Summary
The proper functioning of eukaryotic organelles is largely dependent on the specific packaging of cargo proteins within transient delivery vesicles. The cytoplasm to vacuole targeting (Cvt) pathway is an autophagy-related trafficking pathway whose cargo proteins, aminopeptidase I and α-mannosidase, are selectively transported from the cytoplasm to the lysosome-like vacuole in yeast. This study elucidates a molecular mechanism for cargo specificity in this pathway involving four discrete steps. The Cvt19 receptor plays a central role in this process: distinct domains in Cvt 19 recognize oligomerized cargo proteins and link them to the vesicle formation machinery via interaction with Cvt9 and Aut7. Because autophagy is the primary mechanism for organellar turnover, these results offer insights into physiological processes that are critical in cellular homeostasis, including specific packaging of damaged or superfluous organelles for lysosomal delivery and breakdown.
Introduction
Macroautophagy is a cellular process responsible for bulk degradation of cytoplasmic components in eukaryotic cells. In this process, cytoplasmic materials are nonselectively sequestered by a double-membrane vesicle, the autophagosome, and transported to the lyso-some/vacuole to be degraded (Klionsky and Ohsumi, 1999). Autophagy (Apg) is induced under starvation conditions. Organelles also undergo selective removal by autophagy when they are damaged or no longer needed (Klionsky and Emr, 2000). In addition, vacuolar resident enzymes exploit an autophagy-related process called the cytoplasm to vacuole targeting (Cvt) pathway (Klionsky and Ohsumi, 1999). Aminopeptidase I (Ape1), a cargo of the Cvt pathway, is synthesized in the cytoplasm as a precursor form (prApe1). The prApe1 propeptide contains vacuolar targeting information (Oda et al., 1996; Segui-Real et al., 1995). After synthesis, prApe1 assembles into a dodecamer (Kim et al., 1997), which is further packaged into a unique structure called the Cvt complex (Baba et al., 1997). This complex is subsequently enwrapped by a double membrane to form the Cvt vesicle. This vesicle then fuses with the vacuole to release the inner single-membrane vesicle, the Cvt body (Baba et al., 1997; Scott et al., 1997). In the last decade, morphological, genetic, and biochemical studies on the yeast Saccharomyces cerevisiae have demonstrated that autophagy, peroxisome degradation (pexophagy), and the Cvt pathway share a common mechanism for biogenesis of the double-membrane vesicles that sequester cargo from the cytoplasm (Klionsky and Ohsumi, 1999; Reggiori and Klionsky, 2002). However, the mechanism used to selectively incorporate cargo into these vesicles has been unclear.
Recently, a novel protein, termed Cvt19, has been identified and characterized as a receptor for the Cvt cargos, prApe1 and α-mannosidase (Ams1) (Hutchins and Klionsky, 2001; Scott et al., 2001). Cvt19 is a peripheral membrane protein. Unlike typical transmembrane-type receptors that achieve multiple rounds of binding and release, Cvt19 is transported to the vacuole along with its cargo and is degraded by vacuolar proteinases. Biochemical and fluorescence microscopy analyses indicate that Cvt19 primarily localizes at a perivacuolar preautophagosomal structure (PAS), a predicted site of vesicle formation, where most components required for autophagy and the Cvt pathway localize (Scott et al., 2001, Suzuki et al., 2001; Kim et al., 2002; Noda et al., 2002). From these results, Cvt19 is suggested to function at the step where the Cvt complex is targeted to the PAS. However, it is not clear how Cvt19 localizes to this site. Although components specific for the Cvt and pexophagy pathways, such as Cvt9, have been identified (Scott et al., 2000; Kim et al., 2001b; Nice et al., 2002), the relationship between Cvt19 and these proteins remains unknown.
In this study, we addressed the molecular mechanism of selective transport of Cvt cargos to the vacuole. The mapping of functional domains in Cvt19 identified separate binding sites for interaction with two cargo proteins (prApe1 and Ams1), the Cvt pathway/pexophagy-specific factor Cvt9 and the Cvt vesicle/autophagosome component Aut7. Using this, together with fluorescence microscopy analyses and affinity isolation, we conclude that the process of selective cargo targeting to the PAS involves a specific subset of Apg/Cvt proteins that act in a particular temporal order to allow the formation of the Cvt complex (Ape1 -Cvt19-Ams1) and its incorporation into sequestering vesicles.
Results
GFP-Ape1 Is Transported to the Vacuole by the Cvt and Autophagy Pathways
Aminopeptidase I (Ape1) is a resident hydrolase that travels to the vacuole via the cytoplasm to vacuole targeting (Cvt) pathway. Its N-terminal propeptide contains vacuolar targeting information and is cleaved off in a vacuolar proteinase A (Pep4)-dependent manner subsequent to delivery (Figure 1A, lanes 2 and 5; Klionsky et al., 1992; Oda et al. 1996). To date, most studies of autophagy have concentrated on analyses of the vesicle-forming machinery. To learn more about the Cvt and Apg pathways, we decided to examine import of the cargo protein precursor Ape1 (prApe1) in vivo. Accordingly, we constructed an N-terminal green fluorescent protein (GFP) fusion with prApe1. The resulting GFP-Ape1 chimera was expressed under the control of the endogenous APE1 promoter from a single-copy plasmid. To verify that this construct reflected the trafficking of the native prApe1 protein, we first examined the processing of GFP-Ape1 in the ape1Δ background. Western blot analysis revealed that about 50% of GFP-Ape1 was processed by cleavage of the prApe1 propeptide, resulting in the production of the mature form of Ape1 (mApe1) and GFP (Figures 1A and 1B, lane 3). Under the same conditions, authentic prApe1 was fully processed (Figure 1A, lane 2). In ape1Δpep4Δ cells, no processing of GFP-Ape1 was observed (Figure 1A, lane 6 and Figure 1B, lane 1). These results indicated that GFP-Ape1 was processed to mApe1 in a vacuolar proteinase-dependent manner, although not as efficiently as the native protein.
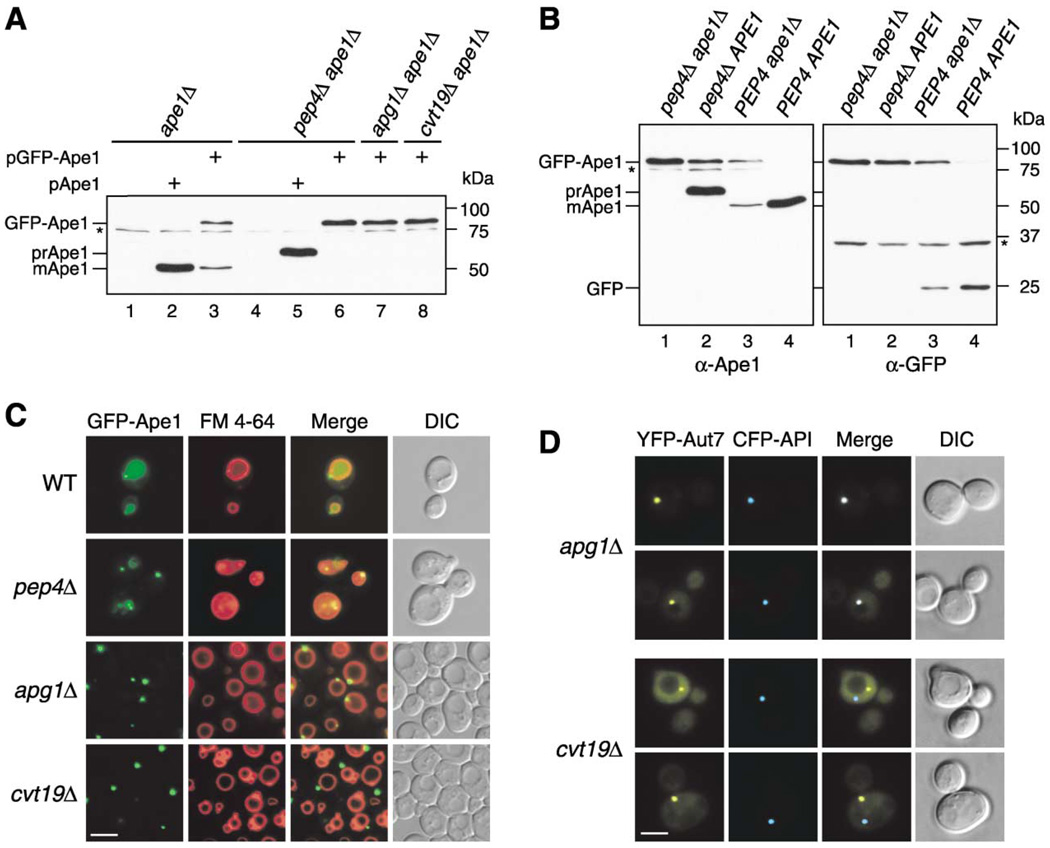
Figure 1. GFP-Ape1 Travels to the Vacuole via the Cvt Pathway.
(A) Cell extracts from ape1 A (lanes 1–3), pep4Δ ape1Δ (lanes 4–6), apg1Δ ape1Δ (lane 7), and cvt19Δ ape1Δ (lane 8) strains containing either empty vector (pRS414; lanes 1 and 4), pApe1 (lanes 2 and 5), or pGFP-Ape1 (lanes 3, 6, 7, and 8) were resolved by SDS-PAGE. Western blots were performed with anti-Ape1 antiserum.
(B) Western blot of cell extracts from pep4Δ ape1Δ (lane 1), pep4Δ (lane 2), ape1Δ (lane 3), and wild-type (lane 4) strains containing pGFP-Ape1 with anti-Ape1 (left panel) and anti-GFP (right panel) antisera. An asterisk (*) indicates nonspecific bands.
(C) Localization of Ape1 by fluorescence microscopy of wild-type, pep4Δ, apg1Δ, and cvt19Δ strains expressing GFP-Ape1. Cells were grown to midlog phase and labeled with FM 4-64 in SCD medium. Bar, 5 µm.
(D) CFP-Ape1 colocalizes with YFP-Aut7 in apg1Δ cells, but not in cvt19Δ cells. The apg1Δ and cvt19Δ cells coexpressing CFP-Ape1 and YFP-Aut7 were grown in SCD medium to midlog phase and examined with fluorescence and DIC microscopy. Bar, 5 µm.
To further confirm whether the processing of GFP-Ape1 was dependent on the Cvt pathway, we next expressed GFP-Ape1 in ape1Δ apg1Δ and ape1Δ cvt19Δ strains. Apg1 is a Ser/Thr protein kinase essential for Cvt vesicle/autophagosome formation (Scott et al., 2000; Kamada et al., 2000). Cvt19 has recently been demonstrated to function as a receptor for cargo proteins that utilize the Cvt pathway (Scott et al., 2001; Leber et al., 2001). In both strains, GFP-Ape1 remained in the unprocessed form (Figure 1A, lanes 7 and 8), indicating that GFP-Ape1 is delivered to the vacuole via the Cvt pathway. Precursor Ape1 assembles into a dodecamer in the cytosol (Kim et al., 1997). Because GFP-Ape1 seemed to be less-efficiently processed than authentic prApe1 in ape1Δ cells, we examined the effect of the presence of endogenous prApe1 on GFP-Ape1 processing. GFP-Ape1 was expressed in wild-type and pep4Δ cells and analyzed by Western blot with both anti-Ape1 and anti-GFP antisera. In pep4Δ cells, the processing of both GFP-Ape1 and endogenous prApe1 was blocked (Figure 1B, lane 2), whereas more than 95% of GFP-Ape1 was converted to mApe1 in the wild-type cells (Figure 1B, lane 4). These results indicated that the presence of endogenous prApe1 facilitated the processing of GFP-Ape1. The GFP-tagging adjacent to prApe1 may affect some function of the propeptide, e.g., the interaction with other proteins (see below; Scott et al., 2001), which can be restored by the formation of an oligomer with native prApe1.
We next investigated the localization of GFP-Ape1 in wild-type and apg/aut/cvt mutant strains. Cells expressing GFP-Ape1 were labeled with the dye FM 4-64, to visualize the vacuole membrane, and examined by fluorescence microscopy. In wild-type cells, the GFP signal was dispersed uniformly in the vacuole with one or a few tiny dot signals in the perivacuolar region (Figure 1C), in agreement with the efficient processing of GFP-Ape1 (Figure 1B). We further confirmed the import of GFP-Ape1 in pep4Δ cells that accumulate Cvt bodies in the vacuolar lumen. In this strain, several GFP dots were seen moving inside the vacuole, indicating GFP-Ape1 is present within the Cvt bodies (Figure 1C). In contrast, in the apg1Δ strain, a single larger dot of GFP-Ape1 appeared in the perivacuolar region, instead of inside the vacuolar lumen (Figure 1C). Because GFP-Ape1 forms a large dot in most apg/aut/cvt mutants (data not shown), it is likely that prApe1 accumulates as a large structure outside the vacuole when the Cvt pathway is blocked. These observations were in agreement with immunoelectron microscopy analyses showing that prApe1 travels to the vacuole as a part of the Cvt complex (Baba et al., 1997). Taken together, these results demonstrated that the vacuolar localization of GFP-Ape1 is Cvt pathway dependent and that GFP-Ape1 serves as a valid marker to follow cargo import through the Cvt pathway.
It has been reported that many proteins required for the Cvt and/or autophagic pathway localize at least transiently to the perivacuolar preautophagosomal structure (PAS), which is thought to be a site of vesicle formation (Kim et al., 2001b; Suzuki et al., 2001; Noda et al., 2002; Nice et al., 2002). Many Apg/Aut/Cvt proteins accumulate at the PAS in apg mutant strains. We examined apg1Δ cells expressing CFP-Ape1 and YFP-tagged Aut7, one of the PAS components (Kim et al., 2001a; Suzuki et al., 2001), by fluorescence microscopy. As shown in Figure 1D, CFP-Ape1 localized to the PAS that was labeled with YFP-Aut7 in the apg1Δ strain. Because the putative receptor protein Cvt19 also localizes to the PAS (Kim et al., 2002), we hypothesized that Cvt19 interacts with the prApe1 dodecamer at the PAS, leading to the formation of the Cvt complex. To test this idea, we examined the localization of GFP-Ape1 in a cvt19Δ strain by fluorescence microscopy. Unexpectedly, GFP-Ape1 still accumulated as a single large dot, but a careful observation revealed that it did not consistently localize in the perivacuolar region (Figure 1C). To determine whether GFP-Ape1 was concentrated at a site other than the PAS in the cvt19Δ strain, we transformed this strain with CFP-Ape1 and YFP-Aut7. As shown in Figure 1D, CFP-Ape1 was not seen at the PAS labeled with YFP-Aut7 in the cvt19Δ strain. This separate localization was also observed in an apg1Δ cvt19Δ strain (data not shown), indicating that cvt19 is epistatic to apg1 in controlling prApe1 localization to the PAS. These results suggested that prApe1 formed a large complex (referred to as the Ape1 complex) independent of Cvt19. However, the Cvt19 protein is required for the recruitment of the Ape1 complex to the PAS, in agreement with its presumed role as a receptor.
The prApe1 Propeptide Is Required for Its Proper Localization and the Interaction with a Receptor Protein
The prApe1 propeptide is essential for prApe1 import to the vacuole but is not required for its oligomerization (Oda et al., 1996; Kim et al. 1997). A secondary-structure prediction suggested that the prApe1 propeptide forms a helix-turn-helix structure and that the first helix (residues 1–18) exhibits the characteristics of an amphipathic α helix, which is critical for membrane association (Oda et al., 1996). Mutations in this region (e.g., deletions of residues 9–11; Δ9–11) prevent prApe1 from associating with the membrane, thus inhibiting its transport to the vacuole. On the other hand, the substitution of pro-line to leucine at position 22 (P22L) causes accumulation of prApe1 on a nonvacuolar membrane structure but also blocks most of prApe1 import to the vacuole (Oda et al., 1996, Scott et al. 1997). We therefore tried to determine the stage in the Cvt pathway that was affected by these mutations in the prApe1 propeptide. We first decided to verify the localization of prApe1 propeptide mutants using GFP-fused constructs in an ape1Δ strain by fluorescence microscopy. The wild-type GFP-Ape1 gave a significant vacuolar lumenal staining with additional small punctate signals in the perivacuolar region (Figure 2A). In contrast, GFP-Ape1Δ9–11 distributed uniformly in the cytosol (Figure 2A), in agreement with previous biochemical analyses. This result suggested that the first amphipathic α helix is involved in the formation of the Ape1 complex. In contrast, GFP-Ape1P22L gave a single large punctate signal away from the vacuole (Figure 2A), suggesting that Ape1P22L was still able to form the Ape1 complex but was defective in localization to the PAS.
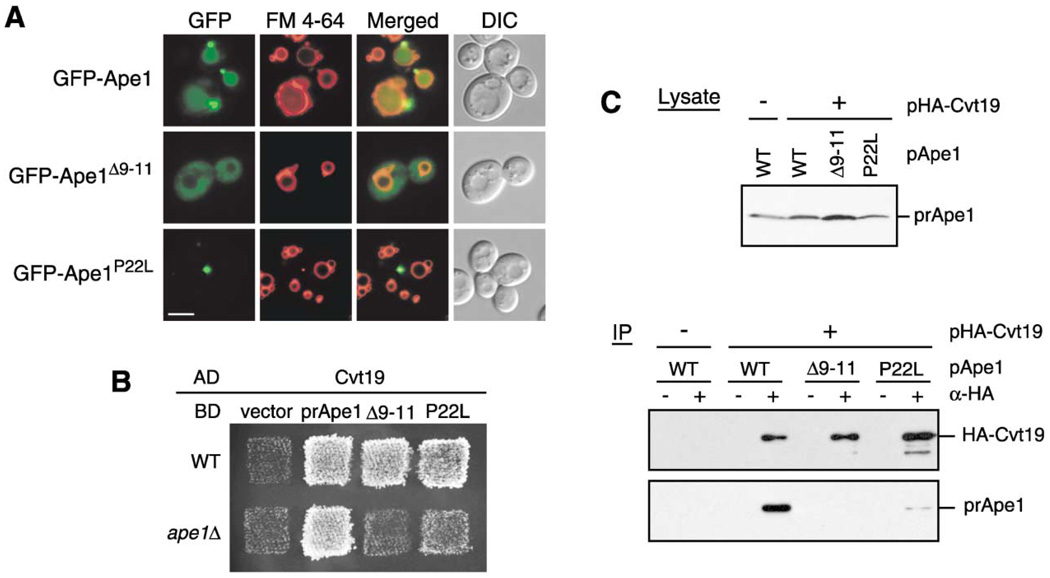
Figure 2. Precursor Ape1 Propeptide Is Required for the Formation of the Ape1 Complex and Interaction With Cvt19.
(A) Localization of the propeptide mutants of GFP-Ape1 by fluorescence and DIC microscopy. The ape1Δ cells harboring pGFP-Ape1, pGFP-Ape1Δ9–11, or pGFP-Ape1P22L were grown to midlog phase and labeled with FM 4-64 in SCD medium. Bar, 5 µm.
(B) Two-hybrid analysis of the physical interaction between Ape1 mutants and Cvt19. Wild-type (PJ69-4A) and isogenic ape1Δ (YTS110) test strains were transformed with the activation domain (AD) plasmid containing CVT19 and the binding domain (BD) plasmid containing wild-type or mutant APE1. Transformants were grown on an SC-Leu-Ura plate and replica plated on an SC-Ade-Leu-Ura plate to assess the interaction-dependent activation of the ADE2 gene. The cells shown here were grown at 30°C for 2 days.
(C) Interaction between prApe1 and Cvt19 is mediated by the prApe1 propeptide. Total lysate from the ape1Δ apg1Δ cells expressing HA-Cvt19 and either wild-type or mutant prApe1 from single-copy plasmids under the control of their own promoters were used for immunoprecipitation with anti-HA antibody, and the immunoprecipitates were analyzed by Western blot with anti-Ape1 and anti-HA antisera (IP). The top panel (Lysate) shows the protein blot of total cell lysate probed with anti-Ape1 antiserum.
The localization of GFP-Ape1 in the cvt19Δ strain is quite similar to that of GFP-Ape1P22L. This observation suggested that the P22L mutation affects the interaction between prApe1 and Cvt19. While previous data have shown that mutations in the prApe1 propeptide affect import, no studies have verified that the defect is due to a block in the interaction with Cvt19. To test this hypothesis, we examined the interaction between prApe1 mutants and Cvt19 by two-hybrid analysis and coimmunoprecipitation. Because of prApe1 homooligomer formation, we utilized an ape1Δ strain (YTS110) as a test strain for the two-hybrid analysis. The ape1Δ cells carrying both Cvt19 and wild-type Ape1 plasmids exhibited growth on an adenine minus plate, indicating that the two proteins could interact (Figure 2B). In contrast, Ape1Δ9–11 displayed no binding activity with Cvt19 in the ape1Δ strain, although it allowed the wild-type test strain (PJ69-4A) to grow on an adenine minus plate (Figure 2B). This result suggested that (1) the interaction between prApe1 and Cvt19 is propeptide-dependent and that (2) an apparent interaction between Ape1Δ9–11 and Cvt19 in a wild-type strain might be mediated by endogenous prApe1. Ape1P22L provided much slower growth than wild-type Ape1 in an ape1Δ strain, suggesting that the P22L mutation severely affected, but did not completely block, the interaction between prApe1 and Cvt19 (Figure 2B). We verified these results by coimmunoprecipitation under native conditions. In this experiment, we used an apg1Δ strain to block the Cvt pathway; an Ape1-Cvt19 complex would be degraded in a wild-type strain as a consequence of its import into the vacuole (Scott et al., 2001; Figure 5A). The ape1Δ apg1A cells were transformed with HA-tagged Cvt19 and wild-type or mutant prApe1 plasmids, and their extracts were used for immunoprecipitation with anti-HA antibody. The wild-type prApe1 was effectively coimmunoprecipitated with HA-Cvt19. In contrast, no prApe1Δ9–11 and only a small amount of Ape1P22L was found to coprecipitate with Cvt19 (Figure 2C), confirming that the interaction of prApe1 and Cvt19 was mediated by the prApe1 propeptide. The reduced association of prApe1 with Cvt19 caused by the P22L mutation would impair the proper localization of the complex to the PAS and explains the phenotype of GFP-Ape1P22L (Figure 2A).
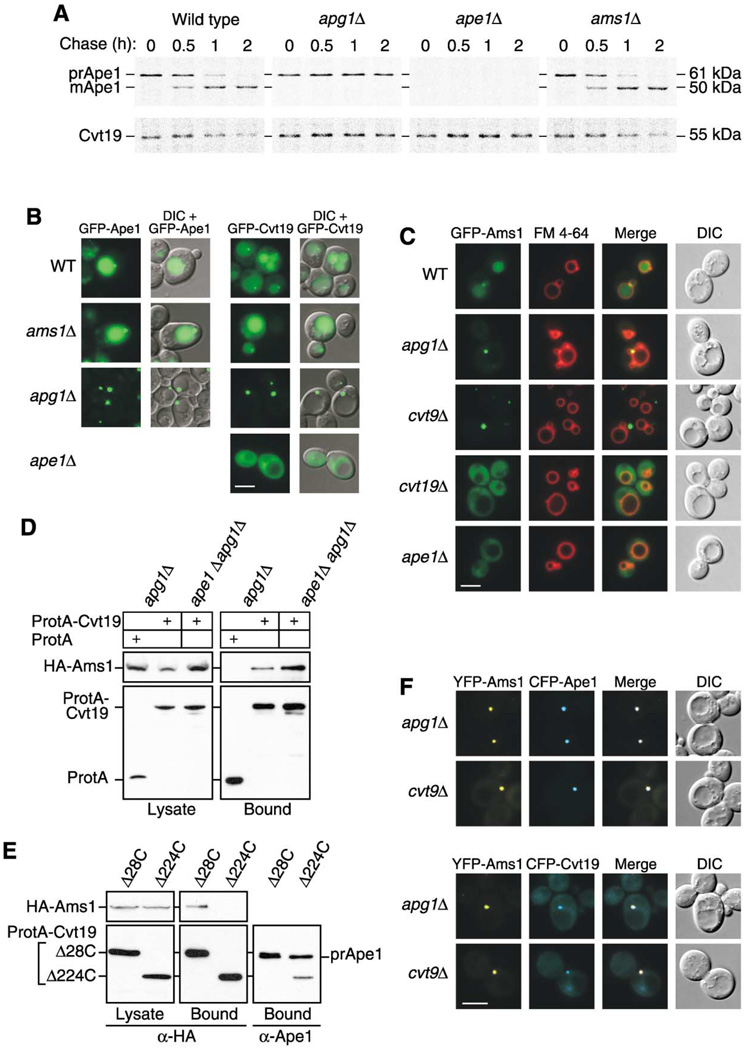
Figure 5. Precursor Ape1 Is Required for the Efficient Transport of Cvt19 and the Cargo Protein Ams1.
(A) Wild-type, apg1Δ, ape1Δ, and ams1Δ cells were pulse labeled for 10 min in SMD medium and chased for the indicated times in SMD medium. Ape1 and Cvt19 were immunoprecipitated from cell extracts and analyzed by SDS-PAGE.
(B) Localization of GFP-Ape1 or GFP-Cvt19 in wild-type, ams1Δ, apg1Δ, and ape1Δ strains by fluorescence and DIC microscopy. Bar, 5 µm.
(C) Localization of GFP-Ams1 in wild-type, apg1Δ, cvt9Δ, cvt19Δ, and ape1Δ strains by fluorescence and DIC microscopy. Cells were grown to midlog phase and labeled with FM 4-64 in SCD medium. Bar, 5 µm.
(D) Ams1 associates with Cvt19, independent of Ape1. The apg1Δ or ape1Δ apg1Δ cells were transformed with pHA-Ams1 and pRS416-CuProtA or pCuProtA-Cvt19. IgG Sepharose was used to precipitate ProtA-Cvt19 from cell lysates. The left and right panels show the protein blots of total cell lysates (Lysate) and the IgG precipitates (Bound), respectively, which were probed with anti-HA antibody.
(E) The Ams1 binding site in Cvt19 is different from the prApe1 binding site. Total lysates from apg1Δ cvt19Δ cells expressing HA-Ams1 and either ProtA-Cvt19Δ28C or ProtA-Cvt19Δ224C were used to precipitate ProtA-Cvt19 proteins with IgG Sepharose. Total cell lysates (Lysate) and the IgG precipitates (Bound) were analyzed by Western blot with anti-HA or anti-Ape1 antisera.
(F) Colocalization of YFP-Ams1 with CFP-Ape1 or CFP-Cvt19 in apg1Δ and cvt9Δ cells by fluorescence and DIC microscopy. Bar, 5 µm.
The Ape1-Cvt19 Complex Is Targeted to the PAS by Cvt9
Autophagy is a nonselective process and enwraps cytoplasm nonspecifically. In contrast, the Cvt pathway and import of prApe1 by autophagy are specific, presumably because of the interaction between prApe1 and Cvt19. However, the domain within Cvt19 that binds prApe1 is not known. Moreover, the precise localization of Cvt19 at the PAS seems to be an important event for selective targeting of prApe1, which allows us to hypothesize that there is a domain in Cvt19 that determines its localization at the PAS. For these reasons, we tried to identify functional domains within the Cvt19 protein. Cvt19 is a 415-amino acid protein containing a predicted coiled-coil domain between amino acids 160 and 187 (Figure 3A). We constructed several Cvt19 N- and C-terminal deletion mutants, e.g., Cvt19Δ15C indicates a mutant lacking the C-terminal 15 amino acids. Because a coiled-coil motif often mediates a protein-protein interaction, we also constructed the Cvt19ΔCC mutant that lacks the 39-amino acid region from residues 153 to 191, containing a coiled-coil domain (Figure 3A). The prApe1 binding domain was first examined by a yeast two-hybrid system. The cvt19Δ test strain with the lacZ gene under the control of a Gal4-dependent promoter was transformed with prApe1 and mutant Cvt19 plasmids, and β-galactosidase activity was measured to assess the interaction. All the Cvt19 proteins containing the coiled-coil domain effectively interacted with prApe1, whereas the ones lacking this domain did not exhibit any interaction (Figure 3A). To verify these results, we performed an affinity isolation experiment. The apg1Δ mutant is defective in the Cvt pathway and autophagy. In an apg1Δ background neither Cvt19 nor prApe1 will be delivered to the vacuole, thus maintaining similar levels of functional and nonfunctional Cvt19 proteins within the cytosol. Lysates were generated from apg1Δ cvt19Δ cells expressing protein A-fused Cvt19 truncates, and Cvt19 proteins were isolated with IgG Sepharose. Western blot analysis of the resulting samples with anti-Ape1 antiserum revealed that only the Cvt19 proteins containing the coiled-coil domain were able to precipitate prApe1 (Figure 3B). These results suggest that the coiled-coil domain of Cvt19 mediates the interaction between prApe1 and Cvt19.
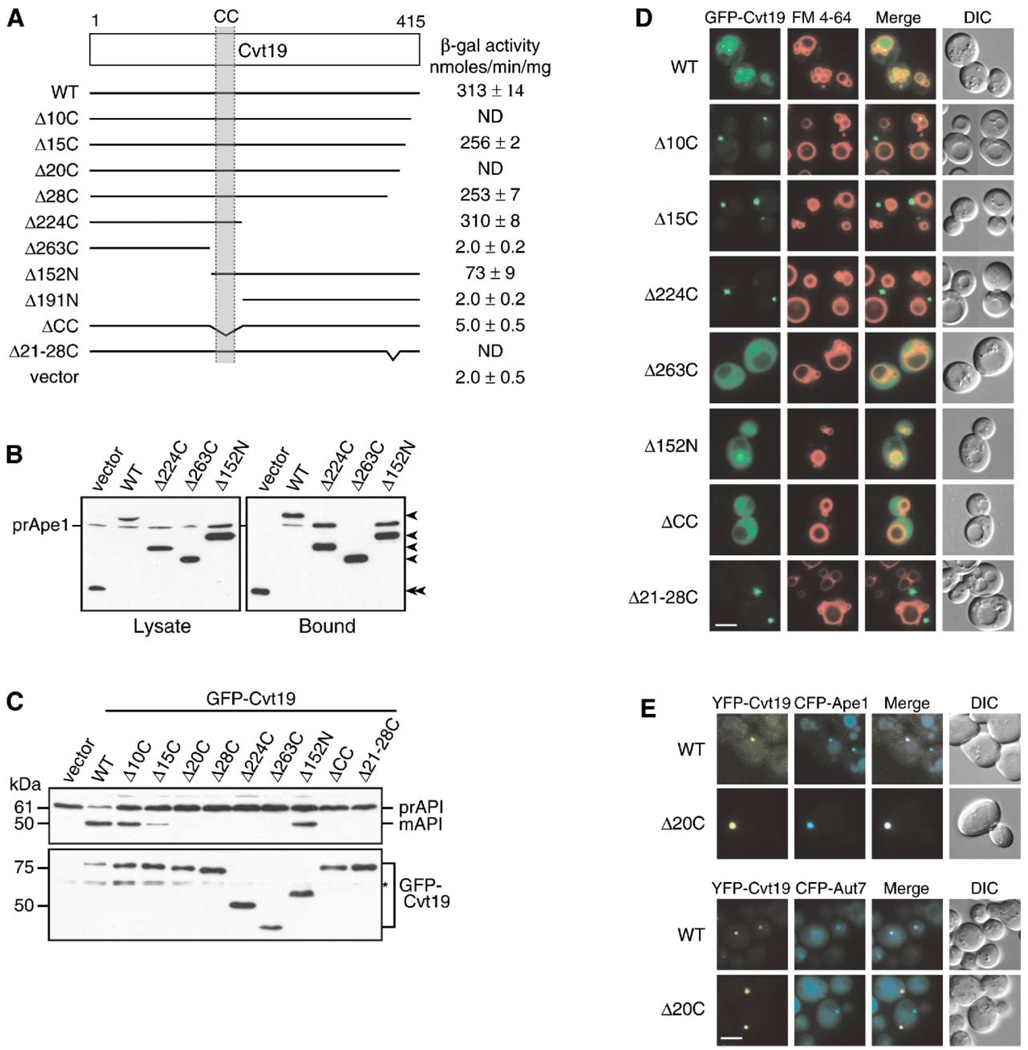
Figure 3. Mapping Functional Domains within Cvt19.
(A) Mapping the prApe1 binding domain by a yeast two-hybrid system. The cvt19Δ test strain (YTS111) was transformed with the binding domain plasmid containing the entire APE1 ORF and the activation domain plasmid containing mutant CVT19. The interaction was assessed by measuring the interaction-dependent induction of β-galactosidase activity. CC, coiled coil; ND, not determined.
(B) Total lysates from the apg1Δ cvt19Δ cells expressing either wild-type or mutant Cvt19 fused with protein A (ProtA-Cvt19) were used to precipitate ProtA-Cvt19 proteins with IgG Sepharose. The left and right panels show the protein blots of total cell extracts (Lysate) and the IgG precipitates (Bound), respectively, which were probed with anti-Ape1 antiserum. Single arrowheads show ProtA-Cvt19 derivatives, and a double arrowhead indicates protein A without fusion. Although Cvt19 mutants containing the coiled-coil domain (Δ10C, Δ15C, Δ20C, Δ28C, Δ21–28C, Δ224C, and Δ152N) were all able to precipitate prApe1, only the results from Δ224C and Δ152N mutants were shown in this figure.
(C) Protein blots of total lysates from the cvt19Δ cells expressing Cvt19 deletion mutants fused with GFP probed with anti-Ape1 (upper panel) and anti-GFP (lower panel) antisera. An asterisk (*) indicates nonspecific bands.
(D) Localization of wild-type and mutant Cvt19 proteins by fluorescence and DIC microscopy of cvt19Δ strains expressing wild-type or mutant GFP-Cvt19. Cells were grown to midlog phase and labeled with FM 4-64 in SCD medium. Bar, 5 µm.
(E) Colocalization of YFP-Cvt19 and YFP-Cvt19Δ20C with CFP-Ape1 and CFP-Aut7 in a cvt19Δ strain by fluorescence and DIC microscopy. Bar, 5 µm.
We decided to extend the analysis by determining the functionality and the localization of the Cvt19 truncates. Accordingly, we constructed N-terminal GFP fusions to Cvt19 and expressed them under the control of the endogenous Cvt19 promoter from a single-copy plasmid in cvt19Δ cells. Western blot analysis of cell extracts with anti-GFP antibody confirmed that the different GFP-Cvt19 mutants had similar expression levels (Figure 3C). The steady-state protein level of GFP-Cvt19 seemed to be lower than those of the deletion mutants (Figure 3C, lower panel), which was probably because of the effective transport of GFP-Cvt19 to the vacuole and subsequent degradation. Moreover, GFP-Cvt19 exhibited enough activity for prApe1 maturation (Figure 3C, upper panel), confirming that the GFP-Cvt19 construct is suitable for this analysis. About 40% of prApe1 was converted to mApe1 in the cells expressing GFP-Cvt19Δ10C, and only a small amount of mApe1 was found in the cells expressing GFP-Cvt19Δ15C or GFP-Cvt19Δ20C (Figure 3C, upper panel). In contrast, GFP-Cvt19Δ28C, GFP-Cvt19Δ224C, GFP-Cvt19Δ263C, and GFP-Cvt19ΔCC failed to complement the defect in prApe1 maturation in cvt19Δ cells (Figure 3C, upper panel). Surprisingly, GFP-Cvt19Δ152N was still partly functional in prApe1 maturation (Figure 3C, upper panel), indicating that a large N-terminal portion of Cvt19 is dispensable, at least for prApe1 transport. Even though many mutants, including Cvt19Δ28C, exhibited a significant interaction with prApe1 (Figures 3A and 3B), prApe1 transport was completely lost in these strains, including cells expressing Cvt19Δ28C. These results suggest that the C-terminal 28 amino acids of Cvt19 might have some function in prApe1 transport other than association with the prApe1 propeptide.
We next determined the localization of the GFP-Cvt19 mutants. The cvt19Δ cells expressing wild-type or mutant GFP-Cvt19 proteins were labeled with FM 4-64 and observed by fluorescence microscopy. As expected from the data presented in Figure 3C, GFP-Cvt19 displayed vacuolar straining together with one or a few tiny dots in the perivacuolar region (Figure 3D). In addition, as shown previously (Kim et al., 2002), the colocalization of YFP-Cvt19 with CFP-Aut7 confirmed that Cvt19 localized at the PAS (Figure 3E, lower panel). The deletion of APG1 resulted in the concentration of GFP-Cvt19 at the PAS, as seen with GFP-Ape1 in the apg1Δ strain (Figure 5B). This result suggests that Cvt19 might be transported to the vacuole together with prApe1 through the PAS. The localization of GFP-Cvt19Δ152N was quite similar to that of wild-type GFP-Cvt19, consistent with its significant activity for prApe1 import (Figure 3D). In contrast to the cells expressing GFP-Cvt19Δ152N, two populations were observed in the cells expressing GFP-Cvt19Δ10C, although the activities for prApe1 import were comparable between these two mutants. About half of the cells expressing GFP-Cvt19Δ10C had slightly GFP-stained vacuoles, with a single prominent dot adjacent to the vacuole, and the rest had a single GFP dot away from the vacuole (Figure 3D). The Δ15C, Δ20C, and Δ224C proteins, containing the putative coiled-coil domain, formed a prominent dot away from the vacuole (Figure 3D). In contrast, GFP-Cvt19Δ263 and GFP-Cvt19ΔCC displayed a dispersed cytosolic localization, suggesting that the punctate localization of Cvt19 depends on the interaction with prApe1 (Figure 3D). Consistent with the data presented in Figures 3A and 3B, the wild-type and YFP-Cvt19 mutants containing the coiled-coil domain, such as YFP-Cvt19Δ20C, colocalized with CFP-Ape1 (Figure 3E, upper panel). However, unlike YFP-Cvt19, YFP-Cvt19Δ20C did not colocalize with the CFP-Aut7 dot (Figure 3E, lower panel). These results suggested that prApe1 and Cvt19 form a complex before recruitment to the PAS. Furthermore, the C-terminal region of Cvt19 is crucial for the proper localization of the complex to the PAS.
Recently, it has been reported that Aut7 physically interacts with Cvt19 (Kim et al., 2002). We hypothesized that this interaction could play an important role in the localization of prApe1 to the PAS and its subsequent vacuolar import. We therefore mapped the Aut7 binding domain within Cvt19 by both a yeast two-hybrid system and a protein A affinity isolation experiment. In the two-hybrid analysis, Aut7 displayed a significant interaction with full-length Cvt19 on the basis of the activation of the ADE2 gene, but did not show significant interaction with any of the Cvt19 C-terminal truncates (Figure 4A). An affinity isolation experiment also indicated that protein A-Aut7 was able to coprecipitate full-length Cvt19, but the efficiencies were severely reduced for the truncated Cvt19 proteins, including Cvt19Δ10C (Figure 4B). These results suggested that the Aut7 binding domain of Cvt19 might be located within, or include, the last 10 C-terminal amino acids. The C-terminally truncated GFP-Cvt19 chimeras were not localized to the PAS (Figure 3D). If this localization resulted from the loss of interaction between Aut7 and Cvt19, the Ape1-Cvt19 complex should not be localized at the PAS in an aut7Δ strain. However, fluorescence microscopy of aut7Δ cells expressing GFP-Ape1 or GFP-Cvt19 revealed that the Ape1-Cvt19 complex seemed to be still localized at the PAS (Figures 4D and 4E), suggesting the existence of another Cvt19 binding partner(s).
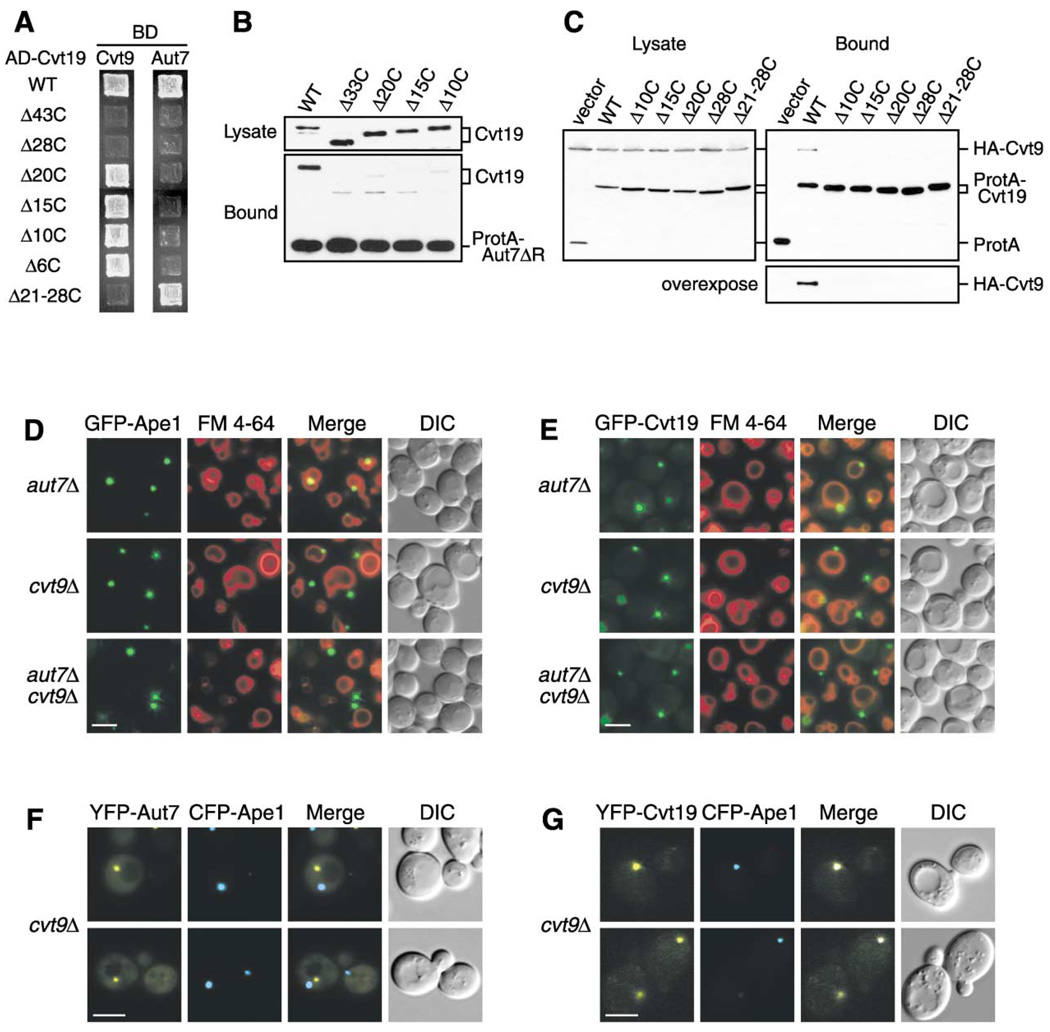
Figure 4. Cvt9 Facilitates the Localization of the Ape1-Cvt19 Complex to the Preautophagosomal Structure.
(A) Mapping of Aut7 and Cvt9 binding domains within Cvt19 by a yeast two-hybrid system. The cvt19Δ test strain (WHY4) was transformed with the binding domain plasmid containing the entire CVT9 or AUT7 ORF (pGBD-Cvt9 or pGBD-Aut7, respectively) and the activation domain plasmid containing wild-type or mutant CVT19. Transformants were selected on SC-Leu-Ura plates (data not shown) and replicated to SC-Ade plates. The figures show cells grown at 30°C for 2 days.
(B) A protein A-Aut7ΔR affinity isolation experiment was performed in lysates from the apg1Δ cvt19Δ strain expressing full-length Cvt19 or the indicated truncate. C-terminal deletion of Cvt19 compromised its coisolation with Aut7.
(C) Cvt9 interacts with the C terminus of Cvt19. Total lysates from apg1Δ cvt19Δ cells expressing HA-Cvt9 and either wild-type or mutant Cvt19 fused with protein A (ProtA-Cvt19) were used to precipitate ProtA-Cvt19 proteins with IgG Sepharose. The left and right panels show the protein blots of total cell lysates (Lysate) and the IgG precipitates (Bound), respectively, which were probed with anti-HA antibody.
(D and E) Localization of GFP-Ape1 (D) or GFP-Cvt19 (E) in the aut7Δ, cvt9Δ, and aut7Δ cvt9Δ strains by fluorescence and DIC microscopy. Bar, 5 µm.
(F and G) Colocalization of CFP-Ape1 with YFP-Aut7 (F) or YFP-Cvt19 (G) in the cvt9Δ strain by fluorescence and DIC microscopy. Bar, 5 µm.
The deletion of AUT7 results in a complete loss of prApe1 transport during vegetative growth, but starvation can partially restore this defect, even though au-tophagy-dependent protein turnover is still defective. The reversal of the prApe1 import defect could result from the formation of small and/or aberrant vesicles, instead of normal autophagosomes, under starvation conditions (Kirisako et al., 1999; Abeliovich et al., 2000). These data indicate that the specific import of prApe1 still occurs in starved aut7Δ cells. In contrast, the cvt9Δ strain shows a relatively normal autophagic protein degradation, whereas only a partial transport of prApe1 is restored by starvation, suggesting that Cvt9 plays an important role in cargo selection in selective autophagy (Kim et al., 2001b). For this reason, we hypothesized that Cvt9 could be another binding partner of Cvt19. To test this possibility, we examined the interaction between Cvt9 and Cvt19 by a yeast two-hybrid system and a protein A affinity isolation experiment. Two-hybrid analysis revealed a significant interaction between Cvt9 and full-length Cvt19 (Figure 4A). Furthermore, Cvt19ΔCC still displayed a significant interaction, indicating that the coiled-coil domain was not necessary for the interaction with Cvt9 (data not shown). We therefore subjected several C-terminal deletion mutants of Cvt19 to two-hybrid analysis and found that Cvt19Δ10C, Cvt19Δ15C, and Cvt19Δ20C were able to interact with Cvt9, whereas Cvt19Δ28C no longer exhibited this interaction (Figure 4A). This result led us to test the Cvt19 mutant lacking the 8-amino acid region from amino acid residues 388 to 395 (Cvt19Δ21-28C). As expected, Cvt19Δ21-28C did not show any interaction (Figure 4A), suggesting that the Cvt9 binding domain might be located within this 8-amino acid region. Because Aut7 was able to associate with Cvt19Δ21-28C (Figure 4A), the binding domains for Aut7 and Cvt9 could be separated (Figure 6A).
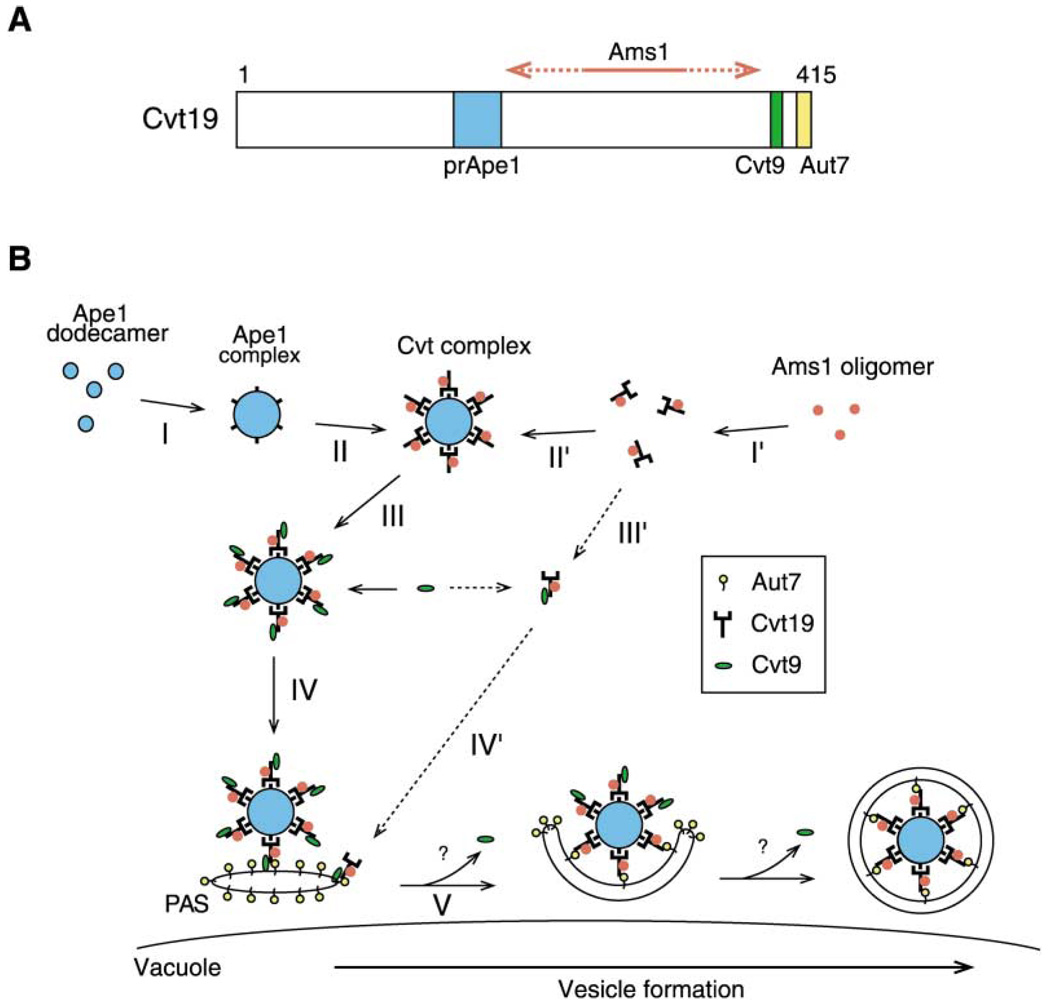
Figure 6. Model for Selective Targeting of Cvt Cargos into the Cvt Vesicle.
(A) Schematic drawing of binding domains in Cvt19. The prApe1 binding domain (blue) is located in the region from amino acid residues 153 to 191, which contains the predicted coiled-coil motif. The 10-amino acid region from 406 to 415 (yellow) and the 8-amino acid region from 388 to 395 (green) are for Aut7 binding and Cvt9 binding, respectively. An exact binding domain for Ams1 has not been determined but is located in a region within amino acid residues 192-387 (red arrow).
(B) After prApe1 is synthesized it immediately forms dodecamers in the cytosol, which are further assembled into an Ape1 complex dependent on its propeptide. Cvt19 then binds to the Ape1 complex, which is also mediated by the prApe1 propeptide. Because Ams1 is able to associate with Cvt19 via a domain distinct from the prApe1 binding domain, Ams1 could be concentrated on the Ape1 complex as a consequence of the Ape1-Cvt19 complex formation. Cvt9 then interacts with the C terminus of Cvt19 to recruit the Ape1-Cvt19-Ams1 complex to the PAS, where Aut7 binds to the extreme C terminus of Cvt19 to ensure the incorporation of the complex into the Cvt vesicle. See text for details.
To extend the analysis, we next performed affinity isolation experiments from the lysates of apg1Δ cvt19Δ cells expressing each protein A-fused Cvt19 derivative and HA-tagged Cvt9. In contrast to the data from the two-hybrid analysis, we were unable to detect a physical interaction between any mutant and HA-Cvt9, whereas wild-type Cvt19 was able to coprecipitate HA-Cvt9 (Figure 4C). This discrepancy between two-hybrid and affinity isolation analyses could result from the difference in the sensitivities of the assays; the recovery of HA-Cvt9 was quite poor, even from the wild-type Cvt19 strain. The microscopy of the cvt9Δ cells expressing GFP-Ape1 or GFP-Cvt19 revealed that both proteins were highly concentrated away from the vacuole (Figures 4D and 4E). Similarly, GFP-Cvt19Δ21-28C accumulated away from the vacuole (Figure 3D). These results strongly suggested that the loss of the PAS localization of Cvt19 deletion mutants resulted from the disruption of the physical interaction between Cvt19 and Cvt9. Furthermore, we confirmed that CFP-Ape1 colocalized with YFP-Cvt19, but not with YFP-Aut7, in the cvt9Δ strain (Figures 4F and 4G). These results indicate that, in the cvt9Δ strain, prApe1 and Cvt19 formed a protein complex that was unable to reach the PAS. Consequently, we concluded that the disruption of the association between Cvt19 and Cvt9 prevented the prApe1-Cvt19 complex from localizing to the PAS. Because GFP-Ape1 and GFP-Cvt19 displayed the same localizations in aut7Δ cvt9Δ cells to those in cvt9Δ cells (Figures 4D and 4E), Cvt9 might function at an earlier step than Aut7 in the recruitment of this complex to the PAS.
The Cargo Protein prApe1 Directs the Cvt19 Receptor and Another Cargo Protein Ams1 toward Incorporation into Cvt Vesicles
Cvt19 is degraded as a consequence of import into the vacuole, and its half-life in ape1Δ cells is four times greater than that in wild-type cells (Scott et al., 2001). This led us to examine whether another cargo protein, α-mannosidase (Ams1), could affect the import of Cvt19. We performed a pulse-chase experiment to monitor the transport of prApe1 and Cvt19 to the vacuole. In ams1Δ cells, however, Cvt19 degradation was comparable to that in wild-type cells (Figure 5A). In addition, the deletion of AMS1 did not lead to any alteration in prApe1 processing (Figure 5A). These results were supported by fluorescence microscopy, which showed normal localization of GFP-Cvt19 and GFP-Ape1 in ams1Δ cells (Figure 5B). Conversely, Cvt19 was stabilized in ape1Δ cells, similar to the result seen in apg1Δ cells (Figure 5A), in agreement with previous studies (Scott et al., 2001). These results indicated that prApe1, but not Ams1, was required for proper import of Cvt19 to the vacuole. To further investigate the effects of prApe1 on Cvt19 import, we compared the localization of GFP-Cvt19 in apg1Δ and ape1Δ cells. As expected from the GFP-Ape1 localization in apg1Δ cells, GFP-Cvt19 was also concentrated at the site adjacent to the vacuole in the apg1Δ strain (Figure 5B). In contrast, in ape1Δ cells, GFP-Cvt19 was distributed uniformly in the cytoplasm (Figure 5B), consistent with the dispersed distribution of GFP-Cvt19Δ263C and GFP-Cvt19ΔCC lacking the prApe1 binding domain (Figure 3D). Altogether, these results suggested that the Ape1 complex concentrated Cvt19 through the interaction between prApe1 and Cvt19.
The transport of Ams1 to the vacuole is also defective in the cvt19Δ strain, demonstrating that Cvt19 is the receptor for both prApe1 and Ams1 (Scott et al., 2001). Because of decreased import of Cvt19 in ape1Δ cells, Ams1 transport to the vacuole could also be reduced. To test this hypothesis, we constructed GFP-Ams1 under the control of the endogenous AMS1 promoter and examined its localization in several mutant strains by fluorescence microscopy. In rich medium, this construct gave a vacuolar staining with a few tiny dots in the perivacuolar region in the wild-type strain (Figure 5C). Induction of autophagy by starvation or rapamycin treatment gave a similar result but made the vacuolar signal brighter (data not shown). In addition, several tiny dots of GFP-Ams1 were observed in the vacuolar lumen in pep4Δ cells (data not shown), suggesting that GFP-Ams1 was incorporated in Cvt vesicles. When Cvt vesicle formation was blocked by deleting APG1, GFP-Ams1 accumulated in the perivacuolar region (Figure 5C). In contrast, the deletion of CVT19 or APE1 caused GFP-Ams1 signals to be dispersed in the cytoplasm (Figure 5C), indicating that prApe1 and Cvt19 are required to localize Ams1 to both the vacuole and the perivacuolar structure. Because GFP-Ams1 still showed a cytosolic distribution in cvt19Δ apg1Δ and ape1Δ apg1Δ strains (data not shown), Cvt19 and prApe1 appear to function in Ams1 transport at an earlier step than Apg1. Although GFP-Ams1 accumulated as a prominent dot in cvt9Δ cells, it did not localize close to the vacuole (Figure 5C). Furthermore, we observed that YFP-Ams1 colocalized with CFP-Ape1 or CFP-Cvt19 in both apg1Δ and cvt9Δ strains (Figure 5F), suggesting that Ams1 is accumulated on the Ape1-Cvt19 complex.
To elucidate whether the accumulation of Ams1 on the Ape1-Cvt19 complex is dependent on the interaction between Ams1 and Cvt19, we performed a protein A affinity isolation experiment from the lysate of the apg1Δ cells expressing protein A-Cvt19 and HA-tagged Ams1. As shown in Figure 5D, HA-Ams1 was copurified with protein A-Cvt19 from the apg1Δ cell lysate. Together with the data from Figure 3, this result suggested the formation of an Ape1 -Cvt19-Ams1 complex. Furthermore, we found that the Ams1-Cvt19 interaction still existed, even in ape1Δ apg1Δ cells, where Ams1 was dispersed in the cytoplasm (Figure 5D), suggesting that Ams1 and Cvt19 could form a cargo-receptor complex that then interacts with the Ape1 complex. If it is possible to form the Ape1 -Cvt19-Ams1 complex, the binding sites for prApe1 and Ams1 within Cvt19 should be located in different places. To test this, we performed the protein A affinity isolation from lysates of the apg1Δ cvt19Δ cells expressing HA-Ams1 and protein A-Cvt19Δ28C or protein A-Cvt19Δ224C. Western blot analysis of the resulting precipitants revealed that protein A-Cvt19Δ28C was able to pull down both prApe1 and Ams1, whereas only prApe1 was precipitated with protein A-Cvt19Δ224C (Figure 5E). This result indicated that Ams1 associated with Cvt19 via a site that is distinct from the prApe1 binding site (Figure 6A).
Discussion
Precursor aminopeptidase I has been used as a marker for both the biosynthetic Cvt pathway and autophagy (Baba et al., 1997). Immunoelectron microscopy and biochemical studies showed that cytosolic prApe1 assembled into a large Cvt complex composed of prApe1 dodecamers (Baba et al., 1997; Kim et al., 1997). Recently, we showed that Cvt19 acts as a receptor through its interaction with the prApe1 propeptide (Scott et al., 2001). Cvt19 also functions as a receptor for a second cargo of the Cvt pathway, Ams1 (Hutchins and Klionsky, 2001; Scott et al., 2001). Along with Cvt19, most of the other proteins of the Cvt and Apg pathways localize to a punctate perivacuolar preautophagosomal structure. However, until now, nothing more has been known about the nature of the interaction between Cvt19 and its cargos or between these proteins and other Apg/Aut/Cvt components. In order to learn more about the mechanism of the Apg/Cvt pathways, we constructed a GFP-Ape1 chimera that could be used to follow the Cvt complex in vivo. GFP-Ape1 was delivered to the vacuole and processed in a Cvt pathway- and Pep4-dependent manner, indicating that it is a valid marker for the Cvt complex.
Our analyses of GFP-Ape1, together with affinity isolation experiments, have allowed us to propose a model for the selective targeting of cargo proteins through the Cvt and autophagy pathways. (1) Precursor Ape1 oligomers assemble into a higher-order structure, the Ape1 complex, in a propeptide-dependent manner. (2) A Cvt19-Ams1 complex concentrates at the Ape1 complex through the interaction between precursor Ape1 and Cvt19. (3) Subsequently, Cvt9 recognizes a C-terminal domain of Cvt19 to target the Ape1-Cvt19-Ams1 complex to the site of Cvt vesicle formation. (4) Finally, the complex is incorporated into the Cvt vesicle through interaction with Aut7.
Assembly and Recognition of the Cargo Complex
Previously we have shown that prApe1 rapidly assembles into a dodecamer in the cytosol and that the prApe1 propeptide contains vacuolar targeting information (Oda et al., 1996; Kim et al., 1997). Formation of the Cvt complex allows Cvt cargos to be effectively packaged by a limited membrane source. However, nothing had been reported about the molecular property and the formation mechanism of the Cvt complex. In this study, we have dissected the process of Cvt complex formation into two steps and found that both steps require the function of the prApe1 propeptide. In the first step, prApe1 dodecamers assemble into the Ape1 complex (Figure 6B, step I). Although no evidence has been provided about whether another protein is required at this step, the cytosolic hsp70 family protein, Ssa1, could be one of the candidates. It was reported that Ssa1 specifically associates with the prApe1 propeptide and is required for the targeting of prApe1 to the Cvt vesicle, but not for assembly into a dodecamer (Silles et al., 2000). In the second step, the propeptide interacts with the Cvt19 receptor, thus defining a function in formation of the Cvt complex (which we define as the Cvt19-cargo complex; Figure 6B, step II). The interaction between prApe1 and Cvt19 is mediated by the predicted coiled coil of Cvt19. Interestingly, the loss of Cvt19’s coiled coil or the deletion of APE1 itself led to the cytosolic localization of Cvt19, indicating that the cargo (Ape1) complex concentrates the receptor (Cvt19) proteins rather than vice versa (Figure 6B, step II). In addition, another Cvt cargo, Ams1, is also concentrated at the Cvt complex as a consequence of the association with Cvt19 (Figure 6B, step I′). A defect in Ape1-Cvt19 complex formation severely affected the import of Ams1 to the vacuole, whereas Ams1 was dispensable for the transport of the Ape1 -Cvt19 complex, indicating that Ams1 might exploit the prApe1 import system to achieve its own effective transport to the vacuole.
Targeting of the Cvt Complex to the Site of Vesicle Formation
Cvt9 localizes to the PAS, the site of Cvt vesicle/autophagosome formation, and is required for the Cvt pathway and pexophagy, but not for nonspecific autophagy, suggesting that Cvt9 plays a role in the recognition of specific cargos (Kim et al., 2001b). Several lines of evidence strongly suggest that Cvt9 is involved in the targeting of the Cvt complex to the PAS and acts like an adaptor protein. (1) Cvt9 physically interacted with the C-terminal region of Cvt19. (2) When the Cvt19 domain responsible for Cvt9 binding was deleted, the Cvt complex was no longer seen at the PAS. (3) Similarly, in the cvt9Δ strain, the Cvt complex did not localize at the PAS. How Cvt9 guides the Cvt complex to the PAS remains to be solved. We propose that Cvt9 forms a stable interaction with the Cvt complex prior to targeting to the PAS (Figure 6B, step III). After the recognition of the Cvt complex by Cvt9, the complex can be targeted to the PAS through the possible interaction between Cvt9 and the PAS components (Figure 6B, step IV). Two candidates for the acceptor on the PAS are Apg1 and/or Apg12 because Cvt9 showed physical interactions with these PAS components (Kamada et al., 2000; Kim et al., 2001b; Ho et al., 2002). We have also mapped a fourth distinct domain within Cvt19 that binds Aut7. After arrival of the Cvt9-Cvt19-cargo complex at the PAS, the Cvt19-cargo complex can be transferred to Aut7 to ensure the incorporation of the Cvt complex into the Cvt vesicle (Figure 6B, step V). Although it is not clear whether Cvt9 is released from the complex at this step, we speculate that it may be released, because GFP-Cvt9 did not reach the vacuole (Kim et al., 2001b).
Mechanism for Selective Autophagy
Macroautophagy, pexophagy, and the Cvt pathway share common machinery for vesicle formation and possess a few specific factors for induction, cargo selection, and supply of the source membrane (Reggiori and Klionsky, 2002). Cvt9 is required for pexophagy in yeast as well as the Cvt pathway, suggesting that pexophagy and the Cvt pathway further share similar mechanisms for cargo selection (Kim et al., 2001b; Scott et al., 2001). In the Cvt pathway, Cvt9 recognizes Cvt19 on the "surface" of the Cvt complex. Because of the size of the Cvt complex, this interaction is similar to recognition of an organelle. Similarly, Cvt9 might be able to recognize some signal on the peroxisome and bring it to the site of vesicle formation. Alternatively, Cvt9 might bind a receptor like Cvt19 that is specific for peroxisomes.
The mechanism of cargo selection in the Cvt pathway provides insight into the selective autophagy of organelles in mammalian cells as well as yeast cells. Excess organelles induced under specific conditions or damaged organelles are specifically removed by autophagy (Luiken et al., 1992; Tolkovsky et al., 2002). The specific removal of damaged organelles may play important roles in preventing premature aging or apoptosis (Brunk and Terman, 2002; LeMasters et al., 1998). Damaged organelles may present an epitope on their surfaces that is recognized by PAS components. Because a Cvt9 counterpart is not found in mammalian cells, other proteins that provide a link between cargo and the PAS components may exist, or a PAS component may directly recognize cargos in mammalian cells.
The majority of the Apg/Aut/Cvt proteins have been localized to the PAS. However, little is known about the function of most of these proteins, and essentially nothing is known about the temporal order of action. We have shown that, in the absence of Cvt19, prApe1 is not able to localize to the PAS. In cells lacking Cvt9, Cvt19 and prApe1 bind each other but, again, do not localize to the PAS. Finally, in aut7Δ cells, the Ape1 -Cvt19-Ams1 complex can localize to the PAS but cannot import into the vacuole. These results indicate that Cvt19 and Cvt9 first act not at the PAS, but at the site of Ape1 complex formation in the cytosol. Furthermore, we have determined a specific order of interaction for Cvt19, Cvt9, and Aut7. Future studies will extend this analysis to determine the way in which Cvt complex formation and binding to the PAS is coordinated with formation of the sequestering vesicle, the hallmark of the Cvt and Apg pathways.
Experimental Procedures
Strains and Media
The yeast Saccharomyces cerevisiae strains used in this study are listed in Table 1. Media used for growing yeast, YPD, synthetic minimal (SMD), synthetic complete (SC), and synthetic with casamino acids (SCD) media are described elsewhere (Adams et al., 1998; Scott et al., 2001; Suzuki et al., 2001). Copper was not added to media, even when expressing genes under the control of the CUP1 promoter.
Table 1.
Yeast Strains Used in This Study
| Strain | Descriptive Name | Genotype | Reference |
|---|---|---|---|
| SEY6210 | wt | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-Δ901 ura3-52 suc2-Δ9 GAL | Robinson et al., 1988 |
| AHY001 | cvt9Δ | SEY6210; cvt9Δ::HIS3 | Kim et al., 2001b |
| SSY31 | cvt19Δ | SEY6210; cvt19Δ::HIS5 S.p. | Scott et al., 2001 |
| THY411 | ams1Δ | SEY6210; ams1Δ::LEU2 | Hutchins and Klionsky, 2001 |
| WHY1 | apg1Δ | SEY6210; apg1Δ::HIS5 S.p. | This study |
| WPHYD7 | aut7Δ | SEY6210; aut7Δ::LEU2 | Kim et al., 2001a |
| YTS107 | ape1Δ | SEY6210; ape1Δ::LEU2 | This study |
| WHY3 | aut7Δ cvt9Δ | SEY6210; aut7Δ::URA3 cvt9Δ::HIS3 | This study |
| WHY2 | ape1Δ apg1Δ | SEY6210; ape1Δ::LEU2 apg1Δ::HIS5 S.p. | This study |
| YTS108 | ape1Δ cvt19Δ | SEY6210; ape1Δ::LEU2 cvt19Δ::HIS5 S.p. | This study |
| TVY1 | pep4Δ | SEY6210; pep4Δ::LEU2 | Gerhardt et al., 1998 |
| YTS11 2 | ape1Δ pep4Δ | TVY1; ape1Δ::HIS5 S.p. | This study |
| YTS106 | apg1Δ cvt19Δ | MATa his3 leu2 trp1 ura3 apg1Δ::LEU2 cvt19Δ::HIS5 S.p. | This study |
| PJ69-4A | wt | MATa trp1-Δ901 leu2-3,112 ura3-52 his3-Δ200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al., 1996 |
| WHY4 | cvt19Δ | PJ69-4A; cvt19Δ::KanMX4 | This study |
| YTS110 | ape1Δ | PJ69-4A; ape1Δ::HIS5 S.p. | This study |
| YTS111 | cvt19Δ | PJ69-4A; cvt19Δ::HIS5 S.p. | This study |
Plasmid Construction
The following method was used to make fluorescent protein or triple haemagglutinin (HA)-tagged constructs for Ape1, Ams1, and Cvt19. The BglII (for APE1 and CVT19) or BamHI (for AMS1) restriction site was introduced just after the initiation codon of each gene on pRS414 or pRS416 with a QuikChange Site-Directed Mutagenesis KIT (Stratagene). The DNA fragment encoding GFP (S65T), EYFP, ECFP, or HA cassette with BamHI or BglII sites on both sides was then ligated to the newly generated BglII or BamHI site of each gene, to make the respective fusion constructs. For the truncated versions of pGFP-Cvt19 and pYFP-Cvt19, the truncated ORFs were PCR-amplified and ligated into pRS416 containing the CVT19 promoter, GFP/YFP, and the CYC1 terminator. For the generation of the protein A-Cvt19 fusion and its deletion versions, a PCR product containing full-length or deleted CVT19 ORF was inserted into the EcoRI/SalI site of pRS416-CuProtA (Kim et al., 2002). For two-hybrid analysis, we used a system developed by James et al. (1996). pGBD-Ape1, pGBD-Aut7, and pGBD-Cvt9 were constructed as bait plasmids by ligating the full-length APE1, AUT7, and CVT9 ORFs, respectively, into pGBDU-C1. The prey plasmid pGAD-Cvt19 or its truncates were constructed by ligating the full-length or the truncated CVT19 ORFs into pGAD-C2. Other plasmids have been described elsewhere (Oda et al., 1996; Kim et al., 2002; Suzuki et al., 2001).
Biochemical Analysis
For native immunoprecipitation, 50 OD units of cells were lysed in 1 ml lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, 1 mM PMSF, amd Complete EDTA-free protease inhibitor [Roche]) with glass beads. After centrifugation at 10,000 × g for 10 min, the resulting supernatant was incubated with 0.6 µg monoclonal anti-HA antibody (Santa Cruz Biotechnology) for 2 hr at 4°C. The immunocomplex was recovered by incubation with protein G-Sepharose 4 Fast Flow (Amersham Pharmacia Biotech) for 1 hr at 4°C. After washing the Sepharose with lysis buffer four times, the bound materials were eluted by boiling the Sepharose in SDS-PAGE loading buffer. The resulting eluate was analyzed by Western blot with anti-Ape1 (Klionsky et al., 1992) or polyclonal anti-HA (Santa Cruz Biotechnology) antisera.
For protein A-Cvt19 affinity isolation, cell extract prepared as described above was incubated with IgG Sepharose 6 Fast Flow (Amersham Pharmacia Biotech) for 1 hr at 4°C. After washing with lysis buffer four times, the bound materials were eluted by boiling the Sepharose in SDS-PAGE loading buffer and analyzed by Western blotting with anti-Ape1 or anti-HA antiserum. Protein A-Aut7ΔR affinity isolation was performed essentially as previously described (Kim et al., 2002).
Pulse-chase analyses and measurements of β-galactosidase activity were performed as previously described (Klionsky et al., 1992; Scott et al., 1996; Adams et al., 1998).
Fluorescence Microscopy
Cells expressing fluorescent protein-fused chimeras were grown in SCD medium to midlog phase. To label the vacuolar membrane, we incubated cells in the medium containing 16 µM N-(3-triethylammo-niumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide (FM 4-64; Molecular Probes) at 30°C for 20 min. After being washed with medium, the cells were incubated in the medium at 30°C for 2 hr. Fluorescence microscopy analysis was performed with a Nikon E-800 fluorescent microscope equipped with a Hamamatsu Orca2 digital camera as previously described (Kim et al., 2002).
Acknowledgments
We would like to thank Dr. Yoshinori Ohsumi for providing pYFP-Aut7 and pCFP-Aut7. We also wish to thank Dr. Fulvio Reggiori and Chao-Wen Wang for helpful discussions and technical advice. This work was supported by National Institutes of Health Public Health Service grant GM53396 to D.J.K.
References
- Abeliovich H, Dunn WA, Jr, Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunk UT, Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur. J. Biochem. 2002;269:1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- Gerhardt B, Kordas TJ, Thompson CM, Patel P, Vida T. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J. Biol. Chem. 1998;273:15818–15829. doi: 10.1074/jbc.273.25.15818. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Scott SV, Oda MN, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang W-P, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 2001a;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001b;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang W-P, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber R, Silles E, Sandoval IV, Mazon MJ. Yol082p, a novel CVT protein involved in the selective targeting of aminopeptidase I to the yeast vacuole. J. Biol. Chem. 2001;276:29210–29217. doi: 10.1074/jbc.M101438200. [DOI] [PubMed] [Google Scholar]
- LeMasters JJ, Nieminen AL, Qian T, Trost LC, Elmore SP, Nishimura Y, Crowe RA, Cascio WE, Bradham CA, Brenner DA, Herman B. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim. Biophys. Acta. 1998;1366:177–196. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- Luiken JJ, van den Berg M, Heikoop JC, Meijer AJ. Autophagic degradation of peroxisomes in isolated rat hepatocytes. FEBS Lett. 1992;304:93–97. doi: 10.1016/0014-5793(92)80596-9. [DOI] [PubMed] [Google Scholar]
- Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Suzuki K, Ohsumi Y. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–235. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD, Klionsky DJ. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J. Cell Biol. 1996;132:999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot. Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm to vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Amino-peptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segui-Real B, Martinez M, Sandoval IV. Yeast amino-peptidase I is post-translationally sorted from the cytosol to the vacuole by a mechanism mediated by its N-terminal extention. EMBO J. 1995;14:5476–5484. doi: 10.1002/j.1460-2075.1995.tb00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silles E, Mazon MJ, Gevaert K, Goethals M, Vandekerckhove J, Leber R, Sandoval IV. Targeting of aminopeptidase I to the yeast vacuole is mediated by Ssa1 p, a cytosolic member of the 70-kDa stress protein family. J. Biol. Chem. 2000;275:34054–34059. doi: 10.1074/jbc.M003514200. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkovsky AM, Xue L, Fletcher GC, Borutaite V. Mitochondrial disappearance from cells: a clue to the role of autophagy in programmed cell death and disease? Biochimie. 2002;84:233–240. doi: 10.1016/s0300-9084(02)01371-8. [DOI] [PubMed] [Google Scholar]