunoprecipitated with anti-HA monoclonal antibodies. Immunoprecipitated RNAs were then used as templates in reverse-transcription reactions with radiolabeled primers specific for the U1, U2, U4, U5 and U6 spliceosomal snRNAs. As shown in Figure 3, substantial amounts of the U4, U5 and U6 snRNAs were found in the immunoprecipitates in the absence of exogenous ubiquitin or in the presence of exogenous wild-type ubiquitin (lanes 3 and 4). When I44A ubiquitin was included, however, little U4 and U6 snRNA was present in the pellet, even though U5 snRNA levels were undiminished (Fig. 3, lane 5). These results indicate that stable U4/U6-U5 triple snRNP (but not the Prp8-containing U5 snRNP) fails to accumulate in the presence of I44A ubiquitin. U4/U6-U5 levels are not affected by I44A ubiquitin lacking the C-terminal tail (data not shown), indicating that conjugation is required for the inhibitory effect of I44A ubiquitin. Endogenous annealed U4/U6 snRNAs were readily detected in the presence of I44A ubiquitin (Supplementary Fig. 5 online), suggesting that the triple snRNP defect is not due to a prior defect in U4/U6 di-snRNP formation or stability.
Figure 3.
I44A ubiquitin blocks spliceosome assembly by interfering with U4/U6-U5 triple snRNP accumulation. Splicing extract from a strain expressing a triple-HA–tagged U5 snRNP component (Prp83HA) was immunoprecipitated (IP) with anti-HA antibodies following the addition of buffer (lane 3), 1 mM wild-type ubiquitin (lane 4) or 1 mM I44A ubiquitin (lane 5). Parallel control immunoprecipitations from an isogenic untagged strain are shown in lanes 6–8. RNAs extracted from the immunoprecipitates were used as templates in reverse-transcriptase reactions with radiolabeled primers specific to the U1, U2, U4, U5 and U6 snRNAs. The mobilities of the primer extension products are indicated on the right. Lane 1, radiolabeled DNA size markers; lane 2, primer extension with total RNA from the splicing extract used in lanes 3–5.
Ubistatin A mirrors the effects of I44A ubiquitin
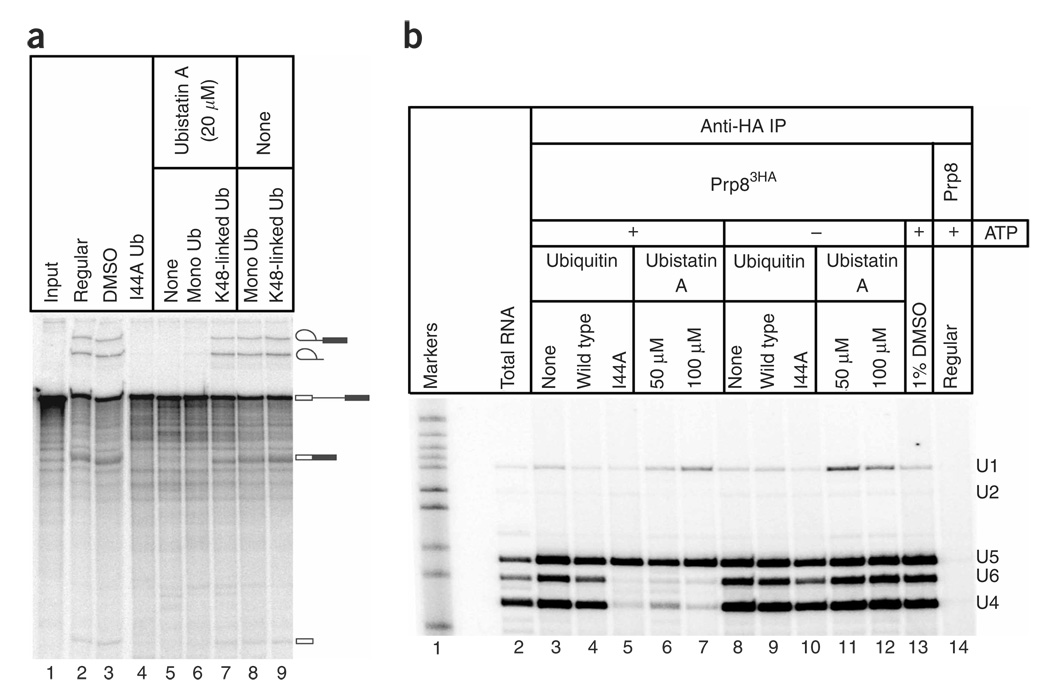
To provide independent evidence of a role for ubiquitin in spliceosome assembly, we sought an alternative approach that did not rely upon the apparent dominant-negative effect exerted by mutant ubiquitin. For this we turned to ubistatin A, a small molecule that inhibits protein degradation by the proteasome33. Ubistatin A binds in the vicinity of the I44-containing hydrophobic surface of ubiquitin33, thereby disrupting ubiquitin’s protein-protein interactions. As anticipated from our earlier results, in vitro splicing was strongly inhibited by ubistatin A solubilized in DMSO (Fig. 4a, lane 5), but not by DMSO alone (lane 3). Nearly complete inhibition was observed in crude splicing extracts at ubistatin A concentrations of 15–20 µM (Fig. 4a, lane 5, and data not shown). The inhibitory effect of ubistatin A could be relieved by preincubation with a three-fold molar excess of K48-linked tetraubiquitin chains (Fig. 4a, lane 7), indicating that the inhibition is in fact due to ubistatin A rather than any potential contaminant. Preincubation with an excess of monoubiquitin was much less effective at blocking ubistatin A’s inhibitory effect (Fig. 4a, lane 6), consistent with the lower affinity of ubistatin A for monoubiquitin than for K48-linked chains33. Ubistatin A does not block ubiquitin conjugation to the one target protein that has been examined thus far33, suggesting that it is more likely to affect the downstream recognition of ubiquitin–target protein conjugates. Although the levels of ubistatin A required to inhibit splicing are approximately 30-fold greater than those required to inhibit proteasomes33, this higher concentration requirement could reflect that (i) ubistatin A is inhibiting an intramolecular interaction in splicing (see below), rather than an intermolecular interaction, and (ii) ubistatin A binds to K48-linked polyubiquitin more tightly than to monoubiquitin or other types of ubiquitin chains, which may be the relevant forms of ubiquitin in the spliceosome (see below). Our observation that ubistatin A inhibits splicing greatly strengthens the conclusion that ubiquitin recognition is important for pre-mRNA splicing.
Figure 4.
The ubiquitin (Ub) binding small molecule ubistatin A recapitulates the inhibitory effects of I44A ubiquitin. (a) Splicing extract was used in 30-min in vitro splicing assays following the addition of I44A ubiquitin (lane 4), 20 µM ubistatin A in DMSO (lanes 5–7) or DMSO alone (lanes 3, 8 and 9). In lanes 6–9, the ubistatin A or DMSO was preincubated with monoubiquitin or K48-linked tetraubiquitin chains before addition to the splicing extract. The mobilities of pre-mRNA, splicing intermediates and splicing products are indicated on the right. (b) Splicing extract from a strain expressing a triple-HA–tagged U5 snRNP component (Prp83HA) was immunoprecipitated (IP) with anti-HA antibodies following the addition of buffer (lanes 3 and 8), 1 mM wild-type ubiquitin (lanes 4 and 9), 1 mM I44A ubiquitin (lanes 5 and 10), DMSO (lane 13), 50 µM ubistatin A in DMSO (lanes 6 and 11) or 100 µM ubistatin A in DMSO (lanes 7 and 12). A parallel control immunoprecipitation from an isogenic untagged strain is shown in lane 14. In lanes 8–12, ATP was depleted after an initial 10-min incubation in the presence of endogenous ATP to permit ubiquitin activation. Extracted RNAs were used as templates in reverse-transcriptase reactions with radiolabeled primers specific to the U1, U2, U4, U5 and U6 snRNAs. The mobilities of the primer extension products are indicated on the right.
To determine whether the inhibitory effect of ubistatin A on splicing was due to a reduction in the triple snRNP, we analyzed U4/U6-U5 levels in the presence of ubistatin A. We immunoprecipitated Prp83HA from splicing extracts and then monitored the co-immunoprecipitation of spliceosomal snRNAs by primer extension. Figure 4b shows that ubistatin A in DMSO caused a dramatic decrease in U4 and U6 snRNA co-immunoprecipitation in the presence of ATP (lanes 6 and 7) whereas DMSO alone had no effect (lane 13). These results corroborate our earlier conclusion that ubiquitin recognition is important for maintaining steady-state triple snRNP levels in the presence of ATP (Fig. 3).
In yeast splicing extracts, the U4, U5 and U6 snRNAs (and their associated proteins) undergo continuous ATP-dependent cycling between the U4/U6-U5 triple snRNP and the dissociated particles4,23,34. We therefore used the co-immunoprecipitation and primer extension assay to test whether ATP was required for the loss of stable U4/U6-U5 triple snRNP in the presence of either I44A ubiquitin or ubistatin A. To deplete ATP after ubiquitin conjugation, we first incubated splicing extract with ubiquitin and endogenous ATP to allow ubiquitin activation; only then did we deplete the reactions of ATP by the addition of glucose. Control experiments confirmed the conjugation of exogenous ubiquitin in the crude extract under these conditions (data not shown). Figure 4b shows that ATP depletion results in stable U4/U6-U5, even in the presence of I44A ubiquitin (lane 10) or ubistatin A (lanes 11 and 12), indicating that the loss of U4/U6-U5 induced by these agents requires ATP. Affinity-purified triple-snRNP particles formed under these conditions are competent to disassemble upon readdition of ATP (see below), demonstrating that they were not inactivated irreversibly by these treatments. These results indicate that I44A ubiquitin and ubistatin A modulate an ATP-dependent phase of the triple-snRNP cycle.
Ubiquitin recognition represses U4/U6-U5 disassembly
Because I44A ubiquitin and ubistatin A decrease U4/U6-U5 levels at steady state (Fig. 3 and Fig 4), the lower U4/U6-U5 levels may result from a reduction in the rate of triple snRNP formation, an increase in the rate of ATP-dependent triple snRNP disassembly or both. Triple snRNP disassembly requires U4/U6 unwinding, a reaction that is promoted by the DExD/H-box ATPase Brr2 (refs. 4,23,35,36) and regulated by the GTPase Snu114 (ref. 4); in its GTP-bound state, Snu114 promotes unwinding and, in its GDP-bound state, represses unwinding. Additionally, U4/U6 unwinding is regulated by the ubiquitin binding protein Prp8 (refs. 3,16,37).
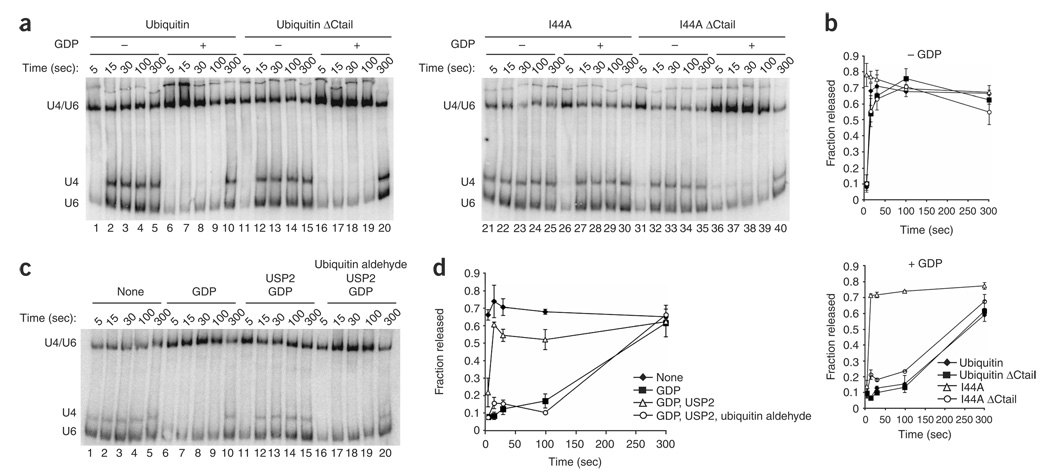
To investigate whether U4/U6 unwinding is downregulated by ubiquitin, we tested whether I44A ubiquitin promoted U4/U6 unwinding in triple snRNP particles affinity-purified using TAP-tagged Brr2. Specifically, we allowed conjugation of I44A mutant or wild-type ubiquitin to targets during a preincubation in extract then purified the triple snRNP using IgG-Sepharose and assayed for U4/U6 unwinding by native gel analysis4. Indeed, the time required for unwinding decreased at least three-fold following preincubation with I44A, as compared to preincubation without ubiquitin or with wild-type ubiquitin (Fig. 5a, lanes 1–5 and 21–25, Fig. 5b and data not shown). We also assayed for derepression of U4/U6 unwinding after repressing unwinding with GDP (ref. 4) and found that pre-incubation with I44A ubiquitin but not wild-type ubiquitin derepressed U4/U6 unwinding (Fig. 5a, lanes 6–10 and 26–30, and Fig. 5b). In contrast, tailless I44A ubiquitin did not derepress U4/U6 unwinding (Fig. 5a, lanes 36–40), indicating that derepression by I44A ubiquitin requires conjugation to a target. These data suggest that an interaction between ubiquitin and a UBD maintains triple snRNP levels by repressing U4/U6 unwinding.
Figure 5.
Conjugated mutant I44A ubiquitin or the deubiquitinating cysteine protease USP2 derepresses U4/U6 unwinding in purified U4/U6-U5 snRNPs. (a) Extract from a strain expressing TAP-tagged Brr2 (yJPS776) was incubated with wild-type or I44A ubiquitin in the presence of ATP to allow conjugation; glucose was then added to deplete ATP, and the U4/U6-U5 particle was immunopurified and assayed for U4/U6 unwinding. Cold-phenol–extracted RNA was fractionated by native gel electrophoresis and the U4 and U6 snRNAs were detected by northern blotting as described4. The mobilities of annealed U4/U6 (corresponding to the triple snRNP) and free U4 and U6 (corresponding to the free U4 and U6 snRNPs) are indicated on the left. Reactions were incubated with ATP; with or without GDP; and with or without wild-type, tailless wild-type (Ubiquitin ΔCtail), mutant (I44A) or tailless mutant (I44A ΔCtail) ubiquitin, as indicated. (b) Data from a and a replicate experiment were quantified and plotted as shown. Error bars represent the range of the two experiments. (c) U4/U6-U5 particles were immunopurified from a Prp28-TAP strain (yJPS1004). The triple snRNP was preincubated with 2.5 µM of the ubiquitin deconjugating enzyme USP2 and in the indicated reaction USP2 itself was preincubated with 2.5 µM ubiquitin aldehyde, a USP2 inhibitor. All reactions were initiated with ATP, and GDP was included or omitted as indicated. U4/U6 unwinding was assayed as described4. (d) Data from c and a replicate experiment were quantitated and plotted as shown. Error bars represent the range of the two experiments.
Because the above experiment does not rule out an indirect effect of I44A, we next tested whether a ubiquitin-UBD interaction repressed U4/U6 unwinding directly. Indeed, using triple snRNPs assembled in vivo and purified using TAP-tagged Prp28 (ref. 4), the time required for U4/U6 unwinding decreased approximately ten-fold in the presence of ubistatin A (Supplementary Results and Supplementary Fig. 6a–d online). Ubistatin A was also able to partially reverse the repression of U4/U6 unwinding by GDP (Supplementary Fig. 6e,f). Moreover, U4/U6 unwinding was similarly derepressed by pretreating purified triple snRNPs with the general deubiquitinating cysteine protease, USP2 (Fig. 5c,d). When U4/U6 unwinding was repressed by GDP, USP2 derepressed U4/U6 unwinding by approximately 20-fold (Fig. 5c). Derepression by USP2 was eliminated by pretreating USP2 with ubiquitin aldehyde, a potent and specific inhibitor of this class of deubiquitinating enzymes38 (Fig. 5c,d). Additionally, the effect of USP2 on U4/U6 unwinding is subject to partial product inhibition by excess free ubiquitin and is inhibited by iodoacetamide, which alkylates cysteines (Supplementary Fig. 7 online). These data confirm that ubiquitin stabilizes the triple snRNP by downregulating U4/U6 unwinding. Furthermore, these data show that strong repression of U4/U6 unwinding requires both GDP and ubiquitin recognition.Most notably, these data demonstrate that triple snRNP dynamics are controlled by ubiquitin directly and imply that an intrinsic component of the triple snRNP is ubiquitinated.
Identification of Prp8 as a ubiquitin conjugate
To identify ubiquitin conjugates within U4/U6-U5 triple snRNP particles, we generated a strain in which Brr2 was TAP-tagged at its C terminus and ubiquitin was His6- and myc-tagged. We affinity-purified U4/U6-U5 particles, denatured the resulting samples in 8 M urea and selected ubiquitin conjugates with Ni2+-NTA resin. The resulting proteins were then identified by MudPIT (multidimensional protein identification technology) analysis. In our first experiment, we recovered the ubiquitin conjugates from the Ni2+-NTA resin by low-pH elution and trichloroacetic acid (TCA) precipitation and then analyzed tryptic digests of the eluate in duplicate using tandem MS. We then repeated the purification and analysis but trypsinized the Ni2+-bound proteins directly on the Ni2+-NTA resin to bypass the low-yield TCA-precipitation step. Only one known splicing factor, Prp8, was reproducibly detected: 8 unique Prp8 peptides (from 10 total spectra combined from duplicate analyses) were identified in the first preparation, and 13 unique Prp8 peptides (from 14 total spectra) were identified in the second preparation. The total number of unique Prp8 peptides from all analyses was 18, which represents 7.6% sequence coverage of this ~280-kDa protein (Supplementary Table 1 online). Notably, no Prp8 peptides were identified in parallel samples from a negative control BRR2-TAP strain expressing untagged ubiquitin. Peptides from Prp43 and Brr2 were also identified at low levels in at least one MS run, but these identifications were not reproducible across all of our MS analyses.
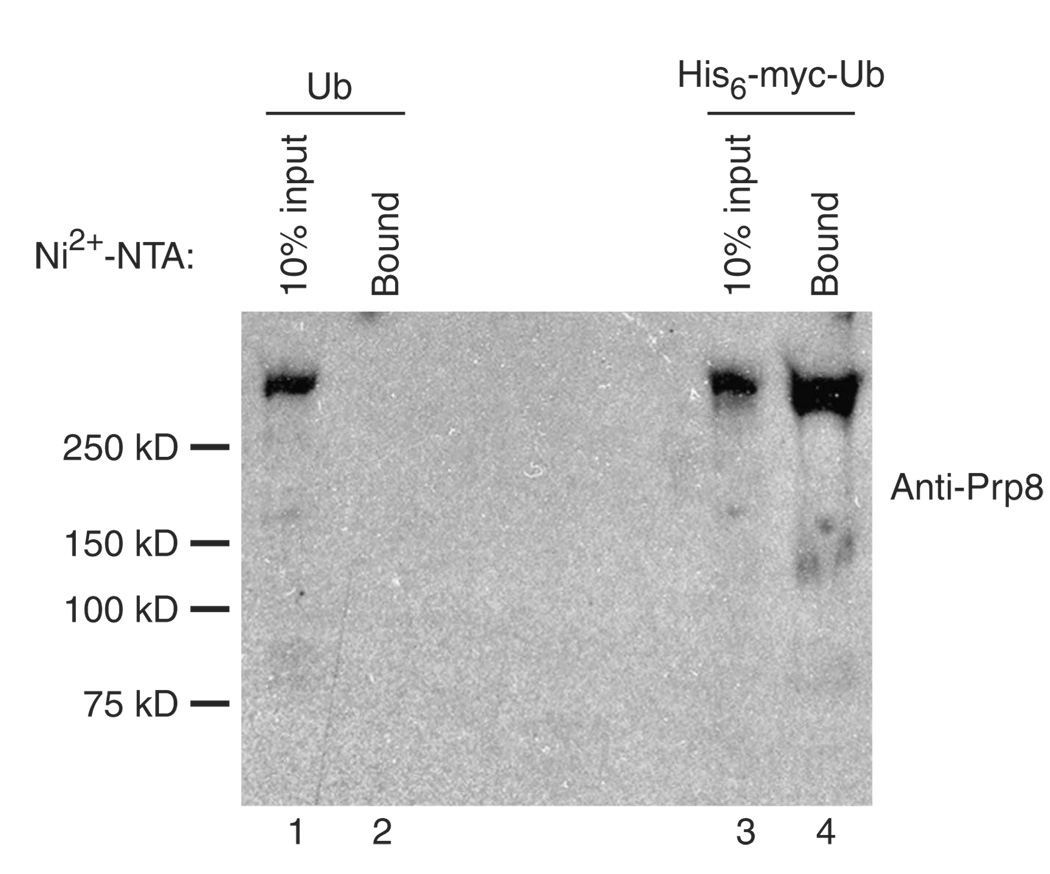
To confirm the ubiquitination of Prp8, we repeated the purification but analyzed the material bound to Ni2+-NTA resin by probing a western blot with antibodies against Prp8 (Fig. 6).With triple snRNPs purified from isogenic strains expressing either tagged or untagged ubiquitin, a protein of the expected mobility was detected (lanes 1 and 3). However, subsequent denaturing purification with Ni2+-NTA resin yielded detectable Prp8 from only the strain expressing tagged ubiquitin (lanes 2 and 4). In contrast, Brr2-TAP, which was detected in purified triple snRNPs, was not observed in either Ni2+-NTA–purified sample (data not shown), establishing the specificity of the purification. These results confirm our conclusion that Prp8 is ubiquitinated within the U4/U6-U5 triple snRNP and (together with our earlier experiments) argue strongly that Prp8 ubiquitination, and the subsequent recognition of the Prp8-ubiquitin conjugate, lead to the repression of Brr2’s ATP-dependent U4/U6 unwinding activity.
Figure 6.
Affinity-purified U4/U6-U5 triple snRNPs contain Prp8-ubiquitin conjugates. U4/U6-U5 triple snRNP particles were immunopurified from a Brr2-TAP strain expressing wild-type ubiquitin (Ub, yJPS1274) or His6-myctagged ubiquitin (His6-myc-Ub, yJPS1275). Following TEV elution, the purified snRNPs were bound to Ni2+-NTA under denaturing conditions. The Ni2+-NTA–bound material (Bound), as well as one-tenth of the TEV eluates that were used in the Ni2+-NTA purification (10% input), were separated on a 4–20% protein gel and subjected to western analysis using antibodies against Prp8. Mobilities of molecular weight markers are given on the left.
DISCUSSION
We have used a new strategy to show that a ubiquitin mutant (I44A) with diminished capacity for protein-protein interactions strongly inhibits pre-mRNA splicing in vitro. This inhibition can be rescued by removing the C-terminal tail of the I44A mutant ubiquitin, indicating that I44A ubiquitin inhibits splicing through its conjugation to a target protein. Further analysis of the splicing defect revealed that I44A ubiquitin diminishes steady-state levels of the U4/U6-U5 triple snRNP, which in turn stalls spliceosome assembly at the B complex (pre-spliceosome). Corroborating these results, ubistatin A, which binds to the I44-containing surface of ubiquitin33, mimics the apparent dominant-negative effect of the mutant ubiquitin. Furthermore, the reduction in U4/U6-U5 triple snRNP levels in both cases can be attributed to an acceleration of U4/U6 unwinding that is likely to result from occlusion of conjugated ubiquitin. Moreover, pretreatment of purified U4/U6-U5 triple snRNPs with the deubiquitinating enzyme USP2 similarly accelerates U4/U6 unwinding, providing further evidence that ubiquitin represses U4/U6 unwinding directly. Indeed, a triple snRNP component (Prp8) that is already implicated in controlling U4/U6 unwinding3,16,37 is ubiquitinated in affinity-purified particles, suggesting a specific mechanism by which ubiquitin represses U4/U6 unwinding. Prp8-ubiquitin conjugates have also been identified, using a distinct methodology, in samples enriched for U4/U6-U5 triple snRNPs (S. Stevens, personal communication).
Our finding that USP2, I44A ubiquitin and ubistatin A each accelerate U4/U6 unwinding strongly suggests that ubiquitin normally serves to suppress U4/U6 unwinding by Brr2, thus stabilizing the triple snRNP under steady-state conditions (Fig. 7). We cannot exclude the possibility that ubiquitin recognition also has a role in promoting triple snRNP formation. Our detection of a Prp8-ubiquitin conjugate, and our previous demonstration of ubiquitin binding activity by Prp8’s Jab1/MPN domain16, suggest a model in which Prp8 establishes an intramolecular interaction between its Jab1/MPN domain and conjugated ubiquitin. Prp8’s affinity for ubiquitin is decreased with the I44A mutant16, and this could readily account for I44A ubiquitin’s ability to disrupt triple snRNP accumulation. Furthermore, mutations in Prp8 that compromise its ubiquitin binding activity also diminish U4/U6-U5 triple snRNP levels16. In some cases, UBDs promote the ubiquitination of the proteins in which they reside, can limit the conjugation reaction to the addition of a single ubiquitin and can probably mediate intramolecular ubiquitin interactions39; such roles are possible for the Jab1/MPN domain of Prp8.
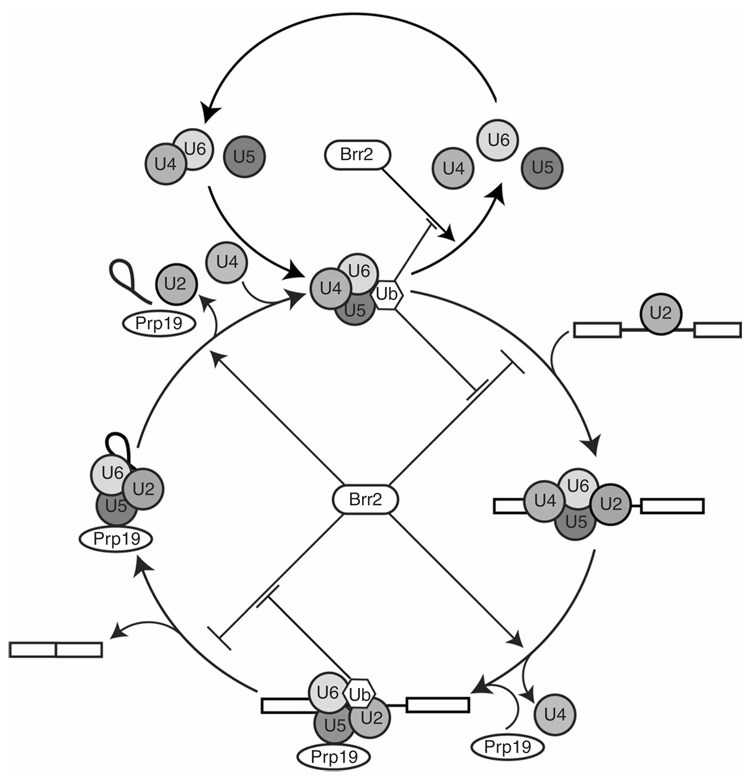
Figure 7.
A model for ubiquitin’s involvement in U4/U6-U5 triple snRNP accumulation and pre-mRNA splicing. The upper cycle depicts U4/U6-U5 triple snRNP assembly and disassembly23. The Brr2 ATPase promotes triple snRNP disassembly by catalyzing the unwinding of the U4 and U6 snRNAs4,23,35,36. Our results indicate that the recognition of a Prp8-ubiquitin conjugate within the U4/U6-U5 triple snRNP suppresses Brr2-catalyzed disassembly. The lower cycle depicts a hypothetical model for ubiquitin’s involvement in the complete pre-mRNA splicing pathway4,23,24. The U4/U6-U5 triple snRNP associates with the U2 snRNP-containing pre-spliceosome, and Brr2 can block this step by catalyzing triple snRNP disassembly; our results indicate that the recognition of a Prp8-ubiquitin conjugate suppresses Brr2 activity at this stage, allowing the triple snRNP to stably engage the pre-spliceosome. Once the spliceosome is assembled, the suppression of Brr2 activity is relieved (perhaps in part by the disruption of ubiquitin recognition), leading to U4/U6 unwinding and catalytic activation of the spliceosome. Upon spliceosome activation, Brr2 activity must once again be suppressed, to inhibit premature spliceosome disassembly probably due to Brr2-catalyzed U2/U6 unwinding4. We speculate that ubiquitin recognition is again involved in Brr2 inhibition at this stage. Once the splicing reaction is complete, the suppression of Brr2 activity is again overcome (perhaps in part by the disruption of ubiquitin recognition), leading to U2/U6 unwinding and spliceosome disassembly (ref. 4; E.C.S. and J.P.S., unpublished data). The points at which ubiquitin enters and exits the triple snRNP and spliceosome cycles are not known and are therefore omitted from the figure for clarity.
We speculate that the formation of an intramolecular interaction between the Jab1/MPN domain of Prp8 and a conjugated ubiquitin could alter the conformation of Prp8 in a manner that diminishes its ability to activate U4/U6 unwinding by Brr2. Numerous subregions of Prp8 and Brr2 (and their human orthologs) have been shown to interact directly20,37,40,41. The regions of Prp8 that interact with Brr2 include the C-terminal portion20,37,40,41 (which contains the ubiquitin-interacting Jab1/MPN domain16) and also an N-terminal domain. Notably, these terminal regions of Prp8 also interact with each other20,40,41. If the temporal modulation of these multiple interactions helps to specify the functional state of the spliceosome, as seems likely3,4,32,37, then the formation and dissolution of ubiquitin–Jab1/MPN domain interactions could account for a subset of these structural and functional switches.
What catalyzes the ubiquitination events that regulate triple snRNP dynamics? Prp19 is unique in that it is both an essential splicing factor and an E3 ubiquitin ligase14,15, although it is possible that other E3 splicing factors remain to be discovered. No in vivo targets of Prp19 E3 ubiquitin ligase activity have been reported, but our evidence that Prp8-ubiquitin conjugates exist within purified triple snRNPs suggests that Prp8 could be a target of Prp19’s E3 ubiquitin ligase activity. Although our studies indicate that ubiquitin recognition helps to maintain U4/U6-U5 levels, previous work has shown that extracts immunodepleted of Prp19-containing complexes are competent for addition of the triple snRNP to the pre-spliceosome42. However, proteins ubiquitinated by Prp19 before depletion could remain in the extract after depletion and could conceivably participate in the maintenance of the U4/U6-U5 triple snRNP. The existence of a postsplicing 35S complex in humans containing Prp19, other components of the Prp19-containing NineTeen Complex (NTC) and U5 snRNP proteins12 raises the possibility that Prp19-mediated ubiquitination of potential U5 snRNP protein targets could occur in this complex during spliceosome recycling, with the ubiquitinated form persisting through the reformation of the U4/U6-U5 triple snRNP.
It is possible that ubiquitin’s role in controlling triple snRNP disassembly is partially redundant with that of Snu114 and its GTPase cycle4, given that both ubiquitin and GDP repress U4/U6 unwinding. Genetic interactions between snu114, prp19, prp8 and sad1 (ref. 43), all of which have been linked to ubiquitin13,15,16,18,19, could (together with our own work) reflect a network of physical and functional interactions that connect Prp19, ubiquitin, the U5 snRNP and splicing complex assembly and disassembly. Potential redundancy between the Prp19 ubiquitin conjugation cycle and the Snu114 GTPase cycle in controlling triple snRNP dynamics43 could help to rationalize several puzzling observations, including (i) the persistence of robust splicing in vitro even after substantial ubiquitin depletion (Fig. 1), (ii) the apparent dispensability of Prp19 during early stages of spliceosome assembly42 and (iii) the inability of GTP and GDP to modulate in vitro splicing activity in whole extracts, despite the well-established in vitro role of the Snu114 GTPase4.
Our discovery that ubiquitin recognition governs triple snRNP disassembly in no way precludes additional roles for ubiquitin in splicing. Several lines of evidence indicate that U4/U6 unwinding in the triple snRNP is mechanistically related to U4/U6 unwinding during spliceosome activation and to U2/U6 unwinding during spliceosome disassembly: all of these events seem to be driven by the Brr2 ATPase4,23,35,36, and all are functionally linked to Snu114 and its guanine-nucleotide binding state4,43–45. It is therefore possible that the role of ubiquitin recognition in inhibiting triple snRNP disassembly is recapitulated later in the splicing cycle: ubiquitin-UBD interactions may continue to repress U4/U6 unwinding during spliceosome assembly until the U4/U6 unwinding stage during spliceosome activation, or it may prevent the premature unwinding of U2/U6 during the catalytic steps until spliceosome disassembly, or both (Fig. 7). A possible role in repressing premature U2/U6 unwinding is especially attractive, given the observation that Prp19 depletion leads to the premature departure of U5 and U6 snRNAs from the spliceosome following U4/U6 unwinding and catalytic activation42. Furthermore, ubistatin A promotes the disassembly of the U2/U6-U5–intron postsplicing complex4 (E.C.S. and J.P.S., unpublished data), and apparent ubiquitin conjugates in post-splicing complexes have been detected in budding yeast (S. Stevens, personal communication). The activities of E3 ubiquitin ligases such as Prp19 are frequently counteracted by deubiquitinating enzymes9, and this may represent a mechanism for derepressing Brr2-dependent spliceosome dynamics.
Dynamic, multicomponent complexes that function in gene expression generally adopt several mechanisms to ensure efficiency and accuracy. The spliceosome is no exception. Our observation that ubiquitin recognition is important for maintaining U4/U6-U5 triple snRNP levels uncovers a discrete stage of spliceosome assembly that is modulated by ubiquitin. This not only points to a previously unrecognized means of regulating the splicing machinery, but also expands the scope of ubiquitin regulation into a new biochemical context.
METHODS
Strains
SUB592 (ref. 27) was used in all experiments except those noted below. In the SUB592 strain, all chromosomal ubiquitin genes were disrupted and a His6-myc-ubiquitin–encoding plasmid was present. SUB280 is isogenic with SUB592, except that the ubiquitin expressed from the plasmid lacks the His6 and myc tags27. In experiments involving the immunoprecipitation of Prp8, we used strains PB2 (ref. 16) or PB592; in the latter, the endogenous PRP8 locus of SUB592 was tagged at the C terminus with a triple-HA tag. To assay triple snRNP disassembly, we used either the Brr2-TAP strain yJPS776 (Fig. 5a,b) or the Prp28-TAP strain yJPS1004 (ref. 4; Fig. 5c,d and Supplementary Fig. 6 and Supplementary Fig 7). For the identification of ubiquitin conjugates, we generated strains yJPS1274 and yJPS1275, which are isogenic with strains SUB280 and SUB592, respectively, except that the endogenous BRR2 gene was TAP-tagged at the C terminus of the open reading frame.
Splicing extracts, ubiquitin depletion and in vitro splicing reactions
Splicing extracts were prepared as described46. For ubiquitin depletion, 100 µl of splicing extract prepared from SUB592 or PB592 was incubated on a Nutator at 4 °C for 45 min with 50 µl of Ni2+-NTA magnetic beads (Qiagen) that had been blocked with BSA; the supernatant was then collected and the procedure was repeated. The depleted extract was analyzed by western blotting using antimyc antibodies (Santa Cruz Biotech). Splicing reactions were done as described previously46, except for the addition of a 10-min incubation at 23 °C in the presence of ubistatin A or purified recombinant wild-type or mutant ubiquitin, before the addition of radiolabeled actin pre-mRNA substrate. The wild-type and mutant ubiquitins were either expressed and purified from Escherichia coli (Supplementary Methods online) or obtained from Boston Biochem, Inc. Ubistatin A was generously provided by the Drug Synthesis & Chemistry Branch of the National Cancer Institute (NSC665534). For splicing rescue experiments with monoubiquitin or K48-linked tetraubiquitin (Boston Biochem, Inc.), 20 µM ubistatin A was incubated with 1 mM monoubiquitin or 60 µM K48 tetraubiquitin in splicing buffer46 for 10 min at 23 °C, followed by an additional 10-min incubation in splicing extract before the addition of radiolabeled actin pre-mRNA.
For additional methods, see Supplementary Methods.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
ACKNOWLEDGMENTS
We thank L. Hicke and D. Finley for strains and plasmids, J. Beggs for anti-Prp8 antibodies and the Drug Synthesis & Chemistry Branch of the National Cancer Institute for ubistatin A. We are grateful to L. Hicke, S. Prakash and members of the Sontheimer and Staley laboratories for advice and discussions, to S. Stevens (University of Texas, Austin) for communicating unpublished results and to M. French, D. Golden, J. Pellino, S. Prakash and J. Preall for critical reading of this manuscript. This work was supported by a US National Institutes of Health grant (GM62264) to J.P.S. and a US National Science Foundation CAREER Award (MCB-0093003) and a US National Institutes of Health grant (GM072830) to E.J.S.
Footnotes
Published online at http://www.nature.com/nsmb/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Will CL, Lührmann R. Spliceosome structure and function. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 369–400. [Google Scholar]
- 2.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn AN, Li Z, Brow DA. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell. 1999;3:65–75. doi: 10.1016/s1097-2765(00)80175-6. [DOI] [PubMed] [Google Scholar]
- 4.Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misteli T. RNA splicing: what has phosphorylation got to do with it? Curr. Biol. 1999;9:R198–R200. doi: 10.1016/s0960-9822(99)80128-6. [DOI] [PubMed] [Google Scholar]
- 6.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 8.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 9.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 11.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makarov EM, et al. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 13.Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 14.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 15.Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellare P, Kutach AK, Rines AK, Guthrie C, Sontheimer EJ. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA. 2006;12:292–302. doi: 10.1261/rna.2152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer A, Mulhauser F, Wersig C, Groning K, Bilbe G. Mammalian splicing factor SF3a120 represents a new member of the SURP family of proteins and is homologous to the essential splicing factor PRP21p of Saccharomyces cerevisiae. RNA. 1995;1:260–272. [PMC free article] [PubMed] [Google Scholar]
- 18.Lygerou Z, Christophides G, Seraphin B. A novel genetic screen for snRNP assembly factors in yeast identifies a conserved protein, Sad1p, also required for pre-mRNA splicing. Mol. Cell. Biol. 1999;19:2008–2020. doi: 10.1128/mcb.19.3.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarova OV, Makarov EM, Luhrmann R. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J. 2001;20:2553–2563. doi: 10.1093/emboj/20.10.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pena V, Liu S, Bujnicki JM, Luhrmann R, Wahl MC. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol. Cell. 2007;25:615–624. doi: 10.1016/j.molcel.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, et al. Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants. Protein Sci. 2007;16:1024–1031. doi: 10.1110/ps.072872007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson CR, et al. Ubiquitin-like protein Hub1 is required for pre-mRNA splicing and localization of an essential splicing factor in fission yeast. Curr. Biol. 2004;14:2283–2288. doi: 10.1016/j.cub.2004.11.058. [DOI] [PubMed] [Google Scholar]
- 23.Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 24.Stevens SW, et al. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 25.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc. Natl. Acad. Sci. USA. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. Distinct functional surface regions on ubiquitin. J. Biol. Chem. 2001;276:30483–30489. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- 27.Spence J, et al. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- 28.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 29.Ecker DJ, et al. Gene synthesis, expression, structures, and functional activities of site-specific mutants of ubiquitin. J. Biol. Chem. 1987;262:14213–14221. [PubMed] [Google Scholar]
- 30.Wilkinson KD, Audhya TK. Stimulation of ATP-dependent proteolysis requires ubiquitin with the COOH-terminal sequence Arg-Gly-Gly. J. Biol. Chem. 1981;256:9235–9241. [PubMed] [Google Scholar]
- 31.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 32.Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verma R, et al. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science. 2004;306:117–120. doi: 10.1126/science.1100946. [DOI] [PubMed] [Google Scholar]
- 34.Stevens SW, et al. Biochemical and genetic analyses of the U5, U6, and U4/U6U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA. 2001;7:1543–1553. [PMC free article] [PubMed] [Google Scholar]
- 35.Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA. 1999;5:959–971. doi: 10.1017/s135583829999012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laggerbauer B, Achsel T, Luhrmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. USA. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hershko A, Rose IA. Ubiquitin-aldehyde: a general inhibitor of ubiquitin-recycling processes. Proc. Natl. Acad. Sci. USA. 1987;84:1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnell JD, Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 2003;278:35857–35860. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- 40.Boon K-L, Norman CM, Grainger RJ, Newman AJ, Beggs JD. Prp8p dissection reveals domain structure and protein interaction sites. RNA. 2006;12:198–205. doi: 10.1261/rna.2281306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S, Rauhut R, Vornlocher HP, Luhrmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA. 2006;12:1418–1430. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 43.Brenner TJ, Guthrie C. Genetic analysis reveals a role for the C-terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics. 2005;170:1063–1080. doi: 10.1534/genetics.105.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartels C, Klatt C, Luhrmann R, Fabrizio P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 2002;3:875–880. doi: 10.1093/embo-reports/kvf172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartels C, Urlaub H, Luhrmann R, Fabrizio P. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 2003;278:28324–28334. doi: 10.1074/jbc.M303043200. [DOI] [PubMed] [Google Scholar]
- 46.Lin RJ, Newman AJ, Cheng SC, Abelson J. Yeast mRNA splicing in vitro. J. Biol. Chem. 1985;260:14780–14792. [PubMed] [Google Scholar]
- 47.Cheng SC, Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.