Abstract
Chromatin remodeling and activation of transcription are important aspects of gene regulation, but these often go awry in disease progression, including during colon cancer development. We investigated the status of global histone acetylation (by measuring H3, H4 acetylation of lysine residues, which also occur over large regions of chromatin including coding regions and non-promoter sequences) and expression of histone deacetylase 2 (HDAC2) in colorectal cancer (CRC) tissue microarrays using immunohistochemical staining. Specifically, HDAC2 and the acetylation of histones H4K12 and H3K18 were evaluated in 134 colonic adenomas, 55 moderate to well differentiated carcinomas, and 4 poorly differentiated carcinomas compared to matched normal tissue. In addition, the correlation between expression of these epigenetic biomarkers and various clinicopathological factors including, age, location, and stage of the disease were analyzed. HDAC2 nuclear expression was detected at high levels in 81.9%, 62.1%, and 53.1% of CRC, adenomas, and normal tissue, respectively (P = 0.002). The corresponding nuclear global expression levels in moderate to well differentiated tumors for H4K12 and H3K18 acetylation were increased while these levels were decreased in poorly differentiated tumors (P = 0.02). HDAC2 expression was correlated significantly with progression of adenoma to carcinoma (P = 0.002), with a discriminative power of 0.74, when comparing cancer and non-cancer cases. These results suggest HDAC2 expression is significantly associated with CRC progression.
Keywords: Global histone acetylation, HDAC2, Colon cancer
Introduction
Colorectal carcinoma is the most common gastrointestinal malignancy. Overall mortality from colorectal cancer has declined since the 1980s. However, this decrease is primarily due to reduced mortality in Caucasians [1-3]. The death rate in African Americans over this period is relatively unchanged. Both genetic and environmental risk factors play an important role in this disease. The over-whelming majority of cases of colorectal cancer are classified as sporadic. However, there is likely some inherited component to sporadic colorectal cancers, since there is an increased risk of cancer in those with a family history of colorectal cancer. In addition, a family history of colorectal adenomas is also a risk factor for developing colon cancer. Studies over the last two decades have identified factors that may protect against colorectal cancer including environmental and socioeconomical factors, exercise, as well as a nutritional diet including fruits, vegetables (low meat and animal fat diet) and fiber.
While it is difficult to determine which of the above factors plays a major role in the observed differences between different ethnic groups, it is becoming clear that both genetic and epigenetic influences are important [4-6]. There is an increasing body of literature evaluating the impact of epigenetic changes on the development of several cancers, including colon cancer [7]. Studies have shown that many colon cancers have global DNA hypomethylation [7, 8]. Relatively few studies, however, have specifically attempted to address the overall impact of chromatin modification, including histone modification, for colorectal cancer risk.
Histone proteins, around which DNA is wrapped, can be chemically modified in several places by the addition of acetyl, methyl, or other groups. Histone acetylation and deacetylation are essential elements along with phosphorylation and methylation of an intricate “histone code” that regulates gene transcription [9, 10]. The reversible process of histone acetylation is controlled by two classes of enzyme, histone acetyltransferases (HAT) and histone deacetylases (HDAC), which catalyze the addition to and removal of acetyl groups on lysine residues in proteins. Hyperacetylation of histones has been associated with transcriptional activation of genes as chromatin becomes accessible to a number of transcription factors and coactivator complexes [11, 12]. Conversely, HDACs induce transcriptional repression by removing these acetyl groups, leading to chromatin condensation [13]. In addition to histones, both HATs and HDACs target non-histone protein substrates, such as transcription factors (e.g., p53) [14, 15] and structural proteins (e.g., α-tubulin) [16]. Deregulated HAT and HDAC activity plays a role in the development of a range of cancers, and inhibitors are being targeted in the clinic at different HDAC family members (i.e., class I, II, and III inhibitors) [17].
Histone modification can effect the activation and repression of oncogenes and tumor suppressor genes. Studies have shown that H3K18 and H4K12 acetylation can be used as predictive biomarkers for cancer recurrence in the prostate [18], and in non-small-cell lung cancer [19]. Alteration of histones (including hypo- and hyperacetylation) is related to promoter methylation of signature genes in cancer which can result in aberrant protein expression [8, 20, 21]. Histone modifications and environmental factors such as smoking, drinking, and obesity are important in colonic malignancy. Colorectal cancer is more advanced in African Americans at the time of diagnosis; however, colonic malignancy and precancerous lesions including adenoma could be detected earlier if diagnosed. Therefore, the acetylation status of genes can be potentially informative in the colonic malignancy process in patients. DNA methylation and histone deacetylation are associated with silencing of APC and DAP kinase gene expression in colorectal cancer (CRC) and gastric cancers [22]. To test the hypothesis that global levels of individual histone modifications and HDAC2 predict tumor outcome in colon cancer patients, we performed immunohistochemical analyses on tissue microarrays, which are specifically designed for high-throughput analysis.
Methods and Materials
Human Colonic Tissues
Archival formalin-fixed paraffin-embedded colonic biopsies were obtained from African American patients undergoing colonoscopy at Howard University Hospital. This study was approved by the Howard University Institutional Review Board (IRB), and the purpose of this study was explained to the patients before colonoscopy. Clinical data collected on each patient included race, gender, associated past medical history, medication use, and family history of adenoma and colonic cancer (Table 1). Ninety-six percent of the tumors were moderate to well differentiated, and 4% were poorly differentiated. Patients were deemed eligible if colonoscopy resulted in a first diagnosis of adenoma or adenocarcinoma, confirmed by histopathology. From a review of the medical records, clinical information was collected and TNM status (TNM Classification of Malignant Tumours) was recorded based upon the American Joint Committee on Cancer (AJCC) staging system [23]. For survival data collection, we defined the vital status (dead or alive) for each of the 59 colon cancer cases in this study based on two different sources—the latest inpatient medical record of cases in Howard University Hospital and the Social Security Death Index website (http://ssdi.rootsweb.com/). For each deceased case the date of death was recorded. In cases with no record of death in both sources, the latest update date of the Social Security Death Index website (10/1/2007) was recorded as the date of follow-up.
Table 1.
Characteristics of adenoma and cancer cases (numbers in parentheses show the percentage unless otherwise noted)
| Characteristic | Adenoma | Cancer | P value |
|---|---|---|---|
| Mean age (SD) | 61.8 (10.5) | 64.9 (15.5) | 0.16 |
| Gender | |||
| Male | 57 (42.5) | 33 (56.9) | |
| Female | 77 (57.5) | 25 (43.1) | |
| Total | 134 | 58 | 0.07 |
| Location | |||
| Right colon | 86 (66.2) | 32 (66.7) | |
| Left colon | 44 (23.8) | 16 (33.3) | |
| Total | 130 | 48 | 0.95 |
Tissue Microarrays and Immunohistochemical Analysis
Tissue microarrays (TMAs) were constructed using a Beecher Instruments MTA-1 tissue arrayer. Each TMA contained tissue from normal, adenoma, and adenocarcinoma, based on a published protocol [24]. At a minimum, duplicate tumor samples were taken from donor tissue blocks. In total, 250 cases were analyzed for all three histone markers; 134 adenomas, 55 moderate to well differentiated carcinomas, and four poorly differentiated carcinomas along with matched adjacent normal tissues (57 samples) were available for control comparisons. A retrospective analysis for outcome assessment was based on detailed clinico-pathological information linked to the TMA specimens. TMA obtained from paraffin-embedded blocks was used for the immunohistochemistry experiments. Sections (5 μm) were mounted on charged glass slides, deparaffinized with xylene for 2 × 10 min and rehydrated using a graded ethanol series. Antigen retrieval was performed by placing the samples in a microwave oven for 12 min, with occasional interruption to avoid tissue degradation by excessive heat. The slides were then treated with hydrogen peroxide, followed by incubation with the primary and secondary antibodies, a streptavidin-biotin complex, an amplification reagent, streptavidin-peroxidase, and substrate-chromogen solution using the Envision system according to the manufacturers’ protocol (DAKO, Carpentaria, CA). The samples were then counterstained with hematoxylin, rinsed with ethanol, dried, and visualized by a light microscopy. Tissue samples to which no primary antibody had been added were used as negative controls. All immunohistochemistry reagents were purchased from DAKO and the antibodies (H3K18, and HDAC2 clone G168-15, 1/10 dilution and H4K12 clone 556349, 1/50 dilution) were purchased from Abcam (San Diego, CA). The slides were read by two pathologists (E.L and M.T.), and the percentage of the nuclear staining was recorded.
Statistical Analysis
Distribution for percentage of stained cells was studied by computing mean and standard deviation (SD). For continuous measures, we first tested the assumption of normal distribution. The analysis of variance (ANOVA) was then applied to test whether the percentage of stained cells differed between normal, adenoma, and cancer samples. P values <0.05 were assumed statistically significant. Then we computed a new categorical variable based on a cut point of 25% of expression in H3K18 or H4K12 showing the level of expression in each of these two markers. This way we have two groups of cases with reasonable sample size. We used chi-square to assess the relationship between co-expression of these markers and pathology. We studied the discrimination power of HDAC2 expression by a receiver operative characteristics curve (ROC). The overall performance of each marker was computed by the area under curve (AUC). We used the Kaplan-Meier method to compute the survival function for all cases. Log-rank test was used to compare the survival function between different demographic variables and histone marker categories (≤50% vs. >50% expression). SPSS15.0 software (Chicago, IL) was used for all statistical analysis.
Results
Global H3, H4 Acetylation and HDAC2 Expression in Progression of CRC
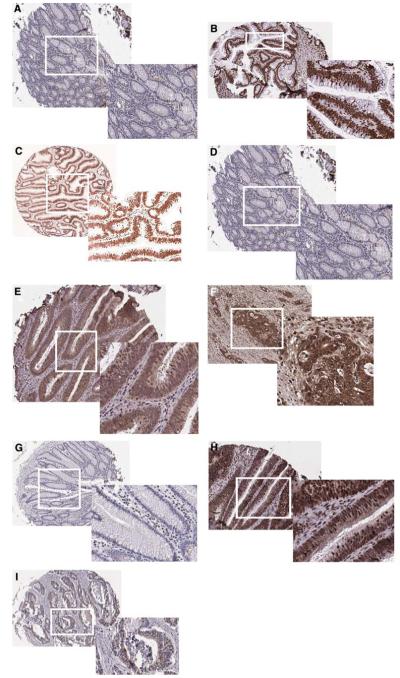
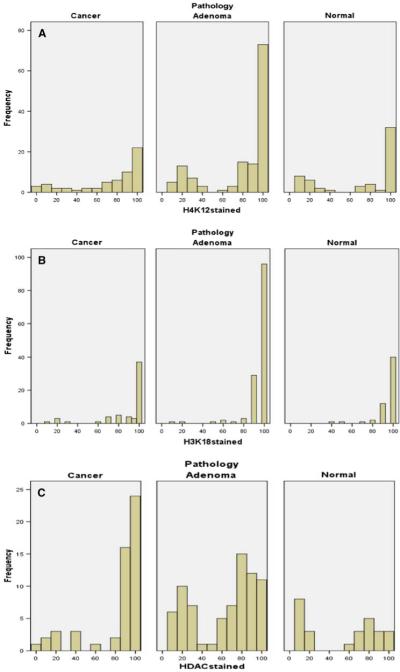
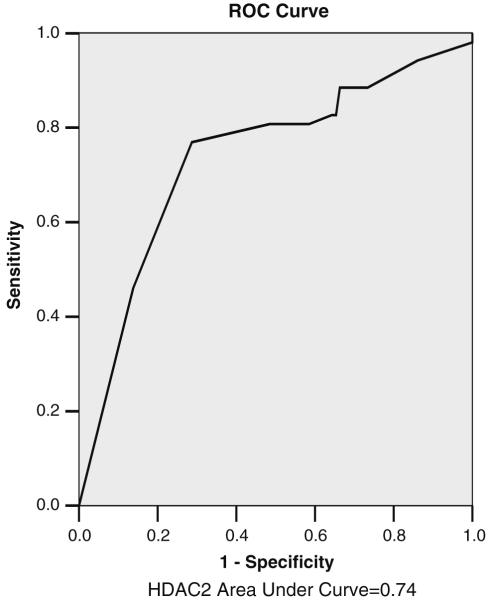
We determined the expression level of H3K18 and H4K12 in TMAs containing 134 adenomas, 55 moderate to well differentiated carcinomas, four poorly differentiated carcinomas, as well as 57 matched adjacent normal tissues. Liver, spleen, tonsil, and kidney tissues were also used as controls in the TMAs. The number of male patients in the adenoma and cancer groups were 57 (42.5%) and 33 (56.9%), respectively. In normal colonic biopsy specimens, we detected low expression for acetylated histones or HDAC2 in epithelial cells at the surface of colonic glands and at the glandular base (Fig. 1a, e, h, respectively). In marked contrast, positive nuclear staining of epithelial cells was observed in adenomas and carcinomas (moderate to well differentiated) for acetylated histones H3 (Fig. 1b, c), H4 (Fig. 1e, f), and HDAC2 (Fig. 1h, i). After calculating the percentage of cells that were positively stained for AcH3, AcH4, and HDAC2, there was no significant difference between cancer, adenoma, and normal specimens with respect to H3K18 expression. For example, H3K18 acetylation was present in 91.3% of adenomas, 86.3% of adenocarcinomas (96% of the tumors were moderate to well differentiated), and 88.4% of adjacent normal cells (Table 2); therefore, H3K18 acetylation was not considered to be informative of tumor stage. On the other hand, global acetylation of H4K12 was highly significant; the percentage of staining was 43.6% in normal, 61% in adenoma and 71.5% in well to moderately differentiated carcinoma (P = 0.002 for the trend) but not in poorly differentiated. HDAC2 expression also was highly significant (P = 0.002), with corresponding numbers of 53.1% (normal), 63.1% (adenoma), and 81.9% (carcinoma, well to moderately differentiated), respectively (Table 2, Fig. 2). In addition, we studied the discriminating power of HDAC2 in distinguishing between cancer and non-cancer diagnosis by ROC, in which the sensitivity and specificity of HDAC2 expression was assessed (Fig. 3). AUC for HDAC2 was 0.74. The additive effect between and among the three markers was also calculated. The correlation coefficient test (Table 3) showed that HDAC2 and H4K12 together may have a meaningful and significant effect in progression of colon cancer.
Fig. 1.
Global histone modifications and HDAC2 expression in human colon moderate to well differentiated cancers. Characteristic nuclear staining of normal colon, adenoma, and adenocarcinoma was determined by immunohistochemistry in tissue microarrays, using antibodies against H3K18 acetylation (a-c), H4K12 acetylation (d-f), and HDAC2 (g-i). Representative sections from H3K18 (c), H4K12 (f), and HDAC2 (i) cancer patients with more than 90% positive staining are shown. Original magnifications ×10, except for enlargements (insets) which are ×20
Table 2.
Percentage of expression of different histone acetylation markers between normal, adenoma, and cancer (parenthesis show standard deviation)
| Normal | Adenoma | Cancer | P value for ANOVA | |
|---|---|---|---|---|
| H3K18 | 88.4 (14.9) | 91.3 (15.8) | 86.3 (24.3) | 0.33 |
| H4K12 | 43.6 (34.3) | 61.0 (32.8) | 71.5 (33.3) | <0.001 |
| HDAC2 | 53.1 (36.5) | 63.1 (31.2) | 81.9 (29.1) | <0.001 |
Fig. 2.
Distribution of staining for acetylated histones and HDAC2 in colon cancers moderate to well differentiated, adenomas, and normal colon. Data are summarized across all 250 tissue samples for H4K12 (a), H3K18 (b), and HDAC2 (c). The y axis shows the fraction of samples with positive staining for the indicated percentage of cells (x axis)
Fig. 3.
Receiver operating characteristic (ROC) plot of HDAC2 expression as a biomarker for cancer diagnosis. The discriminating power of HDAC2 in distinguishing between cancer and non-cancer diagnosis is presented graphically in an ROC curve, with the sensitivity and specificity of HDAC2 expression (HDAC2 antibody). AUC for HDAC2 was 0.74
Table 3.
Correlation table (Spearman correlation coefficient)
| Samples | Histones |
|||
|---|---|---|---|---|
| H4K12 | H3K18 | HDAC2 | ||
| Cancer | H4K12 | |||
| H3K18 | 0.15 | |||
| HDAC2 | 0.40* | 0.06 | ||
| Adenoma | H4K12 | |||
| H3K18 | 0.16 | |||
| HDAC2 | 0.12 | 0.10 | ||
| Normal | H4K12 | |||
| H3K18 | 0.04 | |||
| HDAC2 | 0.22 | 0.27 | ||
| Total | H4K12 | |||
| H3K18 | 0.15 | |||
| HDAC2 | 0.38* | 0.14 | ||
P < 0.01
Acetylated H3, H4, and HDAC2 Expression, Differentiation, and Tumor Stage
Among 59 CRC cases available for analysis, the number of subjects with stage I, II, III, and IV were 14 (24.5%), 20 (35.1%), 21 (36.8), and 2 (3.5%), respectively, with two missing. In general, there were no statistically significant differences for age, sex, anatomic location (Table 1), H4K12, H3K18 acetylation, and HDAC2 expression with tumor stage (data not shown). Co-expression of H4K12 and HDAC2 was found in 80 and 86% of patients with stage II and III disease, respectively; however, this was not significant. The corresponding nuclear global expression levels in moderate to well differentiated tumors for H4K12 and H3K18 acetylation were increased while these levels were decreased in poorly differentiated tumors (P = 0.02).
H3K18, H4K12 Acetylation and HDAC2 Expression Lack Correlation with Survival
Survival curves were calculated using the Kaplan-Meier method. The follow-up was completed in 55 cancer cases (93%), and among them, 25 (46%) were deceased in a follow-up period from 3 days to 123.4 months. The median follow-up time was 27.7 months. The median survival (95% CI) for CRC cases was 61.2 (24.5-97.8) months. One year survival was 78%, 2 year 66%, 5 year was 52%, and 10 year survival was 17%. Higher age at diagnosis was a poor prognostic factor in our patients (Table 4). We did not observe any significant difference in survival between the percentage nuclear staining of H3K18, H4K12, and HDAC2. The pattern of the survival in samples with alteration of H3, H4, and HDAC2 staining was similar (data not shown). In addition, the cut point of 50% staining for either H3 or H4 marker was not a good predictive marker for survival (Table 5).
Table 4.
The median (95% CI) survival time (in months) by demographic factors
| Factor | Median survival (95% CI) | P for log rank |
|---|---|---|
| Gender | 0.13 | |
| Male | 79.51 (26.9-132.2) | |
| Female | 41.3 (9-0-23.6) | |
| Age | 0.001 | |
| <60 | 107 (89.9-124.2) | |
| ≥60 | 40.6 (25.8-55.4) | |
| Tumor location | 0.38 | |
| Right | 37.5 (19.8-55.2) | |
| Left | 91.9 (0.0-197.4) | |
| Stage | 0.72 | |
| 0, 1 | 44.2 (38.2-50.2) | |
| 2 | 73.8 (0.0-155.3) | |
| 3,4 | 37.5 (13.0-62.0) |
Table 5.
The median (95% CI) survival time (in months) compared within H3K18, H4K12 acetylation and HDAC2 expression
| Factor | Median survival (95% CI) | P for log rank |
|---|---|---|
| H3K18 staining | 0.16 | |
| ≤50% | 11.4 (0.0-37.9) | |
| >50% | 61.15 (18.0-104.3) | |
| H4K12 staining | 0.79 | |
| ≤50% | 44.2 (11.6-76.8) | |
| >50% | 73.8 (9.6-138.1) | |
| HDAC staining | 0.89 | |
| ≤50% | 44.1 (19.7-71.5) | |
| >50% | 73.8 (2.6-145.1) |
Discussion
We investigated the association between pathological stage at diagnosis, acetylation of H3, H4 and expression of HDAC2, and whether or not the histone modification and chromatin changes in CRC during tumorigenesis including colon adenoma and cancer. We collected tissue from 59 colon carcinoma and 134 adenoma cases. The data presented here help us to understand the temporal development of colon cancer as a function of changes in global acetylation of H3 and H4, and expression of HDAC2. We have provided evidence that changes in bulk histone modifications of CRC cells are predictive of disease progression from adenoma to carcinoma. The mechanistic basis of such changes are currently unclear but may be related to the altered expression and/or global activities of various histone-modifying enzymes. The variability in the levels of any one modification was not sufficient for predicting outcome. However, in combination, these changes proved to be indicative of the tumor risk in patients with colon adenoma. Considering the substantial number of modifications on histones, it is possible that information on global patterns of other modification sites will help with the further classification of all patients. Hence, the utility of immunohistochemistry, combined with the availability of an extensive set of antibodies, provide such opportunity. We counted the number of stained cells vs. the staining intensity since the complete or partial loss of either H3 or H4 expression cannot reflect the expression status of H3, H4 or HDAC2. For example, when we counted the number of cells in a field of 100, this was highly representative of the expression compared to the general intensity of the signals [25]. The associations between the disease and the markers were identified by percent of the cell being positive for the markers compared to staining intensity analyzed by immunohistochemistry (IHC). The reproducibility and validation are much stronger for the value of the staining vs. the intensity, which is why we chose the percentage value to establish the correlation of expression with the progression of the colorectal cancer [25]. The exact mechanisms following histone modification in human cancers remains unclear. Several mechanisms have been proposed, including increasing enzymatic activity of DNA methyltransferase and imbalance between histone acetylation and deacetylation [26, 27]. Recent studies show that here is a close association between histone acetylation and DNA methylation of some tumor suppressor genes in gastrointestinal carcinogenesis [28, 29]. The association of acetylated histone with a methylated promoter region has been demonstrated in a number of tumor suppressor genes such as p21 (WAF1/CIP1), hMLH1, p16INK4a, and p14ARF [30, 31]. Similarly, our results demonstrated a close association between HDAC2 and colorectal cancer progression.
A recent study showed the frequent down-regulation of SIN3A as a potential tumor suppressor candidate gene with frequent loss of heterozygosity (LOH) and polymorphisms associated with up-regulation of HDAC activity in non-small cell lung cancers [32]. This is consistent with our results; however, we did not observe any correlation between the MSI and histone modification including HDAC2. There are four class of HDAC, of which class I and II are the most important since they are components of large transcriptional co-repressor complexes [33]. The inhibitor of class I and II HDACs, such as SAHA, inhibits the enzymatic activity for these two class, hence inducing growth arrest, differentiation, and apoptosis in cell lines derived from a range of malignancies including colon cancer [34]. Our study showed the discriminating power of HDAC2 in distinguishing between cancer and non-cancer diagnosis by ROC in which the sensitivity and specificity of HDAC2 expression was assessed and AUC for HDAC2 was 0.74 (Fig. 3). Therefore, the role of the HDAC2 inhibitor is important in the treatment of CRC, which implies a physiological role for these proteins in the maintenance of colon cell proliferation and survival, and inhibition of cell differentiation. Functionally, HDACs regulate gene expression by at least two mechanisms. First, upon recruitment to DNA by sequence-specific transcription factors, they catalyze the deacetylation of specific lysine residues in DNA-bound core histone proteins. Lysine deacetylation of histones confers a positive charge on histone proteins, which among other effects, enhances their affinity for negatively charged DNA. A consequence of this alteration in nucleosome conformation is reduced accessibility of the transcriptional regulatory machinery to the DNA template, resulting in transcriptional alteration, and this is why we see the increase of both global acetylation and HDAC2 expression [35, 36]. The presence of many HDACs and the role of individual HDACs that target specific lysine residues within histone tails have not been extensively explored. However, preferential acetylation of H3K18 and H3K9 following knockdown of HDAC1 and HDAC3, respectively, has been reported in Hela cells, suggesting this may be a possibility [37]. Therefore it is important to check whether the other marker, such as H3K9 or H4K16, is over-expressed. We are planning to study this in the near future.
The acetylation of H3 or H4 decreased in the poorly differentiated cancer cells while the net or global expression in moderate to well differentiated cancer cells increased. With the low number of poorly- and well-differentiated samples, we were unable to do meaningful analysis between groups. There was a moderate correlation between the HDAC2 plus H4K12 and the cancer. This connection between HDAC2 and H4K12 is complex, since multiple histone acetyl transferase exist. Therefore, the HAT activity is important to establish a better correlation between HDAC2 and H4 or H3 acetylation. The survival data show that there is no difference between outcome in stage I and stage IV disease and acetylation of H3, H4 and HDAC2 expression, and this may be the result of insufficient sample size.
The exact manner in which histone modification affects promoter activation or repression (through methylation) needs further investigation, possibly by using selective HDAC inhibitors or agents that target methylation of colon cancer signature genes, such as APC [38-40]. In human colon cancer cells, inhibition of HDAC activity by dietary agents such as sulforaphane was accompanied by global increases in histone H3 and H4 acetylation [21], coupled with localized histone hyperacetylation on the promoter of the p21 gene [21]. This may cause the changing balance between proliferation and apoptosis, and block colon tumor progression. One hallmark of human cancers is the loss of monoacetylation and trimethylation of histone H4 [41]. Selective agents are being sought that might target abnormal patterns of histone modification as a means of destroying cancer cells. The differentiation status of the cancer is important in regard to the chromatin modification, and hence, future studies will expand the current work using a wide-genome array-based technique to study histone modification and HDAC2 expression in colorectal adenoma and cancer with different levels of differentiation. The results to date are encouraging because they demonstrate that aberrant expression of HDAC2 frequently occurs in patients with colorectal cancer, providing potential biomarkers for use in future clinical trials.
Acknowledgments
This work was supported in part by grants A102681 and CA122959 from the National Cancer Institute.
Contributor Information
Hassan Ashktorab, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC, USA & Howard University College of Medicine, Washington, DC, USA.
Kevin Belgrave, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC, USA & Howard University College of Medicine, Washington, DC, USA.
Fatemeh Hosseinkhah, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC, USA & Howard University College of Medicine, Washington, DC, USA.
Hassan Brim, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC, USA & Department of Pathology, Howard University College of Medicine, Washington, DC, USA.
Mehdi Nouraie, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC, USA & Howard University College of Medicine, Washington, DC, USA.
Mikiko Takkikto, Tissue Array Research Program, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD, USA.
Steve Hewitt, Tissue Array Research Program, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD, USA.
Edward L. Lee, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC, USA & Department of Pathology, Howard University College of Medicine, Washington, DC, USA
R. H. Dashwood, The Linus Pauling Institute, Oregon State University, Corvallis, ORUSA
Duane Smoot, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC, USA & Howard University College of Medicine, Washington, DC, USA.
References
- 1.Howe HL, Wingo PA, Thun MJ, et al. Annual report to the nation on the status of cancer (1973 through 1998), featuring cancers with recent increasing trends. J Natl Cancer Inst. 2001;93:824–842. doi: 10.1093/jnci/93.11.824. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Troisi RJ, Freedman AN, Devesa SS. Incidence of colorectal carcinoma in the US: an update of trends by gender, race, age, subsite, and stage, 1975-1994. Cancer. 1999;85:1670–1676. [PubMed] [Google Scholar]
- 4.Ashktorab H, Smoot DT, Carethers JM, et al. High incidence of microsatellite instability in colorectal cancer from African Americans. Clin Cancer Res. 2003;9:1112–1117. [PubMed] [Google Scholar]
- 5.Ashktorab H, Smoot DT, Farzanmehr H, et al. Clinicopathological features and microsatellite instability (MSI) in colorectal cancers from African Americans. Int J Cancer. 2005;116:914–919. doi: 10.1002/ijc.21062. doi:10.1002/ijc.21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carethers JM. Racial and ethnic factors in the genetic pathogenesis of colorectal cancer. J Assoc Acad Minor Phys. 1999;10:59–67. [PubMed] [Google Scholar]
- 7.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. doi:10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 8.Tzao C, Sun GH, Tung HJ, et al. Reduced acetylated histone H4 is associated with promoter methylation of the fragile histidine triad gene in resected esophageal squamous cell carcinoma. Ann Thorac Surg. 2006;82:396–401. doi: 10.1016/j.athoracsur.2006.03.066. doi:10.1016/j.athoracsur.2006.03.066. [DOI] [PubMed] [Google Scholar]
- 9.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. doi:10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 10.Marks PA, Richon VM, Breslow R, et al. Histone deacetylase inhibitors as new cancer drugs. Curr Opin Oncol. 2001;13:477–483. doi: 10.1097/00001622-200111000-00010. doi:10.1097/00001622-200111000-00010. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. doi:10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein BE, Schreiber SL. Global approaches to chromatin. Chem Biol. 2002;9:1167–1173. doi: 10.1016/s1074-5521(02)00265-x. doi:10.1016/S1074-5521(02)00265-X. [DOI] [PubMed] [Google Scholar]
- 13.Cress WD, Seto E. Histone deacetylases, transcriptional control, and cancer. J Cell Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Colosimo AL, Yang XJ, et al. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol Cell Biol. 2000;20:5540–5553. doi: 10.1128/mcb.20.15.5540-5553.2000. doi:10.1128/MCB.20.15.5540-5553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Tseng M, Perdreau SA, et al. Histone H2AX is a mediator of gastrointestinal stromal tumor cell apoptosis following treatment with imatinib mesylate. Cancer Res. 2007;67:2685–2692. doi: 10.1158/0008-5472.CAN-06-3497. doi:10.1158/0008-5472.CAN-06-3497. [DOI] [PubMed] [Google Scholar]
- 16.Hubbert C, Guardiola A, Shao R, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. doi:10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 17.de Ruijter AJ, van Gennip AH, Caron HN, et al. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. doi:10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. doi:10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 19.Barlesi F, Giaccone G, Gallegos-Ruiz MI, et al. Global histone modifications predict prognosis of resected non small-cell lung cancer. J Clin Oncol. 2007;25:4358–4364. doi: 10.1200/JCO.2007.11.2599. doi:10.1200/JCO.2007.11.2599. [DOI] [PubMed] [Google Scholar]
- 20.Kondo Y, Shen L, Yan PS, et al. Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proc Natl Acad Sci USA. 2004;101:7398–7403. doi: 10.1073/pnas.0306641101. doi:10.1073/pnas.0306641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin Cancer Biol. 2007;17:363–369. doi: 10.1016/j.semcancer.2007.04.001. doi:10.1016/j.semcancer.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satoh A, Toyota M, Itoh F, et al. DNA methylation and histone deacetylation associated with silencing DAP kinase gene expression in colorectal and gastric cancers. Br J Cancer. 2002;86:1817–1823. doi: 10.1038/sj.bjc.6600319. doi:10.1038/sj.bjc.6600319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sobin LH, Fleming ID, Union Internationale Contre le Cancer and the American Joint Committee on Cancer TNM Classification of malignant tumors, fifth edition (1997) Cancers. 1997;80:1803–1804. doi: 10.1002/(sici)1097-0142(19971101)80:9<1803::aid-cncr16>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 24.Hewitt SM. The application of tissue microarrays in the validation of microarray results. Methods Enzymol. 2006;410:400–415. doi: 10.1016/S0076-6879(06)10020-8. doi:10.1016/S0076-6879(06)10020-8. [DOI] [PubMed] [Google Scholar]
- 25.Zlobec I, Terracciano L, Jass JR, et al. Value of staining intensity in the interpretation of immunohistochemistry for tumor markers in colorectal cancer. Virchows Arch. 2007;451:763–769. doi: 10.1007/s00428-007-0466-8. doi: 10.1007/s00428-007-0466-8. [DOI] [PubMed] [Google Scholar]
- 26.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. doi:10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 27.Ono S, Oue N, Kuniyasu H, et al. Acetylated histone H4 is reduced in human gastric adenomas and carcinomas. J Exp Clin Cancer Res. 2002;21:377–382. [PubMed] [Google Scholar]
- 28.Yasui W, Oue N, Ono S, et al. Histone acetylation and gastrointestinal carcinogenesis. Ann N Y Acad Sci. 2003;983:220–231. doi: 10.1111/j.1749-6632.2003.tb05977.x. [DOI] [PubMed] [Google Scholar]
- 29.Mitani Y, Oue N, Hamai Y, et al. Histone H3 acetylation is associated with reduced p21(WAF1/CIP1) expression by gastric carcinoma. J Pathol. 2005;205:65–73. doi: 10.1002/path.1684. doi:10.1002/path.1684. [DOI] [PubMed] [Google Scholar]
- 30.Fahrner JA, Eguchi S, Herman JG, et al. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–7218. [PubMed] [Google Scholar]
- 31.Baylin SB, Esteller M, Rountree MR, et al. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. doi:10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki H, Ouchida M, Yamamoto H, et al. Decreased expression of the SIN3A gene, a candidate tumor suppressor located at the prevalent allelic loss region 15q23 in non-small cell lung cancer. Lung Cancer. 2008;59:24–31. doi: 10.1016/j.lungcan.2007.08.002. doi:10.1016/j.lungcan.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- 34.Kim YS, Tsao D, Siddiqui B, et al. Effects of sodium butyrate and dimethylsulfoxide on biochemical properties of human colon cancer cells. Cancer. 1980;45:1185–1192. doi: 10.1002/1097-0142(19800315)45:5+<1185::aid-cncr2820451324>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 35.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. doi:10.1016/S0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 36.Ammanamanchi S, Freeman JW, Brattain MG. Acetylated sp3 is a transcriptional activator. J Biol Chem. 2003;278:35775–35780. doi: 10.1074/jbc.M305961200. doi:10.1074/jbc.M305961200. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Wharton W, Yuan Z, et al. Activation of the growth-differentiation actor 11 gene by the histone deacetylase (HDAC) inhibitor trichostatin A and repression by HDAC3. Mol Cell Biol. 2004;24:5106–5118. doi: 10.1128/MCB.24.12.5106-5118.2004. doi:10.1128/MCB.24.12.5106-5118.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong Y, Dowdy SC, Podratz KC, et al. Histone deacetylase inhibitors decrease DNA methyltransferase-3B messenger RNA stability and down-regulate de novo DNA methyltransferase activity in human endometrial cells. Cancer Res. 2005;65:2684–2689. doi: 10.1158/0008-5472.CAN-04-2843. doi:10.1158/0008-5472.CAN-04-2843. [DOI] [PubMed] [Google Scholar]
- 39.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1079–1080. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 40.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. doi:10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 41.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. doi:10.1038/ng1531. [DOI] [PubMed] [Google Scholar]