Abstract
Post-translational modifications of histones are the subject of intensive investigations with the aim of decoding how they regulate, alone or in combination, chromatin structure, genomic stability, and gene expression. Major epigenetic programming events take place during gametogenesis and fetal development and are thought to have long-lasting consequences on adult health. Epidemiological and experimental studies have pointed toward maternal nutrition as a major player during prenatal development in influencing disease susceptibility later in life. Although the mechanisms underlying such observations are not well elucidated, epigenetic alterations of histones by particular maternal diets might be of central importance. Moreover, as much as dietary sources can influence epigenetic programming during pregnancy, they have started to be implicated in cancer chemoprevention, via the targeting of reversible epigenetic deregulations at the level of the histones.
Keywords: epigenetics, histone code, diet, fetal development, cancer, HDAC, HAT
INTRODUCTION
Although an understanding of how epigenetic mechanisms drive the expression of the genome has only recently stirred up the scientific community, reversible histone modifications were discovered about 50 years ago. Together with DNA methylation, they are involved in the control of chromatin assembly, DNA replication and repair, chromosome segregation, and gene expression. Epigenetic mechanisms have been implicated in critical processes, such as gametogenesis, fetal development, and maintenance of early established patterns of gene expression throughout life. The epigenetic information carried by DNA and histone modifications appears to be influenced significantly by environmental factors, including nutrition, and might explain how maternal diets can have lifelong consequences on health and disease risk. In the meantime, nutritional intervention has proven to be a powerful tool in the battle against cancer and other chronic diseases. In the epigenetic arena, research has mainly focused on diet-related changes in DNA methylation status, but the present review highlights recent observations on histone manipulation by dietary agents.
EPIGENETIC LANGUAGE: THE “HISTONE CODE”
In eukaryotic nuclei, a complex of DNA and histones efficiently packages the heritable genetic information in the form of chromatin. Specific patterns of chromatin modification regulate protein interactions with DNA. Transcriptionally active regions of the genome reside in euchromatin, whereas inactive regions reside in heterochromatin. The presence of euchromatic and heterochromatic regions reflects the regulation of chromatin structure by epigenetic mechanisms. Epigenetic regulations also control gene expression and chromosome condensation and segregation during mitosis and meiosis. Alterations in chromatin structure are intimately related to changes in chromatin function and are regulated by specific chromatin remodeling enzymes often grouped in multiprotein complexes. Nucleosomes are composed of 147 bp of DNA wrapped around an octamer of core histone proteins, namely H2A, H2B, H3, and H4 (81). Another histone called H1 is located at the outer surface of the nucleosome and serves as an anchor to fix DNA to the nucleosome. Acetylation, biotinylation, methylation, phosphorylation, ubiquitination, SUMOylation, and ADP ribosylation are among the different types of chemical modifications mainly located within the N-terminal tails of core histones. Over the past few years, the list of histone modifications and histone-modifying complexes has continued to grow, as well as the intricacy of what is now commonly designated as the histone code. The histone code works together with the DNA methylation code to ensure an efficient silent state of chromatin and, from time to time and in a gene-specific way, to allow genes to be transcriptionally expressed. The appropriate on/off switching of genes during cell cycle progression and development requires the relief of repressive mechanisms concomitant with the establishment of a new epigenetic profile. Silencing of chromatin has been diversely related to germline development, X-inactivation, stem cell identity, cell cycle regulation, and mitosis/meiosis. Thus, a description of the most common patterns of DNA and histone modification may help in decoding epigenetic mechanisms implicated in chronic diseases such as cancer.
Epigenetic Mechanisms of Gene Silencing
Historically, epigenetic mechanisms often focused on the methylation of DNA by DNA methyltransferases (DNMT1, 2, 3a, 3b, and 3L), involving the addition of methyl groups to cytosines within dinucleotide CpG islands. More recently (Figure 1), the methylation of histones has been found to constitute a quite stable covalent alteration, but requiring careful interpretation with respect to the specific amino acid residue modified. Methylation at lysines 4 or 79 of histone H3 (H3K4 or H3K79) correlates with active transcription, whereas methylation at H3K9, H3K27, H4K20, and H1K26 are found typically in transcriptionally silent regions (34). Moreover, this methylation code is further complicated by the possibility that a lysine residue can carry one to three methyl groups, which generates differential effects on chromatin structure and transcriptional regulation. For example, monomethylated H3K27 has been found broadly distributed throughout euchromatin, whereas H3K27 di- and trimethylation were missing from active promoters (121).
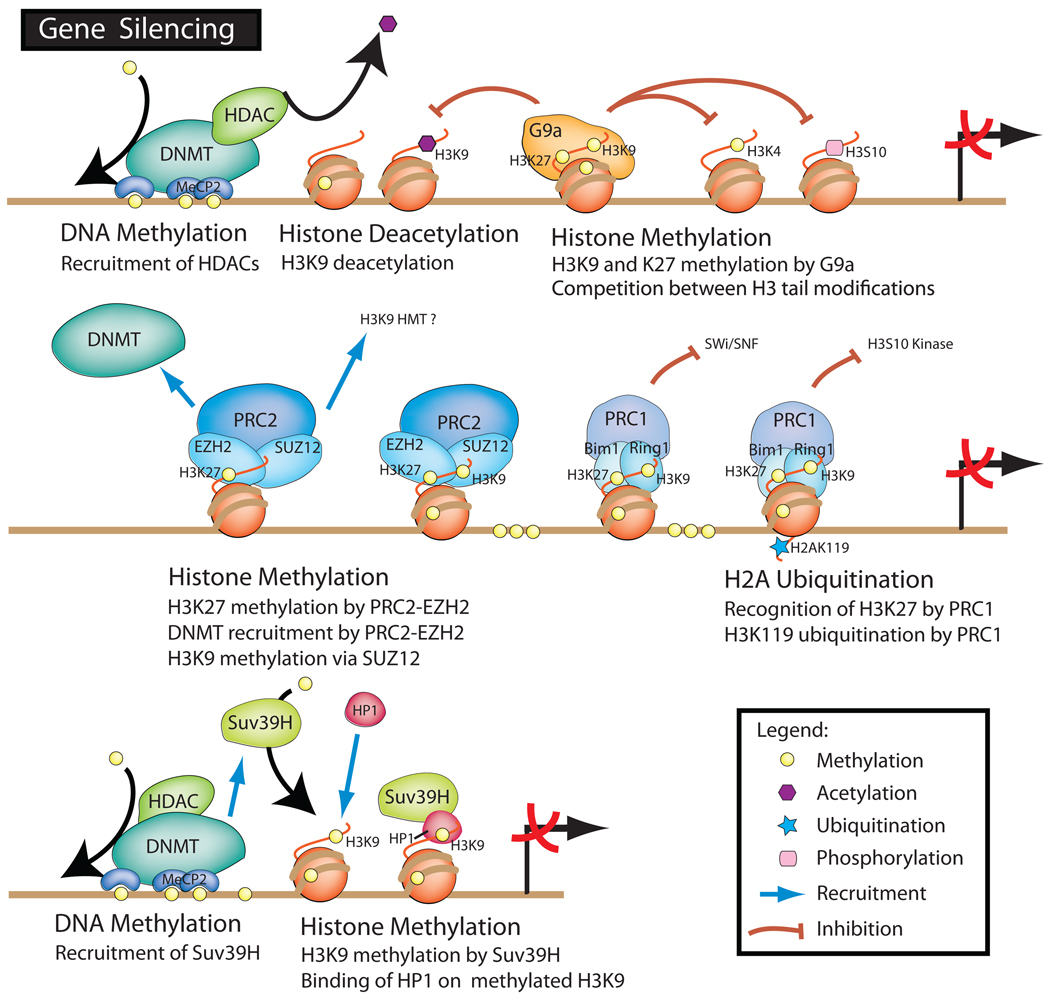
Figure 1.
Interactions between histone-modifying enzymes and histone modifications associated with the silencing of gene expression. DNA methylation catalyzed by DNA methyl transferases and methyl-binding domain proteins can recruit histone deacetylases and histone methyl transferases that further repress gene transcription. DNMT, DNA methyltransferase; HDAC, histone deacetylase; HP, histone-binding protein; PRC, Polycomb repressive complexes; Suv, suppressor of variegation; SWI/SNF, switch/sucrose nonfermentable.
The addition of methyl groups to histone tails is catalyzed by lysine-specific histone methyltransferases (HMTs). Most HMTs contain a SET domain referring to three proteins originally discovered in the fruit fly Drosophila melanogaster, namely suppressor of variegation [Su(var)3–9], enhancer of zeste (Ez), and trithorax (34). Pericentromeric and centromeric heterochromatin is characterized by an enrichment of trimethylated H3K9. The establishment of H3K9 hypermethylation at these regions is catalyzed by the HMTs Suv39h1 and Suv39h2 (mentioned later as Suv39H), counterparts of Drosophila Su(var)3–9 (74). The impairment of Suv29H function and the loss of H3K9 trimethylation reveal their essential role in heterochromatin organization and chromosome segregation during mitosis and meiosis (96). Methylated H3K9 at centromeric heterochromatin is well known to serve as a recognition mark for the recruitment and docking of heterochromatin protein 1 (HP1). The HP1 family, which includes HP1α, HP1β, and HP1γ, is evolutionarily conserved in fungi, plants, and animals, and was first discovered through studies in Drosophila (LPH1). All three HP1 family members are encoded by distinct genes, and are characterized by a chromodomain that binds specifically to di- and trimethylated H3K9.
Maintenance of a heterochromatic structure also requires DNMT1 and DNMT3a, recruited by Suv39h1, which interacts specifically with HP1β (43). The presence of trimethylated H3K9 in transcriptionally repressive heterochromatin prevents the establishment of other histone modifications, such as phosphorylation of H3S10 (96, 100). Consequently, the acetylation of H3K14 by the histone acetyltransferase (HAT) GCN5 (19), normally facilitated by the presence of phosphorylated H3S10, was also inhibited by H3K9 methylation (100). Specific interactions with the methylated CpG-binding protein MeCP2 might also be required for HP1 recruitment at heterochromatic regions (2). Alterations in the level of CpG methylation due to DNMT1 (36) or DNMT3a/3b knockout (47) also resulted in the depletion of di- and trimethylated H3K9, and a concomitant increase in H3K9 acetylation.
Although HP1α and HP1β are exclusively heterochromatic, HP1γ is found in both heterochromatin and euchromatin, which suggests that HP1 may also display different functions depending on the chromatin environment. Accordingly, the H3K9 methylation mark spreads beyond the boundary element of heterochromatin and contributes to transcriptional silencing in neighboring euchromatin. While Suv39H methylates H3K9 at (peri)centromeric chromatin, another SET domain protein, called G9a histone methyltransferase, targets H3K9 at transcriptionally active euchromatin (116). As with Suv39H−/− mice, the depletion of G9a in ES cells altered the methylation of H3K9 as well as other modifications of the neighboring residues in H3 tails. H3S10 phosphorylation and H3K14 acetylation were not affected, and the absence of G9a allowed HAT and H3K4-specific methyltransferase to target specific lysines in histone H3 (116).
Interestingly, whereas Suv39H specifically targets lysine 9, G9a also methylates lysine 27 of histone H3 (115). Methylated H3K27 prevails in silent chromosome regions together with methylated H3K9, and is recognized by chromodomain-containing proteins, such as HP1 and Polycomb repressive complexes (PRCs) (16). Two Polycomb complexes, PRC1 and 2, were first implicated in the hypermethylation of H3K27 associated with long-term transcriptional silencing of the Drosophila homeotic genes (89). The PRC2 complex contains several core protein components, including EED, related to the Drosophila homologue ESC (Extra Sex Combs), SUZ12 (Drosophila Suppressor of Zeste 12 [Su(Z)12]), EZH2 (Drosophila Enhancer of Zeste (E(z)), and the histone-binding proteins RbAp48/46 (Drosophila Nurf55) (108). H3K27 methylation appeared to be specifically triggered by the SET domain-containing subunit EZH2, whereas SUZ12 has been recently implicated in H3K9 trimethylation in human fibroblasts (27). Moreover, the depletion of SUZ12 decreased the level of trimethylated H3K9 without affecting trimethylated H3K27, and altered HP1α distribution. Because the loss of trimethylated H3K9 was not caused by alterations in Suv39H recruitment, the authors (27) suggested that SUZ12 might recruit H3K9 methyltransferases other than Suv39H or inhibit a potential H3K9 demethylase.
Transcriptional silencing mediated by PRC2 was also related to direct recruitment of DNMT1/3a/3b (126). This might explain how EZH2 overexpression can promote aberrant DNA methylation-associated gene silencing in tumors (107). Although PRC2 seems to be involved in the initiation of silencing, PRC1 may recognize methylated H3K27 marks (84) and ensure the maintenance of a silent state of chromatin imposed by DNA and histone methylation (53). PRC1 might, indeed, inhibit mating type switching/sucrose nonfermenting (SWI/SNF) chromatin remodeling and induce ubiquitination of H2AK119 (42, 88). Recent data indicate that PRC1 subunits Ring1B and Bim1 possess an ubiquitin E3 ligase activity targeting specifically H2A. Cao et al. (15) demonstrated that H3K27 trimethylation and H2A ubiquitination cooperated to silence homeotic genes by Polycomb complex groups. H2A ubiquitin status was also related to cell cycle progression (61). Ubiquitinated H2A might prevent H3S10 phosphorylation by Aurora B kinase, which is required for correct condensation of chromatin during the G2/M transition (22).
Epigenetic Mechanisms of Gene Activation
Methylation at CpG islands may sterically interfere with the binding of transcription factors by limiting their accessibility to DNA and, as a result, is usually associated with gene silencing. Conversely, DNA hypomethylation in promoter regions typically correlates with gene activation. DNA demethylation induced in DNMT3a/3b−/−cells was associated with a major loss of dimethylated H3K9 and a progressive acetylation of H3K9 (47). The transfer of an acetyl group to histone lysine residues is usually associated with transcriptional gene activation and is catalyzed by HATs, whereas its removal involves histone deacetylases (HDACs) (Figure 2). HATs have been divided into three main families: Gcn5-related N-acetyltransferase (GNAT), adenoviral E1A-associated protein 300kDa and cAMP-response element-binding protein (p300/CBP), and MOZ, Ybf2/Sas3, Sas2 and TIP 60 (MYST) (137).
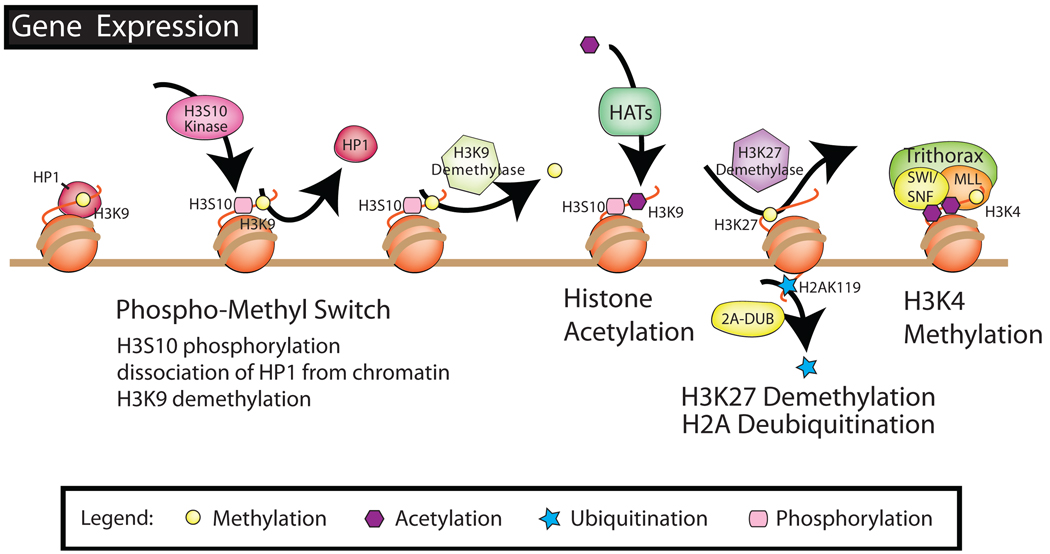
Figure 2.
Interactions between histone-modifying enzymes and histone modifications associated with the activation of gene expression. Gene activation requires the release of histone deacetylases, histone methyl transferases, and other histone-binding protein such as HP1, and the recruitment of chromatin-remodeling complexes including histone acetyl transferases and histone methyl transferases (such as trithorax). HATs, histone acetyltransferases; HP, histone-binding protein; MLL, mixed-lineage leukemia; SWI/SNF, switch/sucrose nonfermentable.
The removal of acetyl groups is under the control of four classes of HDACs. Class I comprises HDAC1-3 and HDAC8, and class II contains HDAC4-7 and HDAC9-10. The third class, NAD-dependent and generally nonresponsive to inhibitors of other classes, comprises the sirtuin family, homologous to yeast sir2 and called SIRT1-7, which has no sequence homology with the other HDACs. A fourth class was recently suggested for HDAC11, and other HDACs previously grouped in class II (48).
HATs and HDACs often exist in multiprotein complexes that facilitate coordination between different histone-modifying enzymes. Consistent with this idea, the acetyltransferase p300/CBP-associated factor has been recently copurified with a histone deubiquitinase. Histone acetylation may then influence H2A deubiquitination at lysine 119 by 2A-deubiquitinase (2A-DUB) by increasing the accessibility of the C-terminal end of H2A, which contains the targeted lysine (138). This is in agreement with the paradigm that acetylation correlates with gene expression, whereas H2A ubiquitination often colocalizes with methylated H3K27 in repressive chromatin.
However, the depletion of 2A-DUB does not seem to trigger other repressive marks, such as methylated H3K9 and H3K27. Acetylation and deubiquitination coincide with the upregulation of histone H1 phosphorylation, which may potentially displace the linker, reduce DNA-nucleosome interactions, and facilitate transcription. Activation of 2A-DUB is apparently independent of another ubiquitinylated histone, H2B, found in active promoters with methylated H3K4 and H3K79. Although not yet investigated in mammals, deubiquitinated H2B is required to maintain silencing of heterochromatic DNA in yeast and plants, whereas heterochromatic-specific histone modifications are destabilized by ubiquitin conjugation (111, 139). Methylation at lysine 4 of histone H3 has also been correlated with histone H3 acetylation and gene expression (91).This modification is under the control of Trithorax, which consists of a SET domain-containing complex mixed-lineage leukemia, as well as other chromatin-remodeling complexes, such as SWI/SNF (108). A recent study revealed that the presence of a methyl group on H3K4 prevented the recruitment of DNMT3L (95). DNMT3L belongs to the DNMT family despite the fact that it lacks the capacity to transfer methyl groups to cytosine in CpG islands. Essential for parental imprinting and transposon silencing, DNMT3L stimulates the recruitment of DNMT3a/3b and HDACs to promoters (32). Nonetheless, it is not known if other transcription-related histone modifications, such as H3S10 or H3K9, also inhibit DNMT3L binding. The phosphorylation of H3S10 by the mitotic kinase Aurora B was at least capable of inducing the dissociation of HP1 from chromatin (39, 54) and facilitating transcription through a phospho/methyl binary switch. The specific back-and-forth interaction between methylated H3K9 and phosphorylated H3S10, turning transcription either on or off, is only one of many examples illustrating the complex interplay between histone modifications, and epigenetic cross-talk with DNA methylation.
DEVELOPMENTAL ORIGIN OF DISEASES
Epigenetic (Re)Programming During Development
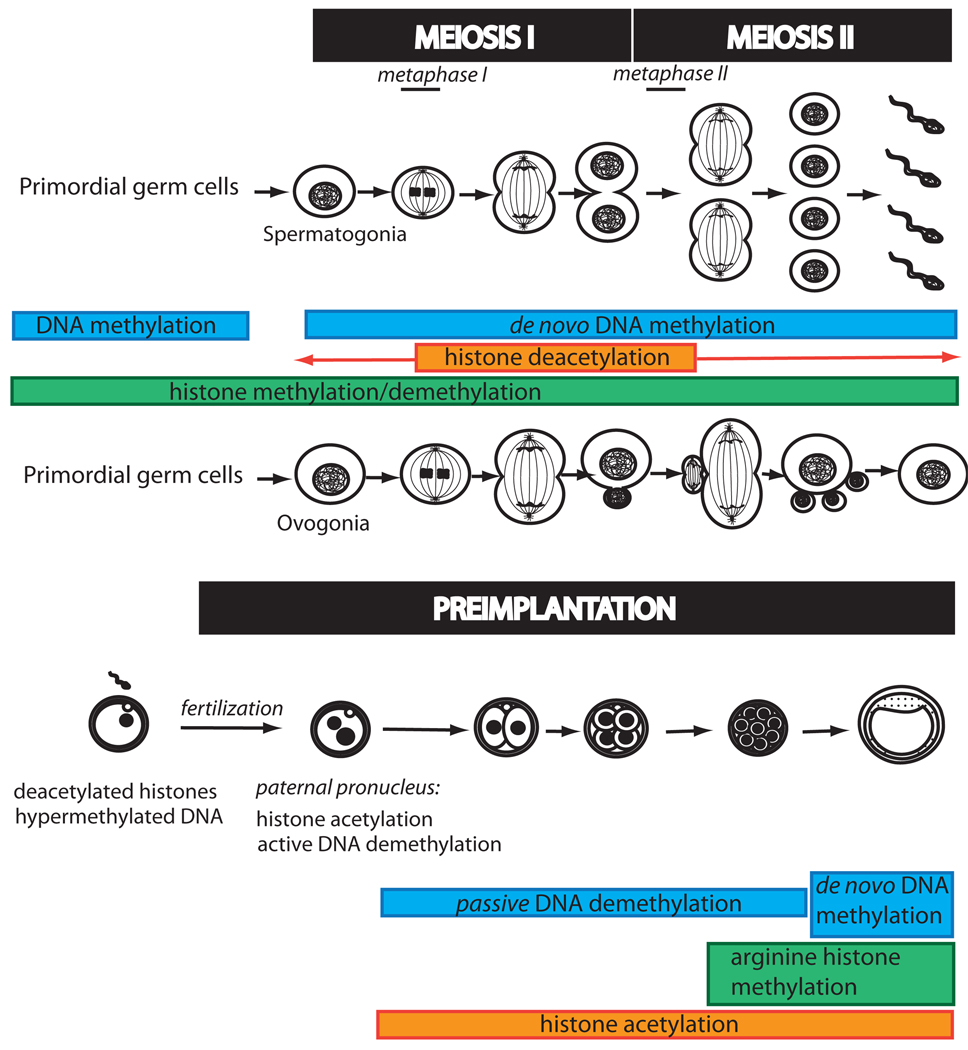
In the lifetime of a mammal, two periods are characterized by extensive epigenetic reprogramming: gametogenesis and, after fertilization, the preimplantation development period (Figure 3).
Figure 3.
Epigenetic modifications associated with gametogenesis, fertilization, and preimplantation developmental steps.
The development of germ cells is a highly ordered process that begins during fetal growth and is completed in the adult. Mammalian meiosis consists of a two-step division process in which one diploid eukaryotic cell divides to generate four haploid gametes (oocyte or sperm). This process implicates a reduction of the genetic material and the introduction of genetic variability through chromosome recombination. Relatively recent evidence has demonstrated that specific chromatin epigenetic remodeling events can orchestrate gametogenesis in a sex-specific way.
Epigenetic reprogramming events play a role very early in the adoption of developmental cell fates with, for example, the establishment of the germ cell lineage from epiblast cells. Later, primordial germ cells (PGCs), developing in the postimplantation embryo, initiate genome-wide epigenetic modifications in the embryonic mouse during their migration from the hindgut region to the genital ridges. A loss of DNA methylation and H3K9me1/2, replaced by increased levels of H3K27me3, characterizes the reprogramming events occurring in PGCs (109).
When the PGCs reach the genital ridges, they are no longer motile and undergo few further rounds of mitosis before upregulating meiosis-specific genes. After female PGCs enter meiosis, they are arrested in prophase I for a long period of time, while male PGCs enter mitotic arrest in G0/G1 prospermatogonia until around birth, when mitosis of spermatogonia is resumed. Under appropriate hormonal stimulation, the oocyte in prophase I successively goes through meiosis I, enters meiosis II, and is again arrested in metaphase II and awaits for fertilization.
The genome of the differentiated oocyte is believed to require epigenetic chromatin remodeling during meiosis and fertilization to produce a totipotent embryo. Changes in DNA methylation (51, 80, 92) and post-translational modifications of histones have been partially characterized, although their biological consequences remain unclear. Specifically, the oocyte at prophase I displays a high level of acetylation at histones H3 and H4, whereas the level dramatically drops at metaphase I, and a complete deacetylation affecting H3K14, H4K12, H4K8, H4K16 (3), and H4K5 (1) was reported later at metaphase II.
Such active and global deacetylation has also been observed in spermatocytes before fertilization, and might result from HDAC activation, whereas HATs appear to be inactivated during gametogenesis (65). As mentioned above, histone acetylation is commonly related to specific conformations of chromatin structure that render the DNA accessible to transcription factors. However, although transcription may decrease during oocyte maturation as the chromatin condenses (30), transcriptional repression is observed long before germinal vesicle breakdown (8) and appears independent of chromatin condensation and deacetylation of histone H3 (28), suggesting an alternate role for deacetylation. Moreover, preventing histone deacetylation with HDAC inhibitors affects neither the chromatin condensation process nor oocyte maturation or further fertilization and preimplantation development. In fact, the deacetylation process may be necessary to maintain the binding of ATR-X, a member of the SNF2 family of helicase/ATPases with chromatin remodeling activity, at centromeric heterochromatin in the mouse oocyte chromosomes at metaphases I and II (29, 83).
The kinetochore is a protein complex attaching the centromeric region of chromosomes to spindle microtubules, ensuring the proper segregation of chromosomes during mitosis and meiosis. ATR-X might play a functional role in chromosome alignment on the spindle microtubules, and may interact with other kinetochore-associated proteins, such as HP1, also localized at centromeric heterochromatin. This suggests that global histone deacetylation during the progression of meiosis may be required for an appropriate recruitment of heterochromatin proteins. Indeed, inadequate histone hyperacetylation was found to interfere with the kinetochore assembly in somatic cell lines, resulting in activating the mitotic checkpoint (101).
If histone deacetylation during meiosis is essential for ATR-X function, and guarantees the proper attachment of chromosomes with the meiotic spindle, it does not exclude the possibility that heterochromatin proteins may be influenced by other epigenetic modifications. Indeed, contrary to rapid histone deacetylation, histone methylation persists through meiotic progression, providing a chromosome environment necessary for HP1 recruitment (12, 67). As mentioned above, poorly aligned chromosomes activate a spindle assembly checkpoint in somatic cells, as well as in male germ cells (14), which delays the cell cycle or induces an arrest, preventing the production of aneuploid progeny. However, such a checkpoint is absent or more permissive in female germ cells, leading to aneuploidy when cells are treated with the HDAC inhibitor trichostatin A (TSA) (3, 29). Similarly, incomplete histone deacetylation during oocyte maturation, observed in aged mice, has been associated with an increased frequency of aneuploidy (3).
Interestingly, TSA might also affect the level of phosphorylated histone H3 at serine 10, which is found closely colocalized with pericentromeric chromatin during metaphase I and anaphase I, where as phosporylated H3S10 is re-distributed uniformly along the chromosomes in the metaphase of the second meiosis (127).
Other histone modifications appear to be of biological significance for genome reprogramming during female gametogenesis. Specifically, methylation of histone arginine residues (H3R17me and H4R3me) was dramatically reduced in metaphase II-arrested oocytes, as well as in the fertilized egg and during the first cleavages of the zygote (106). Methyl arginine marks often associate with decondensed chromatin and activation of gene expression, and removal of arginine methylation coincides with chromatin condensation during meiosis. G9a HMT (114) and Suv39H (74), catalyzing mono-, di-, and trimethylation, as well as Meisetz, a meiosis-specific H3K4 trimethyl-transferase (52), were also involved in the progression through meiosis I. Also, reduced levels of mono- and dimethyl H3K9 in G9a HMT knockdown male germ cells did not correlate with gene derepression or reactivation of retrotransposons, but rather with improper synapsis formation leading to cell death. After completion of synapsis, the methyl mark might be actively removed in wild-type male germ cells by the activation of JHDM2A, a specific H3K9 demethylase (114, 134). As with DNA methylation (92), the prevailing view concerning histone modifications during gametogenesis points to alternative functions in germ cells versus somatic cells for regulation of gene expression.
Major epigenetic events happen upon fertilization when gametic marks have to be replaced by the embryonic pattern characteristic of totipotency. Immediately after fertilization, the paternal genome, condensed and packaged with protamines, undergoes a rapid hyperacetylation as a consequence of a protamine-to-histone exchange (1). Moreover, immediately after histone incorporation, during the pronuclear (one-cell) stage, the paternal genome undergoes active DNA demethylation, while the maternal pronucleus arrested in metaphase II resumes meiosis. Later, both parental genomes experience a passive demethylation related to DNA replication during the preimplantation period up to the morula stage. Although asymmetric, epigenetic changes observed in both pronuclei are believed to be required for restoration of totipotency in the fertilized egg. However, this wave of demethylation excludes specific sequences, such as imprints, retrotransposons, and centromeric heterochromatin, although the mechanisms are still unknown.
H3K9 methylation in the maternal genome also participates in epigenetic asymmetry between the parental pronuclei during the first step of development (78, 104). Specifically, these epigenetic differences may underlie differences in transcriptional activity between the paternal and maternal genome (4). Key observations here are possible utilizing the technique of somatic cell nuclear transfer (SCNT). SCNT consists of the transfer of a somatic nucleus into an enucleated oocyte for the purpose of creating genetically identical animals. The success of such a technique greatly depends on the appropriate reprogramming of the epigenetic marks during conversion of the somatic nuclei into a totipotent state in the oocyte cytosol. Recent evidence shows that histone modifications have a role in primordial epigenetic reprogramming, since the efficiency of SCNT was greatly improved by the use of TSA (102). Acetylation and phosphorylation of specific histone lysines, as well as the lack of methylated H3K9 (58), driven in part by the maternal protein nucleoplasmin, may be responsible for sperm genome decondensation (117).
Successive cell divisions are characterized by gradual loss of DNA methylation due to the exclusion of DNMT1o (embryonic DNMT), whereas phosphorylation (58) and arginine methylation of histones (106) may fluctuate during the cell cycles. The first lineage commitment at the blastocyst stage, as well as de novo methylation, is identified with the formation of the trophectoderm and the inner cell mass. It is not yet clear to what extent histone modifications participate in cell commitment in specific cell lineages, although their function is not restricted to DNA replication (120). Nevertheless, a recent study reported that stem cell differentiation required exposure to a differentiation signal that may regulate a particular gene expression involved in mesoderm selection through increases in H3S10 phosphorylation and K14 acetylation (73).
Parental Imprinting
Parental imprinting is an epigenetic form of gene regulation consisting of about 100 genes for which only one parental allele is expressed. Although there is no definitive description of the mechanisms involved, it has been hypothesized that imprinting of particular developmental genes is required to prevent conflict between maternal and paternal investment in growth and development of the offspring (86). Indeed, the parental conflict theory of imprinting is based upon the observation that paternally imprinted genes are involved in growth of the fetus, while maternally imprinted genes promote development of extraembryonic tissues. The establishment of gene imprinting is preceded by erasure of parental imprints, which takes place in PGCs, and consists of genome-wide DNA and histone demethylation (affecting also nonim-printed genes) (109), although the exact mechanisms are unclear. The acquisition of imprints responsible for sex-specific expression usually occurs during gametogenesis. However, in the case of IGF2R encoding the receptor of insulin-like growth factor 2, the imprinting marks in the mouse appear to be established earlier, during ovocyte growth (13). Similarly, the paternally imprinted gene H19 acquires most of its methylation during the prenatal window and appears fully methylated before the end of prophase I (pachytene phase) (26).
Despite the fact that DNA methylation is considered as the main (if not the sole) mastermind in silencing of the imprinted allele, recent reports reveal that H3K27 methylation (135) and histone hypoacetylation (18, 49) accompany DNA methylation in silencing one parental allele, while the constitutively active allele is enriched in methylated H3K4 (103). Likewise, induction of expression of the normally silent paternal H19 allele by active deacetylation using TSA has also confirmed the importance of HDACs in the silencing process (112). These data, along with other reports (60), suggest that histone modifications may participate in targeting DNMTs to soon-to-be imprinted alleles. Once imprinting is established in the germ line, the silenced marks are maintained throughout the gametogenesis process and beyond, after fertilization and preimplantation,in spite of global demethylation affecting the embryo genome until the blastocyst stage. The mechanisms protecting imprintings, which are still to be elucidated, might involve specific combinations of histone modifications and be strictly dependent on development.
Imprinting genes have been involved in several diseases, including cancers, particularly because there is no protection against recessive mutations. However, losses of imprints are also often observed in cancers (23) and have revealed that, although somehow stable, imprinting marks can be influenced by environmental factors, such as nutrition. In the same genomic region as H19, IGF2 encodes the insulin-like growth factor 2 and is a maternally imprinted gene under control of differentially methylated regions containing CpG islands that are methylated at the maternal allele. Abnormal biallelic IGF2 expression was induced and stabilized in kidneys of mice fed a diet restricted in methyl donors during one to two months postweaning. Interestingly, no changes in DNA methylation were found at the differentially methylated regions governing IGF2 expression, suggesting that other mechanisms, such as histone modifications, could have been targeted by the diet (129). This is consistent with the observation that specific fatty acids in milk modify the expression of chromatin remodeling factors in the small intestine of preweaning rodents (85). Another study reported how easily and quickly manipulations of rodent diets disrupt the balance of epigenetic modifications regulating metabolic genes (55). These studies highlighted the well-documented impact of unbalanced, restricted, or enriched diets with specific compounds (methyl donors, carbohydrates, fatty acids, etc.) on metabolic homeostasis later in life, caused by alterations of epigenetic marks, including imprints. More research is warranted to determine how maternal diet modifies the fetal epigenome and the subsequent risk of developing diseases later in life (fetal origin of adult diseases).
Involvement of Epigenetics in the Fetal Origin of Adult Diseases
The theory of fetal programming of disease (metabolic imprinting) emerged from the idea that in utero nutritional conditions affect epigenetic patterns of gene transcription or silencing normally established early and retained throughout life. This raises the possibility that the laying down of aberrant epigenetic marks in utero might increase disease susceptibility in adulthood. Compelling epidemiological evidence, as well as data from animal studies, have linked maternal diet with susceptibility to metabolic disorders (obesity, glucose intolerance, type II diabetes) and related diseases (atherosclerosis, cardiovascular diseases) (128).
Influence of Maternal Diet on Fetal Programming
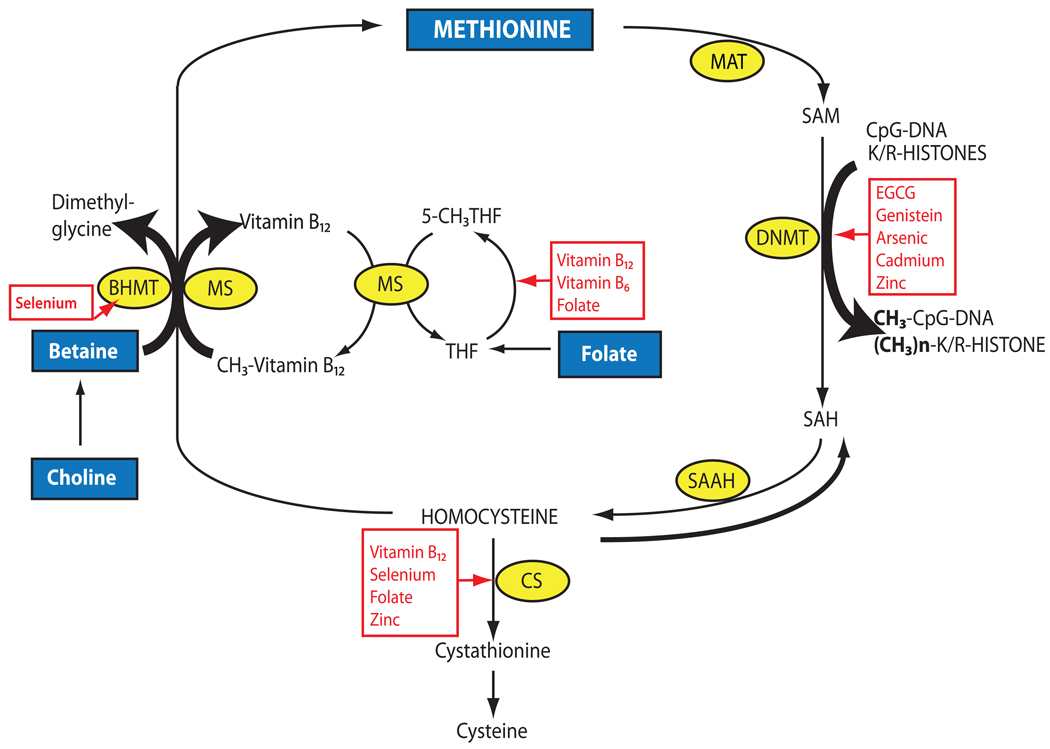
As mentioned above, feeding animals with diets enriched or restricted in methyl donors affects the epigenome, and consequently influences gene expression. S-adenosyl-l-methionine (SAM) is the universal methyl donor for methyltransferases and is exclusively provided by folate-mediated one-carbon metabolism. Many dietary factors, including folate, methionine, choline, betaine, and vitamins B2, B6, and B12, contribute to the production of SAM. Other nutritional sources containing dietary catechols, phytochemicals, and metals may also modulate the methionine-homocysteine cycle, alter methyltransferase activities, and affect the SAM/S-adenosyl-l-homocysteine (SAH) ratio and, eventually, DNA methylation (Figure 4). Dietary restriction in methyl donors (no folate and choline, plus low methionine) as well as genetic polymorphisms in folate metabolism have been associated with abnormal DNMT expression, global DNA hypomethylation, and increased cancer risk (38, 46).
Figure 4.
Overview of the methionine-homocysteine-folate-B12 cycle, which provides methyl donors for methyltransferases. Nutritional regulations of the cycle by phytochemicals and metals are shown in red. BHMT, betaine homocysteine; CS, cystathionine synthase; DNMT, DNA methyltransferase; EGCG, epigallocatechin-3-gallate; MAT, methionine adenosyl transferase; MS, methionine synthase; SAAH, S-adenosyl-L-homocysteine hydrolase; SAH, S-adenosyl-L-homocysteine; SAM, S-adenosyl-L-methionine; THF, tetrahydrofolate.
The first example of maternal diet enriched in methyl donors affecting the phenotype of offspring through epigenetic alterations was reported by Wolff et al. (133) and relates to the mouse model Agouti viable yellow (Avy). During epigenetic reprogramming in PGCs, some transposable elements, such as intracisternal A particles (IAPs), may escape demethylation and remain methylated (69). However, the example of the mouse Agouti Avy revealed that these regions enriched in CpG could be targeted by nutritional factors. Indeed, in the Avy model, the Agouti gene has been mutated by insertion of an IAP sequence into the 5′ upstream region of the A allele, causing constitutive expression of the gene. While the expression of the wild-type Agouti gene (a) is restricted to the skin and gives a brown coat color (wild-type phenotype), the constitutively active mutant Avy is expressed in all tissues and gives a yellow pigmentation to the coat. Interestingly, the phenotype of individuals from the same inbred strain Avy/a exhibited a coat color phenotype varying from yellow to brown, some being mottled to different extents, depending on the degree of IAP methylation. Moreover, IAP hypomethylation was related to mouse obesity and increased susceptibility to diabetes and tumorigenesis. In contrast, fully methylated IAP is associated with a wild-type phenotype. In addition, this so-called metastable epiallele was sensitive to dietary methyl donors in the pregnant mother’s diet. Dietary supplementation with folate, vitamin B12, choline, and betaine shifted the coat color to brown and prevented the early onset of obesity.
Although there is no such example in the human genome, maternal dietary exposure was clearly proven to be a determinant in modulating the susceptibility to diseases in adult life (128). To date, few studies can attest categorically that disease predisposition resulting from restrictive maternal diets is related solely to epigenetic deregulation rather than organ malformation or vascular dysfunction caused by fetal malnutrition (119).
Glucocorticoid excess in mice and humans causes symptoms similar to those described in the “metabolic syndrome”, i.e., obesity, hypertension, insulin resistance, hyperlipidemia, and hyperglycemia. In particular, upregulation of hepatic peroxisome proliferator-activated receptor α (PPARα) by glucocorticoids stimulates fatty acid oxidation and facilitates metabolic disorders. The maternal environment (130), including diet (75), has been found to be critical for imprinting the promoter of genes involved in glucocorticoid metabolism. Specifically, protein restriction in the maternal diet was associated with upregulation of hepatic genes encoding the glucocorticoid receptor (GR) and PPARα of adult offspring. Hypomethylation of the GR promoter possibly is related to a decreased level of DNMT1 activity and reduced binding of MeCP2, which is associated with an increase of gene transcription (76).
To date, attention has been focused primarily on the methylation status of DNA irrespective to the histone code. Interestingly, analysis of GR promoter revealed, as expected, high levels of acetylated histones (H3K9 and H4K9) and methylated H3K4, and low levels of dimethylated H3K9. As discussed elsewhere (113), it is difficult to attribute the consequences of a maternal protein-restricted diet to direct alterations of one-carbon metabolism, although folate supplementation can prevent GR increase induced by the maternal diet (75). However, these results establish a link between maternal diet and altered gene expression in adult offspring through epigenetic regulation and, eventually, its impact on energy homeostasis, assuming enhancement of glucocorticoid responsiveness can be correlated with increased GR levels.
TARGETING THE EPIGENOME TO FIGHT CANCER
The growing interest in cancer epigenetics came from the demonstration that epigenetic modifications are involved in tumor development and progression, creating a cancer-predisposing “epigenotype” that is potentially modifiable. Indeed, the reversibility of epigenetic chromatin modifications by synthetic and natural compounds has opened new avenues in cancer chemoprevention research.
Aberrant Histone Modifications in Tumors
In addition to the accumulation of genetic mutations (i.e. changes in DNA sequence), cancer etiology is characterized by aberrant patterns of DNA methylation. Typically, the CpG islands that are mainly located in the 5′ regulatory region of genes appear unusually hypermethylated in tumors. Such promoter DNA hypermethylation is an efficient alternate mechanism to mutations for silencing genes encoding DNA repair proteins and tumor suppressors. Global genome DNA hypomethylation, on the other hand, results in gene activation and chromosomal instability (31). Because the histone code and DNA methylation cooperate to organize chromatin, abnormal promoter CpG hypermethylation in cancer cells is accompanied by a combination of histone modifications commonly associated with repressed promoters: hypoacetylated histones H3 and H4 (41), hypomethylated H3K4 (110), and hypermethylated H3K27 (66) and H4K20 (41). Contrary to the enrichment of methylated H3K9 in silenced heterochromatin, methylated H3K9 has been located in active promoters of various genes in human cancer cells (121, 131).
Two main epigenetic pathways have been implicated in the transformation of a normal cell into a tumor cell. One consists of a cell de-differentiation process, with alteration or loss of the epigenetic patterning of differentiationspecific genes during a deprogramming process. This may eventually allow cells to regain assets that are specific to pluripotency, such as proliferation. A second possibility is a shift of the pluripotent stem cells into transformed cells. In the latter case, recent studies have highlighted that stem cell pluripotency is regulated by the existence of a bivalent pattern of histone modifications in regulatory regions of particular genes (6, 7). Indeed, the colocalization of permissive histone modifications, such as dimethylated H3K4 and acetylated H3K9, with repressive modifications, such as trimethylated H3K27, suggests that specific genes in stem cells are maintained silent but in a transcriptionally permissive state, allowing their rapid derepression along with lineage induction. The H3K27 methylation mark might help to maintain low gene expression, and be quickly removed to facilitate gene expression and commit the cell to one specific fate.
Although the absence of PRC2, along with loss of H3K27 methylation mark, is able to cause premature expression of lineage-specific genes (7), the maintenance of such a histone marker triggers other repressive histone marks (126) that prevent lineage commitment, thus maintaining pluripotency. The recent discovery of similar bivalent patterns accompanied by other repressive marks in promoters of tumor suppressor genes (93) might favor the inappropriate hypermethylation and silencing of these genes during tumorigenesis.
As chromatin remodels during cancer development, aberrant activities of several histone-modifying molecules occur (37). For example, class I HDACs are over-expressed early in the cancer process (57, 132), and Suv39H knockout mice are predisposed to develop B-cell lymphomas (96). Interestingly, targeting HDAC activity successfully derepresses genes in cancer cells and is becoming a novel therapeutic tool. Therapeutic agents, such as SAHA (Vorinostat), have been identified as potent HDAC inhibitors and are currently being evaluated in clinical trials (45). In addition, HDAC inhibitors cooperate with other therapeutics agents, such as the DNMT inhibitor Decitabine (5-aza-2′-deoxycytidine), to improve the epigenetic anticancer strategy (44, 122).
Epigenetic Reprogramming by Dietary Agents
Investigations on the mechanism(s) of nutrientgene interactions are needed to understand how diet modulates the risk of developing chronic diseases, such as cancer. To date, certain dietary agents have revealed an ability to target specific nuclear receptors, whereas others may preferentially affect the epigenome. An excellent example of cooperation between a diet-derived agent targeting a nuclear receptor and HDAC inhibitors has been described in treatment strategies for acute promyelocytic leukemias (APLs). Abnormal targeting of HDAC, HMT, and DNMT to promoters governed by retinoic acid (RA), a vitamin A derivative, characterizes these diseases. APLs are caused by reciprocal chromosomal translocations involving the promyelocytic leukemia (PML) gene and the gene encoding the nuclear receptor of RA, called RARα. The fusion protein PML/RARα, as well as the wild-type receptor, inhibits transcription by binding to RAR-responsive elements in the promoter of target genes. Gene repression is achieved through the recruitment of several chromatin modifiers: the complex HDAC-NCoR (5), histone methyltransferases Suv39H (17) and PRC2 (125), as well as DNMT (33) and MBD1 (124). Contrary to the wild-type receptor, PML/RARα appears insensitive to physiological concentrations of RA and thus maintains a stable repression of target genes (such as RARb2) involved in hematopoietic differentiation. Complete remission of patients with APL is achieved by treatment with pharmacological doses of RA. RA binding to the RAR moiety leads to release of HDAC proteins, and transcriptional coactivators associated with HAT may then be recruited. However, some RA-resistant leukemia cells failed to respond to RA alone and required cooperation with other agents. Treatment of RA-refractory APL blasts with RA plus the HDAC inhibitors sodium phenylbutyrate, TSA, depsipeptide, and valproic acid, as well as demethylating agents, restored RA sensitivity and cell differentiation (20).
Although HDACs are candidates for anti-cancer drug design, a number of natural compounds found in the human diet can influence HDACs and the acetylation status of histones (Table 1). Three major dietary compounds have proven to induce histone acetylation as well as growth arrest and/or apoptosis in a number of cancer cell lines (reviewed in 25). It is now three decades since the alterations of gene expression induced by butyrate, an end product of the gut fermentation of dietary fibers, were first attributed to changes in histone acetylation (123). Recently, organosulfur compounds from garlic, such as diallyl disulfide, allyl mercaptan, and S-allylmercaptocysteine, as well as the isothiocyanates sulforaphane and 6-methylsulfinylhexyl isothiocyanate from cruciferous vegetables, demonstrated a capacity to alter histone acetylation and/or HDAC activity in vivo and in vitro. Class III HDACs (sirtuins) have garnered much attention after they were implicated in increasing life span and delay aging-related diseases (including cancer) (118). Activation of sirtuins by dietary polyphenols, such as resveratrol from red wine, mimics the increase of life span by food restriction (56). A recent study showed that resveratrol stimulates mitochondrial function and prevents metabolic disorders in mice fed a high-fat diet. The effect of resveratrol was mediated by the activation of PGC1α in a SIRT1-dependent way (68). This finding highlights the need for additional research on specific dietary agents and wholefoods that may affect life span and cancer risk in mammals.
Table 1.
Natural and/or dietary compounds modulating histone acetylation and/or HDAC/HAT activities
| Dietary components | Examples of food/plant sources | References |
|---|---|---|
| S-allylmercaptocysteine | Garlic (Allium sativum L.) | (72) |
| 6-methylsulfinylhexyl-isothiocyanate | Japanese horseradish (wasabi) | (87) |
| Allyl mercaptan | Garlic (Allium sativum L.) | (70) |
| Anacardic acid | Cashew nut | (10) |
| Butein | Rhus verniciflua (stems) | (56, 99) |
| Butyrate | Dietary fiber fermentation | (24) |
| Copper | Ubiquitous | (62, 77) |
| Curcumin | Curcuma longa (tumeric roots) | (11, 79) |
| Diallyl disulfide (DADS) | Garlic (Allium sativum L.) | (35, 71, 82) |
| Dihydrocoumarin | Melilotus officinalis (sweet clover) | (94) |
| Fisetin | Rhus toxicodendron (leaves) | (56, 99) |
| Garcinol | Garcina indica (fruit) | (9) |
| Isoliquiritigenin | Glycyrrhiza glabra (licorice) | (56, 99) |
| Luteolin | Sweet red pepper, celery, parsley | (99) |
| Nickel | Ubiquitous | (63, 136) |
| Piceatannol | Blueberries | (99) |
| Psammapin A | Marine sponges | (64) |
| Quercetin | Apple, tea, onion, nuts, berries | (56, 99) |
| Resveratrol | Red grapes, wines, eucalyptus, spruce | (56, 99) |
| Sulforaphane | Broccoli, broccoli sprouts | 90 |
| Theophylline | Black and green tea | (21, 59) |
HDAC inhibitors have a broad spectrum of targets, including genes encoding DNA-and histone-modifying enzymes (40). However, the impact of HDAC inhibitors on epigenetic modifications is, at least in part, the consequence of the inter-dependence between histone modifications. Recently, Nightingale et al. (91) demonstrated that the degree of H3K4 methylation (mono-, di-, or trimethylation) was dependent on the extent of acetylation on the same histone tail. Subsequently, the use of butyrate in various cancer cell lines not only correlated with histone H3 acetylation but also stimulated the deposition of methyl marks at H3K4 by mixed-lineage leukemia (MLL) (91). A comparable result was reported when histone H4 hyperacetylation, induced in butyrate-treated cells, was found to prevent trimethylation at lysine 20 (105).
Although more recently investigated, histone methyltransferases could potentially be targeted by dietary agents, specifically those identified as modulators of one-carbon metabolism and methionine-homocysteine cycle (Figure 4). The impact of dietary methyl donors on DNMTs has already been examined, but very few studies have focused on HMTs, although they should also be affected by alterations of the SAM/SAH ratio. Recently, the impact of a methyl-deficient diet on histone methylation in rodent liver has been investigated (97). As mentioned above, it is believed that such a diet promotes hepatocarcinogenesis in rodents by inducing DNA demethylation at centromeric heterochromatin and subsequent chromosome instability (46). The authors specifically reported a loss of H4K20 trimethylation at a DNA repetitive element along with the depletion of H4K20-specific histone methyltransferase (Suv4-20h1) in both preneoplastic tissues and tumors. These data and others (98) confirm that methyl-donor deficiency may rapidly affect histone marks, inducing the intensive chromatin remodeling implicated in the development of neoplasia. Therapeutic strategies using dietary methyl donors may prove to be useful at preventing carcinogenesis or halting tumor growth (50).
CONCLUSIONS
Rules of the histone code, together with DNA methylation status, remain to be fully characterized in normal tissues and cancers. Thus, there is considerable uncertainty about the precise impact of dietary factors on epigenetic programming early in life, as well as on epigenetic deprogramming during tumorigenesis. In the future, a better understanding of the mechanisms underlying the influence of nutrition on the epigenome,and subsequently on the expression of the genome, may help to influence dietary habits and behaviors. A key aspect of this work, which raises much excitement, is the potential reversibility of aberrant epigenetic modifications in cancer cells. We are only beginning to understand how epigenetic manipulations by dietary compounds can help us to live a healthier and longer life.
SUMMARY POINTS.
Combinatorial interactions between histone tail modifications, DNA methylation, and histone- and DNA-modifying enzymes constitute epigenetic mechanisms regulating chromatin structure and function.
Extensive epigenetic re/deprogramming events occur during gametogenesis, fetal development, and pathologies such as cancer, rendering these processes potentially more susceptible to nutritional influences.
Metabolic disorders as well as cardiovascular diseases may have their origins in fetal life. It is hypothesized that epigenetic deregulations caused by environmental factors, including maternal nutrition, may induce long-lasting consequences for adult health.
It is likely that nutrition affects gene expression and disease susceptibility through epigenetic mechanisms. The best example is dietary manipulations of folate metabolism that affect DNA methylation status and gene silencing. Dietary agents such as butyrate, sulforaphane, and garlic organosulfur compounds inhibit HDAC activity and de-repress epigenetically silenced genes in cancer cells. More research is needed on diet, epigenetics events, and cancer prevention.
Future research is needed to better understand the involvement of histone modifications in the regulation of chromatin structure and gene expression, and to identify the nature of metabolic pathways that are controlled by nutrition through epigenetic mechanisms, both in early life and adulthood.
ACKNOWLEDGMENTS
The authors gratefully acknowledge Stephen R. Lawson, Tracy L. Oddson, and David E. Williams for proofreading the manuscript and Daniel Schwartz for assistance in developing the figures. Research by the authors is supported in part by NIH grants CA90890, CA65525, and CA122959.
Abbreviations
- H3
histone H3
- Histone code
hypothesis based on the observation that different post-translational histone modifications function together in establishing and maintaining specific chromatin structures, and regulating gene expression. The different combinations of these modifications constitute a code dictating particular chromatin configurations and functions
- HMTs
histone methyltransferases
- HATs
histone acetyltransferases
- PRCs
Polycomb repressive complexes
- SWI/SNF
mating type switching/sucrose nonfermenting
- HDACs
histone deacetylases
- TSA
trichostatin A
- SCNT
somatic cell nuclear transfer
- DNMTs
DNA methyltransferases
- Fetal (or developmental) origin of adult diseases
A concept that implies the risk of developing chronic diseases during adulthood is directed by the nutrition supply to the baby during prenatal development. Specifically, perturbations in fetal nutrition are hypothesized to affect the epigenetic regulation of genes (so-called epigenetic marks) during critical developmental periods
- Epigenetics
an ensemble of processes that stably change the expression of genes through chemical modifications of chromatin (DNA and histones) and without changing the DNA sequence
- APLs
acute promyelocytic leukemias
- RA
retinoic acid
- PML
promyelocytic leukemia
Footnotes
DISCLOSURES
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal N, Hardt T, Brero A, Nowak D, Rothbauer U, et al. MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 2007;35:5402–5408. doi: 10.1093/nar/gkm599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiyama T, Nagata M, Aoki F. Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. Proc. Natl. Acad. Sci. USA. 2006;103:7339–7344. doi: 10.1073/pnas.0510946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 5.Atsumi A, Tomita A, Kiyoi H, Naoe T. Histone deacetylase 3 (HDAC3) is recruited to target promoters by PML-RARalpha as a component of the N-CoR corepressor complex to repress transcription in vivo. Biochem. Biophys. Res. Commun. 2006;345:1471–1480. doi: 10.1016/j.bbrc.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 6.Attema JL, Papathanasiou P, Forsberg EC, Xu J, Smale ST, et al. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc. Natl. Acad. Sci. USA. 2007;104:12371–12376. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, et al. Chromatin signatures of pluripotent cell lines. Nat. Cell. Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 8.Bachvarova R. Gene expression during oogenesis and oocyte development in mammals. Dev. Biol. 1985;1:453–524. doi: 10.1007/978-1-4615-6814-8_11. (N.Y. 1985) [DOI] [PubMed] [Google Scholar]
- 9.Balasubramanyam K, Altaf M, Varier RA, Swaminathan V, Ravindran A, et al. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J. Biol. Chem. 2004;279:33716–33726. doi: 10.1074/jbc.M402839200. [DOI] [PubMed] [Google Scholar]
- 10.Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. Small molecule modulators of histone acetyltransferase p300. J. Biol. Chem. 2003;278:19134–19140. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- 11.Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 12.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 13.Bao S, Obata Y, Carroll J, Domeki I, Kono T. Epigenetic modifications necessary for normal development are established during oocyte growth in mice. Biol. Reprod. 2000;62:616–621. doi: 10.1095/biolreprod62.3.616. [DOI] [PubMed] [Google Scholar]
- 14.Burke DJ. Complexity in the spindle checkpoint. Curr. Opin. Genet. Dev. 2000;10:26–31. doi: 10.1016/s0959-437x(99)00040-4. [DOI] [PubMed] [Google Scholar]
- 15.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 17.Carbone R, Botrugno OA, Ronzoni S, Insinga A, Di Croce L, et al. Recruitment of the histone methyltransferase SUV39H1 and its role in the oncogenic properties of the leukemia-associated PML-retinoic acid receptor fusion protein. Mol. Cell. Biol. 2006;26:1288–1296. doi: 10.1128/MCB.26.4.1288-1296.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carr MS, Yevtodiyenko A, Schmidt CL, Schmidt JV. Allele-specific histone modifications regulate expression of the Dlk1-Gtl2 imprinted domain. Genomics. 2007;89:280–290. doi: 10.1016/j.ygeno.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 20.Claus R, Lubbert M. Epigenetic targets in hematopoietic malignancies. Oncogene. 2003;22:6489–6496. doi: 10.1038/sj.onc.1206814. [DOI] [PubMed] [Google Scholar]
- 21.Cosio BG, Tsaprouni L, Ito K, Jazrawi E, Adcock IM, et al. Theophylline restores histone deacetylase activity and steroid responses in COPD macrophages. J. Exp. Med. 2004;200:689–695. doi: 10.1084/jem.20040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, et al. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 24.Davie JR. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003;133 doi: 10.1093/jn/133.7.2485S. 2485–93S. [DOI] [PubMed] [Google Scholar]
- 25.Davis CD, Ross SA. Dietary components impact histone modifications and cancer risk. Nutr. Rev. 2007;65:88–94. doi: 10.1111/j.1753-4887.2007.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 26.Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum. Mol. Genet. 2000;9:2885–2894. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- 27.de la Cruz CC, Kirmizis A, Simon MD, Isono K, Koseki H, et al. The polycomb group protein SUZ12 regulates histone H3 lysine 9 methylation and HP1 alpha distribution. Chromosome Res. 2007;15:299–314. doi: 10.1007/s10577-007-1126-1. [DOI] [PubMed] [Google Scholar]
- 28.De La Fuente R, Viveiros MM, Burns KH, Adashi EY, Matzuk MM, et al. Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev. Biol. 2004;275:447–458. doi: 10.1016/j.ydbio.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 29.De La Fuente R, Viveiros MM, Wigglesworth K, Eppig JJ. ATRX, a member of the SNF2 family of helicase/ATPases, is required for chromosome alignment and meiotic spindle organization in metaphase II stage mouse oocytes. Dev. Biol. 2004;272:1–14. doi: 10.1016/j.ydbio.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Debey P, Szollosi MS, Szollosi D, Vautier D, Girousse A, et al. Competent mouse oocytes isolated from antral follicles exhibit different chromatin organization and follow different maturation dynamics. Mol. Reprod. Dev. 1993;36:59–74. doi: 10.1002/mrd.1080360110. [DOI] [PubMed] [Google Scholar]
- 31.Deng G, Nguyen A, Tanaka H, Matsuzaki K, Bell I, et al. Regional hypermethylation and global hyomethylation are associated with altered chromatin conformation and histone acetylation in colorectal cancer. Int. J. Cancer. 2006;118:2999–3005. doi: 10.1002/ijc.21740. [DOI] [PubMed] [Google Scholar]
- 32.Deplus R, Brenner C, Burgers WA, Putmans P, Kouzarides T, et al. Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Res. 2002;30:3831–3838. doi: 10.1093/nar/gkf509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 34.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Druesne N, Pagniez A, Mayeur C, Thomas M, Cherbuy C, et al. Diallyl disulfide (DADS) increases histone acetylation and p21(waf1/cip1) expression in human colon tumor cell lines. Carcinogenesis. 2004;25:1227–1236. doi: 10.1093/carcin/bgh123. [DOI] [PubMed] [Google Scholar]
- 36.Espada J, Ballestar E, Fraga MF, Villar-Garea A, Juarranz A, et al. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J. Biol. Chem. 2004;279:37175–37184. doi: 10.1074/jbc.M404842200. [DOI] [PubMed] [Google Scholar]
- 37.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 38.Fang JY, Xiao SD. Folic acid, polymorphism of methyl-group metabolism genes, and DNA methylation in relation to GI carcinogenesis. J. Gastroenterol. 2003;38:821–829. doi: 10.1007/s00535-003-1207-7. [DOI] [PubMed] [Google Scholar]
- 39.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 40.Fiskus W, Pranpat M, Balasis M, Herger B, Rao R, et al. Histone deacetylase inhibitors deplete enhancer of zeste 2 and associated polycomb repressive complex 2 proteins in human acute leukemia cells. Mol. Cancer Ther. 2006;5:3096–3104. doi: 10.1158/1535-7163.MCT-06-0418. [DOI] [PubMed] [Google Scholar]
- 41.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 42.Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol. Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 43.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, Yang H, Rosner G, et al. Phase 1/2 study of the combination of 5-aza-2′-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108:3271–3279. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Manero G, Yang H, Bueso-Ramos C, Ferrajoli A, Cortes J, et al. Phase I study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid, SAHA) in patients with advanced leukemias and myelodysplastic syndromes. Blood. 2008;111:1060–1066. doi: 10.1182/blood-2007-06-098061. [DOI] [PubMed] [Google Scholar]
- 46.Ghoshal K, Li X, Datta J, Bai S, Pogribny I, et al. A folate-and methyl-deficient diet alters the expression of DNA methyltransferases and methyl CpG binding proteins involved in epigenetic gene silencing in livers of F344 rats. J. Nutr. 2006;136:1522–1527. doi: 10.1093/jn/136.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert N, Thomson I, Boyle S, Allan J, Ramsahoye B, et al. DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J. Cell. Biol. 2007;177:401–411. doi: 10.1083/jcb.200607133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Gregory RI, Randall TE, Johnson CA, Khosla S, Hatada I, et al. DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol. Cell. Biol. 2001;21:5426–5436. doi: 10.1128/MCB.21.16.5426-5436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guruswamy S, Swamy MV, Choi CI, Steele VE, Rao CV. S-adenosyl l-methionine inhibits azoxymethane-induced colonic aberrant crypt foci in F344 rats and suppresses human colon cancer Caco-2 cell growth in 3D culture. Int. J. Cancer. 2008;122:25–30. doi: 10.1002/ijc.23031. [DOI] [PubMed] [Google Scholar]
- 51.Hata K, Kusumi M, Yokomine T, Li E, Sasaki H. Meiotic and epigenetic aberrations in Dnmt3 L-deficient male germ cells. Mol. Reprod. Dev. 2006;73:116–122. doi: 10.1002/mrd.20387. [DOI] [PubMed] [Google Scholar]
- 52.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 53.Hernandez-Munoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol. Cell. Biol. 2005;25:11047–11058. doi: 10.1128/MCB.25.24.11047-11058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 55.Honma K, Mochizuki K, Goda T. Carbohydrate/fat ratio in the diet alters histone acetylation on the sucrase-isomaltase gene and its expression in mouse small intestine. Biochem. Biophys. Res. Commun. 2007;357:1124–1129. doi: 10.1016/j.bbrc.2007.04.070. [DOI] [PubMed] [Google Scholar]
- 56.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 57.Huang BH, Laban M, Leung CH, Lee L, Lee CK, et al. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell. Death Differ. 2005;12:395–404. doi: 10.1038/sj.cdd.4401567. [DOI] [PubMed] [Google Scholar]
- 58.Huang JC, Lei ZL, Shi LH, Miao YL, Yang JW, et al. Comparison of histone modifications in in vivo and in vitro fertilization mouse embryos. Biochem. Biophys. Res. Commun. 2007;354:77–83. doi: 10.1016/j.bbrc.2006.12.163. [DOI] [PubMed] [Google Scholar]
- 59.Ito K, Lim S, Caramori G, Cosio B, Chung KF, et al. A molecular mechanism of action of theophylline: induction of histone deacetylase activity to decrease inflammatory gene expression. Proc. Natl. Acad. Sci. USA. 2002;99:8921–8926. doi: 10.1073/pnas.132556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyl-transferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 62.Kang J, Lin C, Chen J, Liu Q. Copper induces histone hypoacetylation through directly inhibiting histone acetyltransferase activity. Chem. Biol. Interact. 2004;148:115–123. doi: 10.1016/j.cbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Kang J, Zhang Y, Chen J, Chen H, Lin C, et al. Nickel-induced histone hypoacetylation: the role of reactive oxygen species. Toxicol. Sci. 2003;74:279–286. doi: 10.1093/toxsci/kfg137. [DOI] [PubMed] [Google Scholar]
- 64.Kim DH, Shin J, Kwon HJ. Psammaplin A is a natural prodrug that inhibits class I histone deacetylase. Exp. Mol. Med. 2007;39:47–55. doi: 10.1038/emm.2007.6. [DOI] [PubMed] [Google Scholar]
- 65.Kim JM, Liu H, Tazaki M, Nagata M, Aoki F. Changes in histone acetylation during mouse oocyte meiosis. J. Cell. Biol. 2003;162:37–46. doi: 10.1083/jcb.200303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, et al. pRB family proteins are required for H3K27 trimethylation and polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 68.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 69.Lane N, Dean W, Erhardt S, Hajkova P, Surani A, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 70.Lea MA, Randolph VM. Induction of histone acetylation in rat liver and hepatoma by organosulfur compounds including diallyl disulfide. Anticancer Res. 2001;21:2841–2845. [PubMed] [Google Scholar]
- 71.Lea MA, Randolph VM, Patel M. Increased acetylation of histones induced by diallyl disulfide and structurally related molecules. Int. J. Oncol. 1999;15:347–352. doi: 10.3892/ijo.15.2.347. [DOI] [PubMed] [Google Scholar]
- 72.Lea MA, Rasheed M, Randolph VM, Khan F, Shareef A, et al. Induction of histone acetylation and inhibition of growth of mouse erythroleukemia cells by S-allylmercaptocysteine. Nutr. Cancer. 2002;43:90–102. doi: 10.1207/S15327914NC431_11. [DOI] [PubMed] [Google Scholar]
- 73.Lee ER, McCool KW, Murdoch FE, Fritsch MK. Dynamic changes in histone H3 phosphoacetylation during early embryonic stem cell differentiation are directly mediated by mitogen-and stress-activated protein kinase 1 via activation of MAPK pathways. J. Biol. Chem. 2006;281:21162–21172. doi: 10.1074/jbc.M602734200. [DOI] [PubMed] [Google Scholar]
- 74.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 75.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J. Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 76.Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, et al. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br. J. Nutr. 2007;97:1064–1073. doi: 10.1017/S000711450769196X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin C, Kang J, Zheng R. Oxidative stress is involved in inhibition of copper on histone acetylation in cells. Chem. Biol. Interact. 2005;151:167–176. doi: 10.1016/j.cbi.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 78.Liu H, Kim JM, Aoki F. Regulation of histone H3 lysine 9 methylation in oocytes and early preimplantation embryos. Development. 2004;131:2269–2280. doi: 10.1242/dev.01116. [DOI] [PubMed] [Google Scholar]
- 79.Liu HL, Chen Y, Cui GH, Zhou JF. Curcumin, a potent antitumor reagent, is a novel histone deacetylase inhibitor regulating B-NHL cell line Raji proliferation. Acta Pharmacol. Sin. 2005;26:603–609. doi: 10.1111/j.1745-7254.2005.00081.x. [DOI] [PubMed] [Google Scholar]
- 80.Lucifero D, La Salle S, Bourc’his D, Martel J, Bestor TH, et al. Coordinate regulation of DNA methyltransferase expression during oogenesis. BMC Dev. Biol. 2007;7:36. doi: 10.1186/1471-213X-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 82.Marcu MG, Jung YJ, Lee S, Chung EJ, Lee MJ, et al. Curcumin is an inhibitor of p300 histone acetyltransferase. Med. Chem. 2006;2:169–174. doi: 10.2174/157340606776056133. [DOI] [PubMed] [Google Scholar]
- 83.McDowell TL, Gibbons RJ, Sutherland H, O’Rourke DM, Bickmore WA, et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc. Natl. Acad. Sci. USA. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mochizuki K, Kawai H, Mochizuki H, Shimada M, Takase S, et al. Fatty acids in component of milk enhance the expression of the cAMP-response-element-binding-protein-binding protein (CBP)/p300 gene in developing rats. Br. J. Nutr. 2007:1–6. doi: 10.1017/S0007114507831680. [DOI] [PubMed] [Google Scholar]
- 86.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 87.Morimitsu Y, Nakagawa Y, Hayashi K, Fujii H, Kumagai T, et al. A sulforaphane analogue that potently activates the Nrf2-dependent detoxification pathway. J. Biol. Chem. 2002;277:3456–3463. doi: 10.1074/jbc.M110244200. [DOI] [PubMed] [Google Scholar]
- 88.Mulholland NM, King IF, Kingston RE. Regulation of Polycomb group complexes by the sequence-specific DNA binding proteins Zeste and GAGA. Genes Dev. 2003;17:2741–2746. doi: 10.1101/gad.1143303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 90.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 91.Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, et al. Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J. Biol. Chem. 2007;282:4408–4416. doi: 10.1074/jbc.M606773200. [DOI] [PubMed] [Google Scholar]
- 92.Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. Developmental acquisition of genome-wide DNA methylation occurs prior to meiosis in male germ cells. Dev. Biol. 2007;307:368–379. doi: 10.1016/j.ydbio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 93.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, et al. A stem-cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olaharski AJ, Rine J, Marshall BL, Babiarz J, Zhang L, et al. The flavoring agent dihydrocoumarin reverses epigenetic silencing and inhibits sirtuin deacetylases. PLoS Genet. 2005;1:e77. doi: 10.1371/journal.pgen.0010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 97.Pogribny IP, Ross SA, Tryndyak VP, Pogribna M, Poirier LA, et al. Histone H3 lysine 9 and H4 lysine 20 trimethylation and the expression of Suv4–20h2 and Suv-39h1 histone methyltransferases in hepatocarcinogenesis induced by methyl deficiency in rats. Carcinogenesis. 2006;27:1180–1186. doi: 10.1093/carcin/bgi364. [DOI] [PubMed] [Google Scholar]
- 98.Pogribny IP, Tryndyak VP, Muskhelishvili L, Rusyn I, Ross SA. Methyl deficiency, alterations in global histone modifications, and carcinogenesis. J. Nutr. 2007;137 doi: 10.1093/jn/137.1.216S. 216–22S. [DOI] [PubMed] [Google Scholar]
- 99.Porcu M, Chiarugi A. The emerging therapeutic potential of sirtuin-interacting drugs: from cell death to lifespan extension. Trends Pharmacol. Sci. 2005;26:94–103. doi: 10.1016/j.tips.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 100.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 101.Robbins AR, Jablonski SA, Yen TJ, Yoda K, Robey R, et al. Inhibitors of histone deacetylases alter kinetochore assembly by disrupting pericentromeric heterochromatin. Cell Cycle. 2005;4:717–726. doi: 10.4161/cc.4.5.1690. [DOI] [PubMed] [Google Scholar]
- 102.Rybouchkin A, Kato Y, Tsunoda Y. Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol. Reprod. 2006;74:1083–1089. doi: 10.1095/biolreprod.105.047456. [DOI] [PubMed] [Google Scholar]
- 103.Sakamoto A, Liu J, Greene A, Chen M, Weinstein LS. Tissue-specific imprinting of the G protein Gsalpha is associated with tissue-specific differences in histone methylation. Hum. Mol. Genet. 2004;13:819–828. doi: 10.1093/hmg/ddh098. [DOI] [PubMed] [Google Scholar]
- 104.Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 2005;280:225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 105.Sarg B, Helliger W, Talasz H, Koutzamani E, Lindner HH. Histone H4 hyperacetylation precludes histone H4 lysine 20 trimethylation. J. Biol. Chem. 2004;279:53458–53464. doi: 10.1074/jbc.M409099200. [DOI] [PubMed] [Google Scholar]
- 106.Sarmento OF, Digilio LC, Wang Y, Perlin J, Herr JC, et al. Dynamic alterations of specific histone modifications during early murine development. J. Cell Sci. 2004;117:4449–4459. doi: 10.1242/jcs.01328. [DOI] [PubMed] [Google Scholar]
- 107.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, et al. Polycomb-mediated methylation on Lys27 of histone H3 premarks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 108.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by Polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 109.Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, et al. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 110.Soejima H, Zhao W, Mukai T. Epigenetic silencing of the MGMT gene in cancer. Biochem. Cell Biol. 2005;83:429–437. doi: 10.1139/o05-140. [DOI] [PubMed] [Google Scholar]
- 111.Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, et al. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447:735–738. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 112.Svensson K, Mattsson R, James TC, Wentzel P, Pilartz M, et al. The paternal allele of the H19 gene is progressively silenced during early mouse development: The acetylation status of histones may be involved in the generation of variegated expression patterns. Development. 1998;125:61–69. doi: 10.1242/dev.125.1.61. [DOI] [PubMed] [Google Scholar]
- 113.Symonds ME, Stephenson T, Gardner DS, Budge H. Long-term effects of nutritional programming of the embryo and fetus: mechanisms and critical windows. Reprod. Fertil. Dev. 2007;19:53–63. doi: 10.1071/rd06130. [DOI] [PubMed] [Google Scholar]
- 114.Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 2007;26:3346–3359. doi: 10.1038/sj.emboj.7601767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 116.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tamada H, Van Thuan N, Reed P, Nelson D, Katoku-Kikyo N, et al. Chromatin decondensation and nuclear reprogramming by nucleoplasmin. Mol. Cell. Biol. 2006;26:1259–1271. doi: 10.1128/MCB.26.4.1259-1271.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 119.Torrens C, Brawley L, Anthony FW, Dance CS, Dunn R, et al. Folate supplementation during pregnancy improves offspring cardiovascular dysfunction induced by protein restriction. Hypertension. 2006;47:982–987. doi: 10.1161/01.HYP.0000215580.43711.d1. [DOI] [PubMed] [Google Scholar]
- 120.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Venturelli S, Armeanu S, Pathil A, Hsieh CJ, Weiss TS, et al. Epigenetic combination therapy as a tumor-selective treatment approach for hepatocellular carcinoma. Cancer. 2007;109:2132–2141. doi: 10.1002/cncr.22652. [DOI] [PubMed] [Google Scholar]
- 123.Vidali G, Boffa LC, Bradbury EM, Allfrey VG. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc. Natl. Acad. Sci. USA. 1978;75:2239–2243. doi: 10.1073/pnas.75.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Villa R, Morey L, Raker VA, Buschbeck M, Gutierrez A, et al. The methyl-CpG binding protein MBD1 is required for PML-RARalpha function. Proc. Natl. Acad. Sci. USA. 2006;103:1400–1405. doi: 10.1073/pnas.0509343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 126.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 127.Wang Q, Wang CM, Ai JS, Xiong B, Yin S, et al. Histone phosphorylation and pericentromeric histone modifications in oocyte meiosis. Cell Cycle. 2006;5:1974–1982. doi: 10.4161/cc.5.17.3183. [DOI] [PubMed] [Google Scholar]
- 128.Waterland RA, Garza C. Potential mechanisms of metabolic imprinting that lead to chronic disease. Am. J. Clin. Nutr. 1999;69:179–197. doi: 10.1093/ajcn/69.2.179. [DOI] [PubMed] [Google Scholar]
- 129.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum. Mol. Genet. 2006;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- 130.Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 131.Wiencke JK, Zheng S, Morrison Z, Yeh RF. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene. 2008;27:2412–2421. doi: 10.1038/sj.onc.1210895. [DOI] [PubMed] [Google Scholar]
- 132.Wilson AJ, Byun DS, Popova N, Murray LB, L’Italien K, et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J. Biol. Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 133.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 134.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 135.Yamasaki-Ishizaki Y, Kayashima T, Mapendano CK, Soejima H, Ohta T, et al. Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol. Cell. Biol. 2007;27:732–742. doi: 10.1128/MCB.01329-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yan Y, Kluz T, Zhang P, Chen HB, Costa M. Analysis of specific lysine histone H3 and H4 acetylation and methylation status in clones of cells with a gene silenced by nickel exposure. Toxicol. Appl. Pharmacol. 2003;190:272–277. doi: 10.1016/s0041-008x(03)00169-8. [DOI] [PubMed] [Google Scholar]
- 137.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 138.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zofall M, Grewal SI. HULC, a histone H2B ubiquitinating complex, modulates heterochromatin independent of histone methylation in fission yeast. J. Biol. Chem. 2007;282:14065–14072. doi: 10.1074/jbc.M700292200. [DOI] [PubMed] [Google Scholar]