Abstract
To survive starvation conditions, eukaryotes have developed an evolutionarily conserved process, termed autophagy, by which the vacuole/lysosome mediates the turnover and recycling of non-essential intracellular material for re-use in critical biosynthetic reactions. Morphological and biochemical studies in Saccharomyces cerevisiae have elucidated the basic steps and mechanisms of the autophagy pathway. Although it is a degradative process, autophagy shows substantial overlap with the biosynthetic cytoplasm to vacuole targeting (Cvt) pathway that delivers resident hydrolases to the vacuole. Recent molecular genetics analyses of mutants defective in autophagy and the Cvt pathway, apg, aut, and cvt, have begun to identify the protein machinery and provide a molecular resolution of the sequestration and import mechanism that are characteristic of these pathways. In this study, we have identified a novel protein, termed Apg2, required for both the Cvt and autophagy pathways as well as the specific degradation of peroxisomes. Apg2 is required for the formation and/or completion of cytosolic sequestering vesicles that are needed for vacuolar import through both the Cvt pathway and autophagy. Biochemical studies revealed that Apg2 is a peripheral membrane protein. Apg2 localizes to the previously identified perivacuolar compartment that contains Apg9, the only characterized integral membrane protein that is required for autophagosome/Cvt vesicle formation.
Cellular homeostasis requires the precise regulation of protein synthesis and organelle biogenesis as well as the degradation of unnecessary proteins and organelles. The degradation profile of the cell is comprised of the selective degradation of proteins that occurs predominantly in the cytosol through the ubiquitin/proteasome pathway, and the degradation of bulk cytoplasm that takes place in the lysosome in mammals and plants and in the vacuole in fungi (1, 2). Autophagy is the major degradation route by which bulk cytoplasm is delivered to the lysosome/vacuole. This pathway is conserved from yeast to human. Although the autophagic machinery only functions at a basal level in nutrient-rich conditions, it plays an essential physiological role in starvation conditions, which are more likely to mimic the natural environment of organisms such as yeast. In yeast, when cells sense nitrogen starvation, cytosol and organelles are nonspecifically sequestered and delivered to the vacuole by the up-regulated autophagic machinery, where they can be recycled and used for critical processes, thus ensuring cell survival (3, 4).
Interestingly, in yeast, autophagy overlaps with the biosynthetic delivery of the vacuolar hydrolase aminopeptidase I (Ape1)1 by the cytoplasm to vacuole targeting (Cvt) pathway. Under vegetative growth conditions, precursor Ape1 (prApe1) is synthesized in the cytosol where it quickly forms a homododecamer (5). These dodecamers subsequently assemble into a higher order complex termed the Cvt complex. The Cvt complex becomes enwrapped by a membrane resulting in a doublemembrane Cvt vesicle (150 nm diameter). The contents of the Cvt vesicles appear to exclude cytosol and are observed as an electron-dense structure by electron microscopy (6). Autophagy is morphologically similar to the Cvt pathway, however, the cytosolic double membrane vesicles (autophagosomes) are much larger (300–900 nm diameter) than Cvt vesicles and they include bulk cytoplasm (7). In either case, the completed cytosolic vesicles then dock and fuse with the vacuole membrane releasing single membrane Cvt or autophagic bodies into the vacuole lumen. During starvation conditions, prApe1 is specifically sequestered, along with bulk cytoplasm, inside the double membrane autophagosome for its delivery to the vacuole. Precursor Ape1 is proteolytically processed to mature Ape1 once it reaches the vacuole by either the Cvt pathway or autophagy. Therefore, prApe1 processing serves as a useful diagnostic marker for studying both the Cvt pathway in nutrient-rich conditions and the autophagy pathway during starvation (8).
Genetic analyses support the morphological data that indicate an overlap between the Cvt pathway and autophagy. A genetic screen that was based on the accumulation of precursor Ape1 under nutrient-rich conditions was used to isolate mutants in the Cvt pathway (9, 10). These cvt mutants exhibited an extensive genetic overlap with apg and aut mutants that were isolated as being defective in autophagy, indicating that the two pathways also share protein components (8 –10). Recently, it has been shown that the selective degradation of excess peroxisomes (pexophagy) occurs by a similar process using the same molecular machinery shared between the Cvt and autophagy pathways (11). The analysis of the molecular components required for these pathways has provided significant insights into the process by which proteins are delivered post-translationally to the vacuole/lysosome.
While many of the details of cytoplasm to vacuole transport remain to be elucidated, some of the key questions have begun to be answered. For example, the Cvt pathway operates under nutrient-rich conditions while autophagy is induced by starvation. Degradation of peroxisomes occurs during adaptation to preferred carbon sources following growth in peroxisome proliferating conditions such as methanol or oleate. The identified mechanism of signal transduction includes a signaling pathway regulated by Tor, a phosphatidylinositol 3-kinase homologue. Tor represses autophagy under nutrient-rich conditions. During starvation or following treatment with rapamycin, Tor kinase activity is inhibited and autophagy is induced (12). The Tor signaling cascade also affects the phosphorylation state of Apg13 and its affinity for the Apg1 kinase (13). Recently, Cvt9 and Apg17 were characterized as additional proteins that associate with the Apg1 kinase (13, 14). Interestingly, Cvt9 is specific for the Cvt pathway and is also needed for pexophagy while Apg17 is specific for autophagy. These findings suggest that a potential protein complex including the Apg1 kinase and various regulatory proteins may determine which of these pathways operates within the cell.
One of the most distinctive features of the Apg/Cvt pathways is the formation of sequestering vesicles that engulf cytoplasm. A major difference between autophagy and the Cvt pathway is that autophagosomes are substantially larger than the Cvt vesicles that form under nutrient-rich conditions. Aut7 is the first identified component that localizes to both Cvt vesicles and autophagosomes. The level of Aut7 is up-regulated under starvation conditions, suggesting that it is a limiting component that is required for Cvt vesicle formation as well as autophagosome expansion (15, 16). The formation and completion of the Cvt vesicle/autophagosome also requires a novel protein conjugation system consisting of Apg5, Apg7, Apg10, Apg12, and Apg16 (17–20). Recent findings suggest that Aut7 membrane association is dependent on the autophagy (Apg) conjugation system as well as a novel membrane lipidation process involving the Aut1 protein (21,22). Finally, Apg9 is the only characterized integral membrane protein required for the vesicle formation/completion step for both the Cvt and autophagy pathways. It is localized at a perivacuolar punctate structure that appears to be distinct from typical plasma membrane and endomembrane marker proteins. Interestingly, the Apg9 protein does not appear to be localized to the Cvt vesicle/autophagosome suggesting that it might mark the site of the donor membrane for vesicle formation (23).
Although some components have been cloned, many questions remain to be answered concerning the Cvt pathway, autophagy, and pexophagy. The identification of additional components that function in these pathways will help us to understand the molecular details of the import process including the origin of the sequestering membrane, the signal transduction cascade that senses the nutritional conditions, the mechanism of vesicle formation, and the subsequent delivery and breakdown of the subvacuolar vesicles. In the present study we have begun a characterization of the Apg2 protein that is required for both the Cvt and autophagy pathways and also for the related process of peroxisome degradation. The apg2Δ strain accumulated the precursor form of Ape1 in a protease-accessible, membrane-associated form, suggesting that Apg2 is required for the completion stage of Cvt vesicle and autophagosome formation. Subcellular fractionation and biochemical studies demonstrate that Apg2 is a peripheral membrane protein that localizes to the previously identified Apg9 compartment. Finally, microscopy experiments indicate that GFPApg2 localization is dependent on the Apg9 protein. This finding is supported by an in vitro pull-down assay that demonstrates a physical interaction between Apg2 and Apg9.
EXPERIMENTAL PROCEDURES
Strains and Growth Media
The strains used in this study are listed in Table I. Yeast cells were grown in rich medium (YPD; 1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal medium (SMD; 0.67% yeast nitrogen base (YNB), 2% glucose, and auxotrophic amino acids and vitamins as needed). Peroxisomes were induced by incubation in synthetic glycerol (0.67% YNB without amino acids, pH 5.5, 3% glycerol, 0.1% glucose) and oleic acid (YTO; 0.67% YNB, 0.1% Tween 40, 0.1% oleic acid) media. Starvation was carried out in synthetic minimal medium lacking nitrogen (SD-N; 0.17% YNB without amino acids, 2% glucose).
TABLE I.
Yeast strains used in this study
| Strain | Genotype | Source or Ref. |
|---|---|---|
| SEY6210 | MATα his3-Δ200 leu2–3,112 lys2–281 trp1-Δ901 ura3–52 suc2-Δ9 GAL | 35 |
| WCGa | MATa his3–11,15 leu2–3,112, ura3 GAL | 36 |
| WSY99 | SEY6210 ypt7Δ::HIS3 | 37 |
| CWY1 | SEY6210 apg2Δ::HIS5 | This study |
| CWY2 | WCGa apg2Δ::HIS5 | This study |
| MT2–4-3 | MATa ura3–52 trp1 leu2–3,112 apg2–1 | 28 |
| VDY101 | SEY6210 apg7Δ::LEU2 | 26 |
| JKY007 | SEY6210 apg9Δ::HIS3 | 23 |
Materials
All restriction enzymes and Vent DNA polymerase were from New England Biolabs, Inc. (Beverly, MA). Tran35S-label was from ICN (Costa Mesa, CA). Ficoll 400 was from Amersham Pharmacia Biotech (Piscataway, NJ). OptiPrep™ was from Accurate Chemical and Scientific Corp. (Westbury, NY). Dynabeads™ M-500 Subcellular were from Dynal Biotech, Inc. (Lake Success, NY). Complete™ EDTA-free protease inhibitor mixture was from Roche Molecular Biochemicals (Indianapolis, IN). ChromPure human IgG was from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). Oligonucleotides were synthesized by Operon (Alameda, CA). The centromeric APG2 plasmid pYCG_YNL242w containing the APG2 gene in pRS416 was purchased from EUROSCARF (Frankfurt, Germany). The pME3 vector, which contains a Schizosaccharomyces pombe HIS5 auxotrophic marker, was a gift from Dr. Neta Dean (State University of New York, Stony Brook, NY). All other reagents were from Sigma-Aldrich.
Antiserum
To prepare antiserum against Apg2, residues 101–333 and 900–1163 encoded by the APG2 open reading frame (ORF) were PCR amplified and fused to a glutathione S-transferase and a maltose-binding protein tag, respectively. The resulting two plasmids were then transformed into the Escherichia coli BL21 strain. Cells were grown in TYE to A600 = 1.0 and then fusion protein expression was induced for 3 h by adding isopropyl-1-thio-β-d-galactopyranoside (final concentration of 0.5 mm). Cells were lysed by sonication to obtain the crude cell lysate followed by column purification procedures as described by the suppliers of the vector fusion systems. The glutathione S-transferase fusion system was from Amersham Pharmacia Biotech and the pMAL protein fusion and purification system was from New England Biolabs, Inc. (Beverly, MA). Antiserum was generated in a New Zealand White rabbit using standard procedures (24). Antisera against Ape1 (25), Apg9 (23), and Fox3 (11) were prepared as described previously. Monoclonal antibodies to alkaline phosphatase (Pho8) and Dpm1 were from Molecular Probes (Eugene, OR). Antiserum against α1,3-mannosyltransferase (Mnn1), Pep12, and phosphoglycerate kinase (Pgk1) were generously provided by Dr. Todd Graham (Vanderbilt University, Nashville, TN), Dr. Scott Emr (University of California at San Diego, La Jolla, CA) and Dr. Jeremy Thorner (University of California, Berkeley, CA), respectively.
Disruption and Cloning of APG2
The apg2Δ strains were made by replacing the APG2 ORF with the HIS5 gene from S. pombe. The HIS5 gene was PCR amplified using the plasmid pME3 as template. The 5′ and 3′ ends of the knockout cassette contains 41 base pairs of flanking region beginning 8 bases after the start codon of the APG2 ORF, and 66 base pairs after the stop codon, respectively. Plasmid pYCG_YNL242w containing the APG2 gene in pRS416 was digested with SpeI and KpnI and a 5.7-kilobase fragment cloned into the SpeI and KpnI sites of pRS426 to generate the multicopy plasmid pAPG2(426).
Plasmids
A plasmid encoding GFPSKL under control of the CUP1 promoter (pCuGFP-SKL (416)) will be described elsewhere.2 The plasmids for expressing protein A and the protein A-Apg9 fusion were prepared by BamHI and SalI digestion of pRS416-ProtA and pRS416-ProtA-APG9, respectively. The corresponding fragments were cloned into the BamHI and SalI sites of pRS425 to generate pRS425-ProtA and pRS425-ProtA-APG9. The plasmid pRS416-ProtA was a gift from Dr. Scott Emr. The construction of pRS416-ProtA-APG9 will be described elsewhere.3
To fuse GFP to the NH2 terminus of Apg2, the APG2 ORF was amplified by PCR using pYCG_YNL242w as the template. The 5′ and 3′ oligonucleotides used for the PCR amplification were 5′-TTGATTTCGACCCGGGGGCATTTTGGTTACCTCAAAATATACAAAAGCG-3′ and 5′- ATATCAAAAATGTCGGTACCAATTTTTACGAATCAGTCCGATTGGACTT-3′, respectively. The 5′ oligonucleotide contains an XmaI site that is in the same reading frame as the end of the GFP ORF of pCuGFP (414), a plasmid in which GFP expression is under the control of the CUP1 copper-inducible promoter (22). The 3′ oligonucleotide contains a KpnI site. The APG2 PCR product was subcloned into pCuGFP(414) by an XmaI-KpnI digest, resulting in the pCuGFPAPG2(414) plasmid.
Cell Viability under Nitrogen Starvation Conditions
To examine the survival of various yeast strains under nitrogen starvation conditions, cells were grown to an A600 = 1.0 in SMD medium, washed once in SD-N, and then resuspended in SD-N to an A600 = 1.0. At the indicated times, aliquots were removed, serially diluted and plated onto YPD plates in triplicate. After 2–3 days incubation, colonies were counted on plates.
Protease Protection Analysis
To examine the protease sensitivity of prApe1 in the apg2Δ, apg7Δ, and ypt7Δ strains, cells were grown to mid-log phase (1.0 A600/ml) and converted into spheroplasts. The spheroplasts were then subjected to osmotic lysis in PS200 lysis buffer (20 mm PIPES, pH 6.8, 200 mm sorbitol) containing 5 mm MgCl2 at a spheroplast density of 20 A600/ml. The lysed spheroplasts were centrifuged at 13,000 × g for 10 min resulting in supernatant (S13) and pellet (P13) fractions. The P13 pellet was resuspended in lysis buffer in the presence or absence of proteinase K (50 µg/ml) and 0.2% Triton X-100. Samples were incubated on ice for 20 min, followed by trichloroacetic acid precipitation, acetone wash, and immunoblot with anti-Ape1 and anti-Pgk1 antisera.
Membrane Flotation Analysis
The procedure for the membrane flotation experiments is a modification of a previously described protocol (26). Spheroplasts from the apg2Δ strain were osmotically lysed in PS200 containing 5 mm MgCl2 at a spheroplast density of 20 A600/ml. The lysate was subjected to a 13,000 × g centrifugation for 10 min, resulting in supernatant (S) and pellet (P) fractions. The pellet fractions, which contained all of the prApe1, were resuspended in 200 µl of 15% Ficoll 400 (w/v) in lysis buffer with or without the addition of 0.2% Triton X-100. The resuspended pellet fractions were overlaid with 1 ml of 13% Ficoll 400 in lysis buffer and then 200 µl of 2% Ficoll 400 in lysis buffer. The resulting step gradients were subjected to a 13,000 × g centrifugation for 10 min at room temperature. Fractions were collected from the top. The first 400 µl was the float fraction (F), the remaining 1 ml was the non-float fraction (NF), and the gradient pellet was considered the pellet fraction (P2). The resulting fractions were trichloroacetic acid precipitated and washed twice with acetone followed by immunoblot with anti-Ape1 antiserum.
Cell Labeling
To examine Apg2 stability and post-translational modification, cells were grown to A600 = 1.0 in SMD. Cells (20 ml) were collected and labeled in 300 µl of SMD with 20 µCi of Tran35S-label for 30 min. The cells were then subjected to a non-radioactive chase in SMD supplemented with 0.2% yeast extract, 4 mm methionine, and 2 mm cysteine at a final density of 2.0 A600/ml. Samples were removed at the indicated time points and precipitated with 10% trichloroacetic acid. Crude extracts were prepared by glass bead lysis and subjected to immunoprecipitation as described previously (9).
Subcellular Fractionation
Wild-type cells (SEY6210) were spheroplasted at an A600 = 1.0 before osmotic lysis in PS200 buffer containing 0 or 5 mm MgCl2. The total cell lysates were then subjected to centrifugation at 13,000 × g for 10 min at 4 °C, resulting in soluble (S13) and pelletable (P13) fractions. The S13 fraction was further centrifuged at 100,000 × g in a Beckman Coulter, Inc. (Fullerton, CA) Optima ™ MAX-E ultracentrifuge using a TLA 100.4 rotor for 30 min at 4 °C to generate the corresponding S100 and P100 fractions. The resulting fractions were trichloroacetic acid precipitated and washed twice with acetone followed by immunoblot with anti-Apg2 antiserum. To further examine the biochemical nature of the Apg2 pelletable fractions, spheroplasts of wild type cells were osmotically lysed in PS200 lysis buffer containing 5 mm MgCl2. The cell lysate was aliquoted and then centrifuged at 100,000 × g for 30 min. The total pellet fraction was collected and resuspended in one of the following buffers: 1 m KCl, 0.1 m Na2CO3 (pH 10.5), 3 m urea, 1% Triton X-100. After incubation at room temperature for 10 min, all samples were separated into supernatant (S) and pellet (P) fractions by centrifugation at 100,000 × g for 30 min. The samples were analyzed by Western blot as described previously (9).
OptiPrep Density Gradient
Wild type cells were grown to mid-log phase and converted into spheroplasts before being subjected to osmotic lysis in PS200 buffer containing 1 mm EDTA, 1 mm MgCl2, and a protease inhibitor mixture (Complete ™ EDTA-free protease inhibitor tablets, 1 mm phenylmethylsulfonyl fluoride, 1 µg/ml leupeptin, and 1 µg/ml pepstatin A). After a pre-clear centrifugation step at 800 × g for 5 min to remove the cell debris, the crude lysate (20 A600 units of cells) was centrifuged at 100,000 × g for 20 min at 4 °C. The resulting total membrane fraction was resuspended in lysis buffer and layered on top of a density gradient consisting of 0–66% OptiPrep in PS200 containing 1 mm EDTA, 1 mm MgCl2, and 1 mm dithiothreitol, and a protease inhibitor mixture. The gradients were subjected to centrifugation at 100,000 × g for 16 h at 4 °C in a TLS 55 rotor. Samples were collected from the top of the gradients into 14 fractions. The fractions were then resolved by SDS-polyacrylamide gel electrophoresis and examined by immunoblot using antiserum or antibodies against Apg2, Dpm1, Mnn1, Pep12, Pho8, and Apg9.
Protein A Pull-down
The multicopy APG2 plasmid, pAPG2(426), was co-transformed into SEY6210 wild type cells with pRS425-ProtA or pRS425-ProtA-APG9. Spheroplasts prepared from transformants were grown to mid-log phase and lysed by Dounce homogenization at a cell density of 30 A600/ml in phosphate-buffered saline lysis buffer with or without 0.5% Nonidet P-40 (1 × phosphate-buffered saline, 200 mm sorbitol, 5 mm MgCl2, ± 0.5% Nonidet P-40, and protease inhibitors). Cell lysates (1 ml) were incubated with 20 µl of Dynabeads M-500 Subcellular cross-linked to human IgG for 2 h at 4 °C. The beads were then washed in phosphate-buffered saline lysis buffer 4 times and eluted with SDS-polyacrylamide gel electrophoresis sample buffer. Apg2 was detected by immunoblot.
Pexophagy Thiolase Assay
The assay was carried out essentially as described previously (11). In brief, cells were grown in YPD to the exponential stage and then shifted to synthetic glycerol for 12 h at 30 °C. A 10 × solution of YP was added to a final concentration of 1X and the culture incubated 4 h at 30 °C. Cells were then shifted to YTO for 19 h to induce peroxisomes. Cells were harvested and washed once in 1 × YNB (0.67% YNB) and SD-N, respectively. After washing, cells were resuspended in SD-N to an A600 = 1.0. At the indicated times, aliquots were removed, trichloroacetic acid precipitated, and then subjected to immunoblot with anti-Fox3 antiserum.
Fluorescence Microscopy
Induction of peroxisomes in cells expressing GFP-SKL was done as described for the pexophagy thiolase assay. Following induction, cells were shifted to SD-N. At 0, 12, and 26 h following incubation in SD-N medium, aliquots were removed and examined by fluorescence microscopy. To examine GFPApg2 localization, cells were grown to midlog phase and viewed directly or shifted to SD-N medium for 4 h prior to examination. Microscopy was performed on a Nikon E-800 fluorescent microscope (Mager Scientific Inc., Dexter, MI). Images were captured by an ORCA II CCD camera (Hamamatsu Corp., Bridgewater, NJ) using Openlab software (Improvision, Inc., Lexington, MA).
To examine induction and stability of Apg2 and GFPApg2, the apg2Δ (CWY1) and apg9Δ (JKY007) strains transformed with a plasmid expressing GFPApg2, and non-transformed apg9Δ and wild type (SEY6210) strains were grown in SMD or shifted to SD-N for 4 h. Protein extracts were prepared and resolved by SDS-polyacrylamide gel electrophoresis. GFPApg2 and Apg2 were detected by Western blot using antiserum against Apg2.
RESULTS
Apg2 Is a Shared Component of the Autophagy and Cvt Pathways
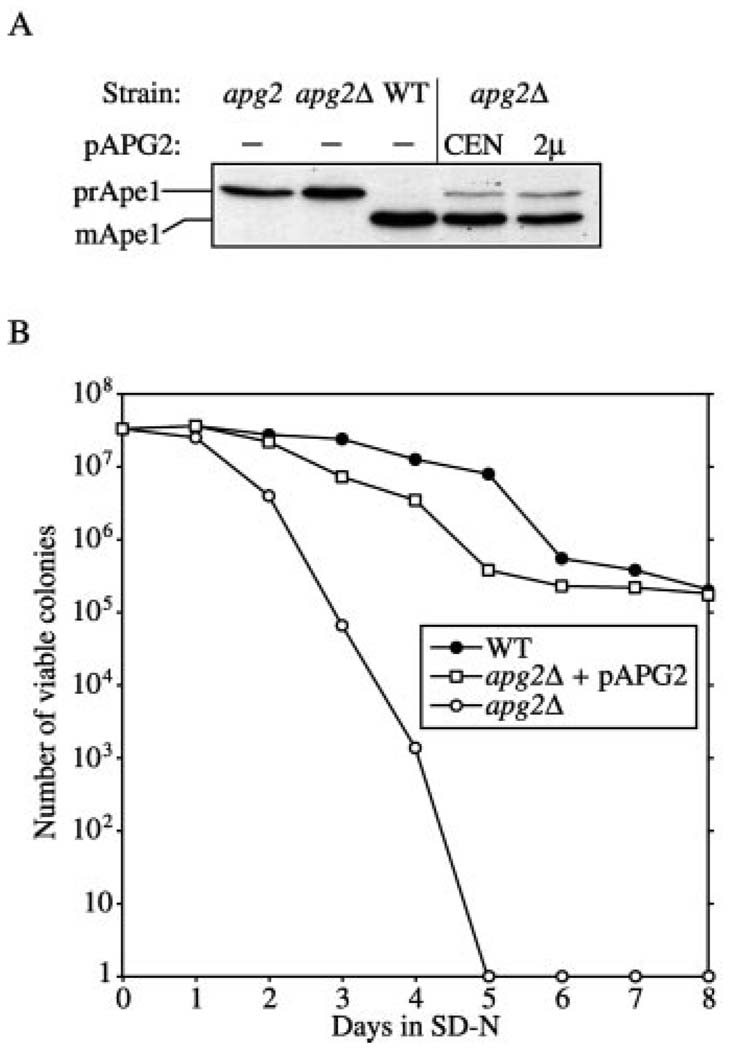
In the methylotrophic yeast Pichia pastoris, growth on methanol results in the proliferation of peroxisomes. Ethanol adaptation results in the degradation of excess peroxisomes through macropexophagy, where the organelles are sequestered within cytosolic vesicles and subsequently delivered to the vacuole (1, 2). In contrast, glucose adaptation induces the direct engulfment and degradation of excess peroxisomes by the vacuole in a process termed micropexophagy (1, 2). Despite the differences in the site of sequestration, micropexophagy also utilizes machinery required for macroautophagy. For example, Gsa7 and Gsa9, proteins involved in the micropexophagy pathway in P. pastoris, are homologous to Apg7 and Cvt9, respectively, that are needed for macroautophagy and the Cvt pathway (14, 26, 27). The gsa11 mutant was isolated based on defects in micropexophagy in P. pastoris.4 The S. cerevisiae homologue of GSA11, YNL242w, encoded an as yet uncharacterized protein. To determine if the corresponding gene product was required for import of prApe1 by the Cvt pathway, we generated a null mutant. The YNL242w chromosomal locus was deleted by one-step gene disruption as described under “Experimental Procedures.” Wild type and ynl242wΔ cells were examined by Western blot to determine the state of Ape1. We found that the ynl242wΔ strain accumulated only the precursor form of Ape1 indicating the requirement of this protein in the Cvt pathway (Fig. 1A).
FIG. 1. Apg2 is required for both the Cvt and autophagy pathways.
A, cloning and characterization of APG2 (YNL242w). Wild type (WT; SEY6210), apg2 (MT2–4-3), apg2Δ (ynl242wΔ; CWY1), and the apg2Δ strain transformed with single copy (CEN; pYCG_YNL242w) or multicopy (2 µm; pAPG2(426)) plasmids encoding APG2 were grown to A600 = 1.0 in SMD. Cells were precipitated with 10% trichloroacetic acid, lysed with glass beads, and analyzed by immunoblot using antiserum against Ape1 as described under “Experimental Procedures.” The APG2 gene complements the prApe1 accumulation phenotype of the apg2Δ strain. B, Apg2 is essential for survival during nitrogen starvation. Wild type (SEY6210), apg2Δ (CWY1), and the apg2Δ strain transformed with an APG2 centromeric plasmid (pYCG_YNL242w) were grown in SMD and shifted to SD-N as described under “Experimental Procedures.” The APG2 gene complements the starvation-sensitive phenotype of the apg2Δ mutant.
To determine if the YNL242w gene complemented any of the previously isolated mutant strains that are defective in prApe1 import, we transformed the cvt and apg mutants with a plasmid encoding this gene. One of the mutants, apg2, displayed complementation of the prApe1 accumulation phenotype when transformed with this plasmid (data not shown). Both the single copy and multicopy plasmids of APG2 rescued the prApe1 processing defect in the apg2 mutant, as well as in the apg2Δ strain (Fig. 1A). The apg2 mutant was originally isolated from a morphological screen for cells that failed to accumulate autophagic bodies under starvation conditions in the presence of phenylmethylsulfonyl fluoride (28). A diploid strain constructed by mating the apg2 mutant with ynl242wΔ was defective for prApe1 import (data not shown) further suggesting that APG2 is allelic with YNL242w. The YNL242w gene was first identified as part of the systematic disruption of yeast ORFs (29). The ynl242w homozygous null strain was found to be defective in sporulation and the gene was named SPO72. Many mutants defective in vacuolar function are also defective in sporulation. Because our studies suggest a more specific role for the YNL242w gene product in transport of proteins from the cytoplasm to the vacuole, we will refer to this ORF as APG2 hereafter in this study.
Mutants that are defective only in the Cvt pathway or that are not needed for the induction or nucleation steps of autophagy show substantial maturation of prApe1 following a shift to starvation conditions. For example, prApe1 is almost completely processed to the mature form in the vac8Δ strain upon induction of autophagy (30). In contrast, mutants that are tightly blocked in autophagosome formation do not show a reversal of the prApe1 phenotype when shifted to starvation media. To determine whether the apg2Δ strain was defective in autophagosome formation, we grew apg2Δ cells in rich medium, shifted the cells to SD-N and examined the prApe1 phenotype. The defect in prApe1 processing in the apg2Δ strain did not reverse when cells were shifted from rich medium to starvation conditions (data not shown), suggesting that autophagy was blocked in this strain. Autophagy provides the essential machinery that prolongs cell viability in nitrogen deprivation conditions. As a further assessment of the autophagic capacity of the apg2Δ strain, we examined the viability of apg2Δ cells under starvation conditions. Both the wild type and apg2Δ strains transformed with a CEN APG2 plasmid showed a high level of viability even after 8 days in SD-N medium (Fig. 1B). In contrast, the apg2Δ strain exhibited a dramatic decrease in cell viability within 5 days in SD-N indicating a severe autophagy defect. From these studies, we conclude that Apg2 is a novel protein required for both the biosynthetic Cvt pathway and the degradative autophagy pathway. The defect in autophagy seen in the apg2Δ strain explains the previously observed sporulation defect of the ynl242w mutant (29).
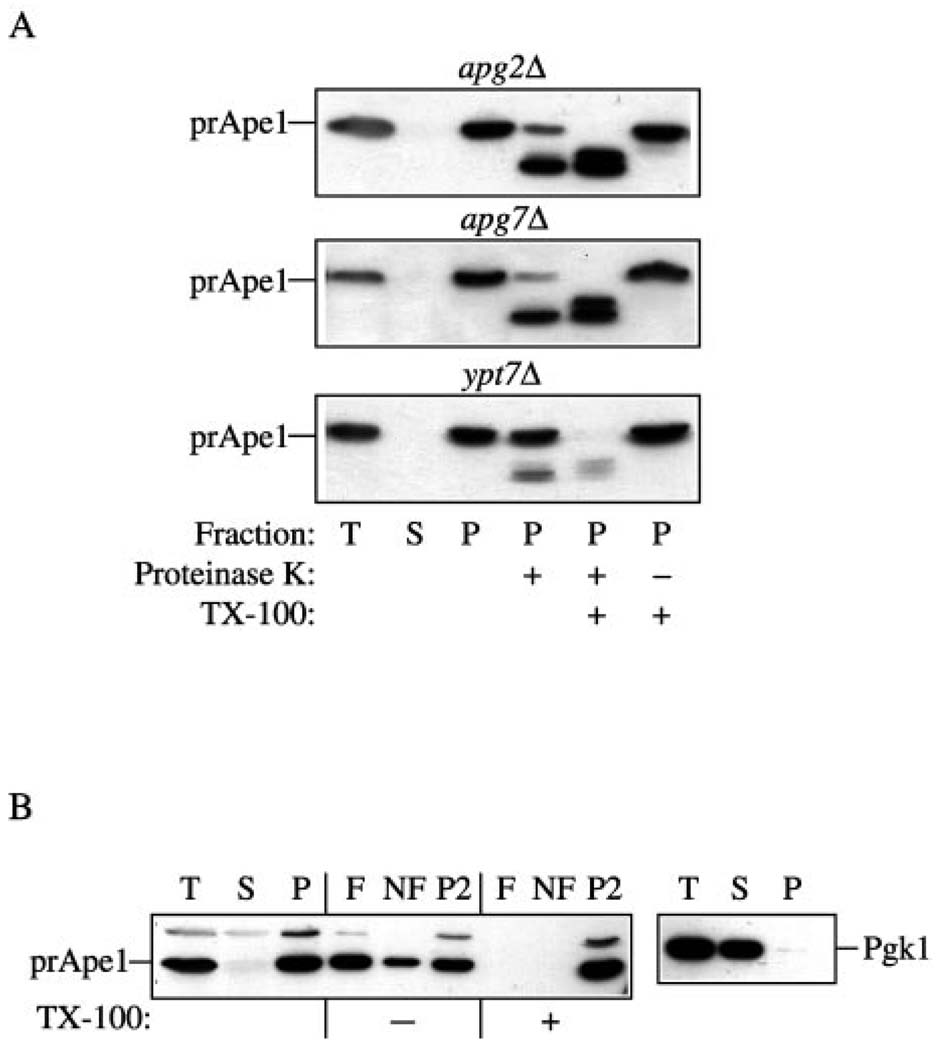
The inability to reverse the prApe1 accumulation phenotype in starvation conditions and the decrease in viability in nitrogen-depleted medium suggested that Apg2 was required for the induction and/or formation of autophagosomes. To gain further insight into the stage of action of Apg2, we examined the state of prApe1 during import through the Cvt pathway. apg2Δ cells were harvested at mid-log phase and subjected to spheroplasting followed by osmotic lysis. The crude cell lysates were separated into a low-speed supernatant fraction (S13), and a pellet fraction (P13) containing prApe1. The P13 fraction was then treated with exogenous protease in the absence or presence of detergent. As a control, we also analyzed the phenotype of prApe1 accumulated in the ypt7Δ and apg7Δ strains in the same experiment. Apg7 is an E1-like (ubiquitin activating) ATPase and functions in the initiation step of the conjugation machinery that is required for both the Cvt and autophagy pathways (26). The apg7Δ strain accumulated membrane-associated, but protease-sensitive precursor Ape1, and thus can be characterized as a protein required for the completion stage of Cvt vesicle and autophagosome formation (Fig. 2A) (26). Ypt7 is a Rab GTPase that plays a role in yeast endosomal transport. Because it is also required for the fusion step for both the Cvt and autophagy pathways, the ypt7Δ strain accumulates protease-resistant prApe1 inside the completed cytosolic vesicle (Fig. 2A) (26). Precursor Ape1 was found in a pelletable fraction in the apg2Δ strain similar to the result seen in the apg7Δ and ypt7Δ strains. As in the apg7Δ strain, prApe1 in the apg2Δ strain was mostly sensitive to exogenous protease. These results suggest that loss of Apg2 causes a defect in the sequestration of prApe1 into completed Cvt vesicles.
FIG. 2. Analysis of the apg2Δ strain indicates that Apg2 is required for the vesicle formation/completion step.
A, precursor Ape1 in the apg2Δ strain is protease-accessible. Spheroplasts isolated from apg2Δ, apg7Δ, and ypt7Δ cells were osmotically lysed (T, total) and centrifuged at 13,000 × g to obtain low-speed supernatant (S) and pellet (P) fractions. The pellet fractions were subjected to protease treatment in the absence or presence of 0.2% Triton X-100 as described under “Experimental Procedures.” B, precursor Ape1 in the apg2Δ strain accumulated in a membrane-associated float fraction. Spheroplasts from the apg2Δ strain were osmotically lysed (T, total) and separated into supernatant (S) and pellet (P) fractions by centrifugation at 13,000 × g for 10 min. The pellet fraction was resuspended in 15% Ficoll 400 in the absence or presence of 0.2% Triton X-100, and overlaid with 13 and 2% Ficoll 400 in a step gradient. The gradients were centrifuged at 13,000 × g for 10 min at room temperature. Three fractions were collected including float (F), non-float (NF) and pellet (P2) fractions as described under “Experimental Procedures.” All samples were trichloroacetic acid precipitated, acetone washed twice, resolved by SDS-polyacrylamide gel electrophoresis, and probed by immunoblot with antiserum against Ape1.
Prior to the completion of Cvt vesicles, prApe1 dodecamers accumulate to form a Cvt complex that interacts with membrane in a magnesium-dependent manner. To further dissect the prApe1 transport defect, we checked the magnesium-dependent pelleting of prApe1 in the apg2Δ strain. Yeast spheroplasts were lysed in the presence of various concentrations of MgCl2, followed by centrifugation as described under “Experimental Procedures.” Precursor Ape1 in the apg2Δ strain exhibited a magnesium-dependent association with a pelletable fraction similar to that in the apg7Δ strain. This result suggests that Apg2 is not needed for the membrane binding of the Cvt complex (data not shown).
We extended the analysis of membrane binding by examining flotation of the protease-sensitive prApe1. In this analysis, spheroplasts are osmotically lysed and the cell lysate is resolved on a Ficoll 400 step gradient. Membrane-associated proteins float to the top layer of the gradient (F), whereas soluble proteins remain in the non-float (NF) fraction, and proteins associated with a large complex are pelleted (P2). The apg2Δ cells were spheroplasted at mid-log phase and then subjected to centrifugation at 13,000 × g to collect a pellet fraction (P). This prApe1-containing pellet fraction was then subjected to the flotation analysis. In the absence of detergent, the majority of prApe1 was located in both the F and P2 fractions (Fig. 2B). However, the prApe1 present in the float fraction was shifted to the P2 fraction in the presence of detergent. These data suggest that a population of membrane-associated prApe1 accumulated in the apg2Δ cells, further indicating that Apg2 is not necessary for binding of the Cvt complex to its target membrane. Taken together, the protease protection and membrane binding experiments suggest that Apg2 plays a role in formation/completion of the sequestering vesicles.
Apg2 Is a Membrane-associated, Peripheral Protein
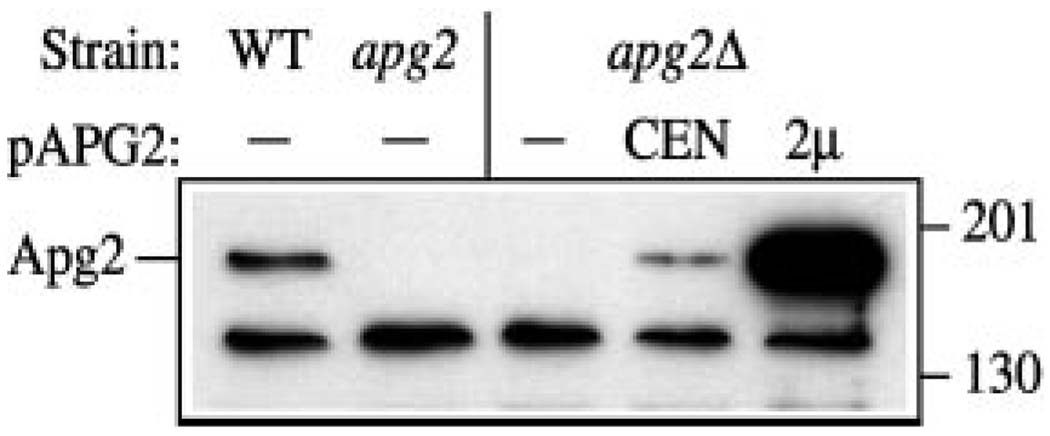
In order to study the biosynthesis of Apg2, we generated polyclonal antiserum against the protein as described under “Experimental Procedures.” Protein extracts were prepared and examined by immunoblot using the anti-Apg2 antiserum (Fig. 3). In the wild type strain, a single predominant band of ~180 kDa was detected, consistent with the expected size of Apg2 based on the deduced amino acid sequence. This band was not detected in the apg2 and apg2Δ strains. In addition, the crude cell lysate derived from apg2Δ cells expressing a multicopy APG2 plasmid showed a greatly increased level of this protein. These results suggest that the serum was specific for Apg2.
FIG. 3. Antiserum against Apg2 specifically detects an ~180-kDa protein.
Antiserum to Apg2 was prepared as described under “Experimental Procedures.” Wild type (WT; SEY6210), apg2 (MT2–4-3), apg2Δ (CWY1), and the apg2Δ strain transformed with single copy (CEN; pYCG_YNL242w) or multicopy (2µ; pAPG2(426)) plasmids encoding APG2 were grown to A600 = 1.0 in SMD. Cells lysates were resolved by SDS-polyacrylamide gel electrophoresis and analyzed with antiserum against Apg2. The positions of molecular weight markers are indicated on the right.
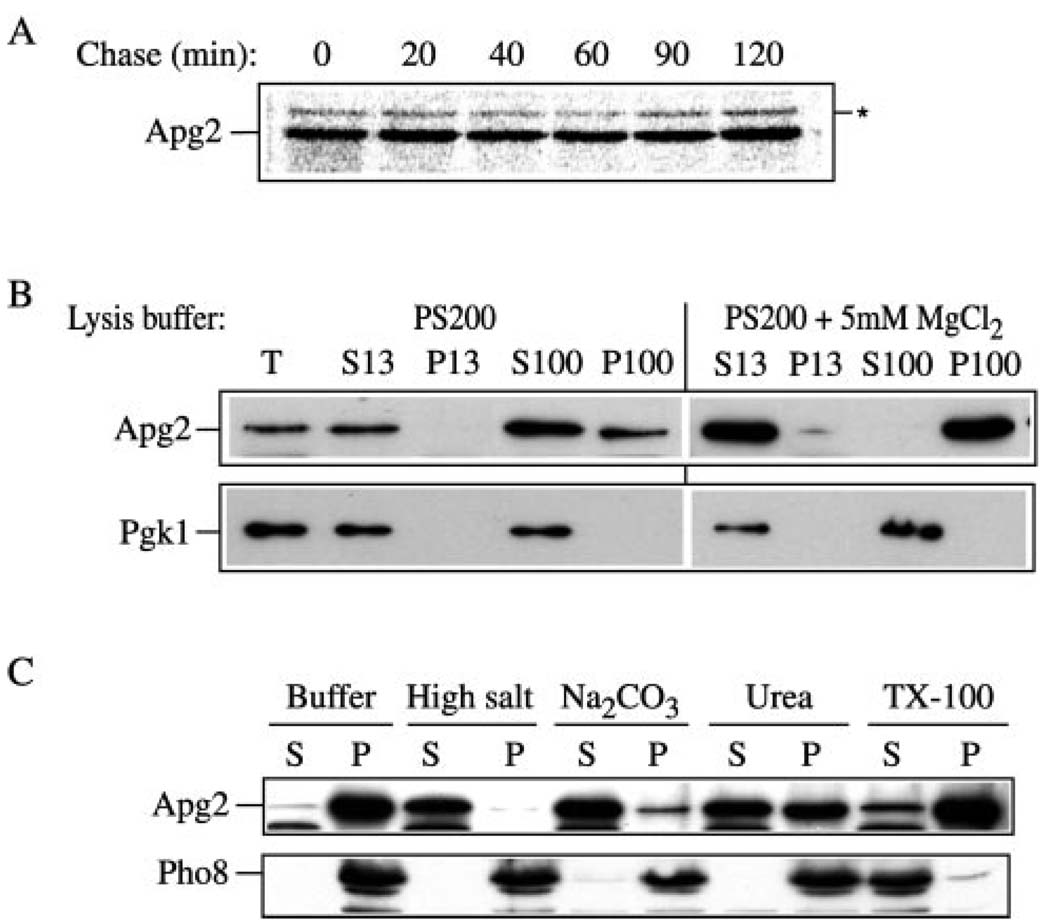
Some of the proteins required for the Apg/Cvt pathways are packed inside of the sequestering vesicles and are delivered to the vacuole where they are degraded following vesicle formation. For example, Aut7 is a vesicle component that is transported to the vacuole inside of autophagosomes/Cvt vesicles and is degraded in a PEP4-dependent manner (15). To gain some insight into the cellular itinerary of Apg2, we examined its stability through a pulse-chase analysis. Wild type cells were labeled with [35S]methionine and subjected to a nonradioactive chase for various lengths of time. At each time point, aliquots were trichloroacetic acid-precipitated and immunoprecipitated with the anti-Apg2 antiserum. Proteins were resolved by SDS-polyacrylamide gel electrophoresis. The Apg2 protein was stable over the course of a 2-h chase (Fig. 4A) suggesting that it was not delivered to the vacuole. In addition, this result suggests that unlike Aut7, Apg2 is not a component of the Cvt vesicle/autophagosome. Furthermore, the protein did not show a clear shift in mass that would suggest any type of post-translational modification such as glycosylation or proteolytic processing indicative of transit through the secretory pathway. The latter result is in agreement with the absence of a signal sequence or transmembrane domain that would allow translocation into the endoplasmic reticulum. In addition to being a vesicle component, Aut7 is induced during autophagy and shows an elevated level of synthesis under starvation conditions (15). In contrast, the steady-state protein level of Apg2 was not substantially changed upon shift to SD-N (Fig. 5C).
FIG. 4. Biosynthesis of Apg2.
A, Apg2 is a stable peripheral membrane protein. Wild type (SEY6210) cells expressing APG2 from a multicopy plasmid were labeled for 30 min, followed by a non-radioactive chase. Crude cell extracts were collected as described under “Experimental Procedures” at the indicated chase time points, and analyzed by immunoprecipitation with antiserum to Apg2. There was no detectable change in the electrophoretic mobility of Apg2 over the 2-h time course. The asterisk marks a background band that cross-reacts with the anti-Apg2 serum. B, Apg2 exists in both soluble and pelletable pools. Wild type (SEY6210) cells were spheroplasted at mid-log phase and osmotically lysed as described under “Experimental Procedures” in PS200 buffer without or with 5 mm MgCl2. The total cell lysate (T) was separated into low-speed supernatant (S13) and pellet (P13) fractions by centrifugation at 13,000 × g. The S13 fraction was further separated by high-speed centrifugation at 100,000 × g for 30 min to generate high-speed supernatant (S100) and pellet (P100) fractions. Fractions were trichloroacetic acid precipitated and examined by immunoblot with antiserum against Apg2 and the cytosolic marker Pgk1. C, biochemical characterization of pelletable Apg2. Spheroplasts from wild type (SEY6210) cells were osmotically lysed as described under “Experimental Procedures.” A total membrane pellet was obtained by highspeed centrifugation (100,000 × g) of the crude cell lysate and resuspended in buffer alone or buffer containing 1 m KCl, 0.1 m Na2CO3 (pH 10.5), 3 m urea, or 1% Triton X-100 and further separated into supernatant (S) and pellet (P) fractions as described under “Experimental Procedures.” Samples were resolved by SDS-polyacrylamide gel electrophoresis and detected by immunoblot with antibodies or antiserum to Pho8 (integral vacuolar membrane protein) and Apg2.
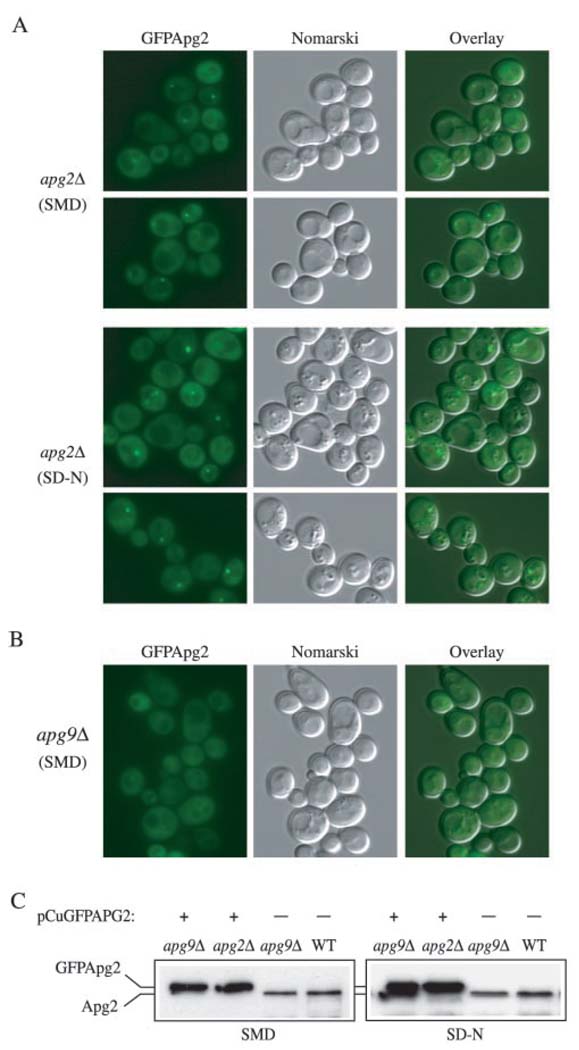
FIG. 5. GFPApg2 localization pattern.
A and B, the apg2Δ (CWY1) and apg9Δ (JKY007) strains were transformed with a plasmid expressing GFPApg2 and grown in nutrient-rich conditions (SMD) or shifted to nitrogen-starvation conditions (SD-N) for 4 h and examined with a Nikon E-800 fluorescence microscope. A, GFPApg2 displays both a cytosolic and punctate, perivacuolar distribution in the apg2Δ strain. B, GFPApg2 displays a largely diffuse, cytosolic pattern in the apg9Δ strain in both SMD (shown) and SD-N (data not shown) conditions. Fluorescent (GFP) panels are shown on the left for each strain, Nomarski panels are shown in the middle, and an overlay of both panels is shown on the right. C, Apg2 and GFPApg2 are not induced under starvation conditions and are stable in the absence of Apg9. The apg2Δ (CWY1) and apg9Δ (JKY007) strains transformed with a plasmid expressing GFPApg2, and the non-transformed apg9Δ and wild type (SEY6210) strains were grown in SMD or shifted to SD-N as described in part A. Protein extracts were prepared and resolved by SDS-polyacrylamide gel electrophoresis. GFPApg2 and Apg2 were detected by Western blot using antiserum against Apg2.
To investigate the site of action for Apg2, we examined the protein by subcellular fractionation. The wild type strain SEY6210 was grown to mid-log phase, and subjected to spheroplasting followed by osmotic lysis in PS200 buffer in the absence or presence of 5 mm MgCl2. The crude cell lysates were then centrifuged at 13,000 × g to generate low-speed supernatant (S13) and pellet (P13) fractions. The S13 fractions were then separated into high-speed supernatant (S100) and pellet (P100) fractions by 100,000 × g centrifugation. The distribution of the abundant cytosolic protein, phosphoglycerate kinase (Pgk1), served as a marker to confirm faithful separation of membrane and soluble proteins. As expected, Pgk1 was recovered in the supernatant fractions in the absence or presence of MgCl2 (Fig. 4B). When we examined the pelletability of Apg2 by lysing spheroplasts in buffers containing 0 or 5 mm MgCl2, we found that the association of Apg2 with the pellet fraction was destabilized in the absence of MgCl2. About 40% of the total Apg2 was still detected in the P100 fraction, however, even when cells were lysed in buffers lacking MgCl2 (Fig. 4B). These data suggest that Apg2 localizes to both a soluble and pelletable fraction in a salt-dependent manner.
The predicted amino acid sequence of Apg2 has no apparent transmembrane domains or site of lipid modification. Because many of the Apg and Cvt proteins are associated with a membrane fraction, we decided to examine the pelletable nature of Apg2 in greater detail. To gain further insight into the nature of its interaction with the membrane, we biochemically examined the stability of membrane binding. Spheroplasts from wild type cells were lysed and centrifuged at 100,000 × g as described under “Experimental Procedures” to collect the total membrane pellet fraction. As a control, we analyzed the vacuolar membrane marker alkaline phosphatase (Pho8). As expected, detergent efficiently solubilized Pho8 whereas salt, pH, and urea treatments retained the protein in the pellet fractions (Fig. 4C). In contrast, the pelletable fraction of Apg2 was stripped from the membrane by 1 m KCl and 0.1 m Na2CO3 (pH 10.5), and was partially removed with 3 m urea (Fig. 4C). However, the majority of Apg2 remained in the pellet fraction fol1owing treatment with 1% Triton X-100, suggesting that Apg2 may interact with a detergent-resistant protein complex. Taken together, the data indicate that Apg2 behaves like a peripheral membrane protein.
Apg2 Localizes to the Apg9 Compartment
To investigate the subcellular localization of Apg2 in vivo, we constructed an NH2-terminal fusion of GFP to Apg2 under control of the regulable CUP1 promoter. GFPApg2 expressed from a centromeric plasmid without copper induction showed partial complementation of the prApe1 processing defect of the apg2Δ strain in SMD medium, but almost complete complementation when cells were shifted to SD-N (data not shown). When expressed in apg2Δ cells, GFPApg2 displayed both a cytosolic and punctuate staining pattern (Fig. 5A). The cytosolic staining pattern of the GFPApg2 fusion protein became more pronounced when GFPApg2 was expressed from a multicopy plasmid or when synthesis from the CUP1 promoter was induced with copper (data not shown). These data suggest that overexpressed GFPApg2 saturated its membrane binding resulting in an increased cytosolic pool of the protein. The punctate pattern of GFPApg2 was localized in proximity to the vacuole (Fig. 5A). Furthermore, the punctate staining was enhanced when cells were shifted to SD-N medium. This increase in signal intensity under starvation conditions is reminiscent of that seen for the Apg7GFP fusion protein (26). To determine whether the increased intensity of the GFPApg2 dot seen in SD-N medium was due to induction of the protein, we examined the level of the protein in extracts prepared from cells grown in either SMD or SD-N medium. The Apg2 and GFPApg2 steady state levels remained relatively constant under both rich and starvation conditions although there was a slight increase seen in SD-N (Fig. 5C). Because Apg2 and GFPApg2 are not induced substantially under starvation conditions, the increased intensity of the punctate dot probably reflects a higher level of recruitment of Apg2 to this perivacuolar structure when autophagy is induced.
The punctate staining pattern displayed by GPF Apg2 is also similar to that seen for other Apg/Cvt proteins including Apg9 (23). Apg9 is the only characterized integral membrane protein that is required for formation/completion of Cvt vesicles and autophagosomes. Because Apg9 is an integral membrane protein we decided to examine whether it was needed for correct localization of Apg2. Accordingly, we examined GFPApg2 in the apg9Δ strain. We found that GFPApg2 displayed a primarily cytosolic localization in the apg9Δ strain in nutrient-rich conditions, suggesting that efficient Apg2 localization is dependent on the Apg9 protein (Fig. 5B). Furthermore, shifting cells expressing GFPApg2 to SD-N medium did not facilitate its perivacuolar, punctate localization in the apg9Δ background (data not shown). In contrast, localization of Apg9GFP did not change in the apg2Δ strain (data not shown) suggesting that Apg9 localization is independent of Apg2. To verify that the loss of punctate signal in the apg9Δ strain was not due to degradation of the GFPApg2 protein, we examined the stability of Apg2 and GFPApg2 in wild type and apg9Δ cells under both rich medium and starvation conditions. Both proteins were stable in the apg9Δ strain under either condition (Fig. 5C). These data suggest that the predominantly cytosolic staining of GFPApg2 in the apg9Δ strain reflects detachment of GFPApg2 from the punctate dot structure and transfer to the cytosol, rather than degradation of the protein.
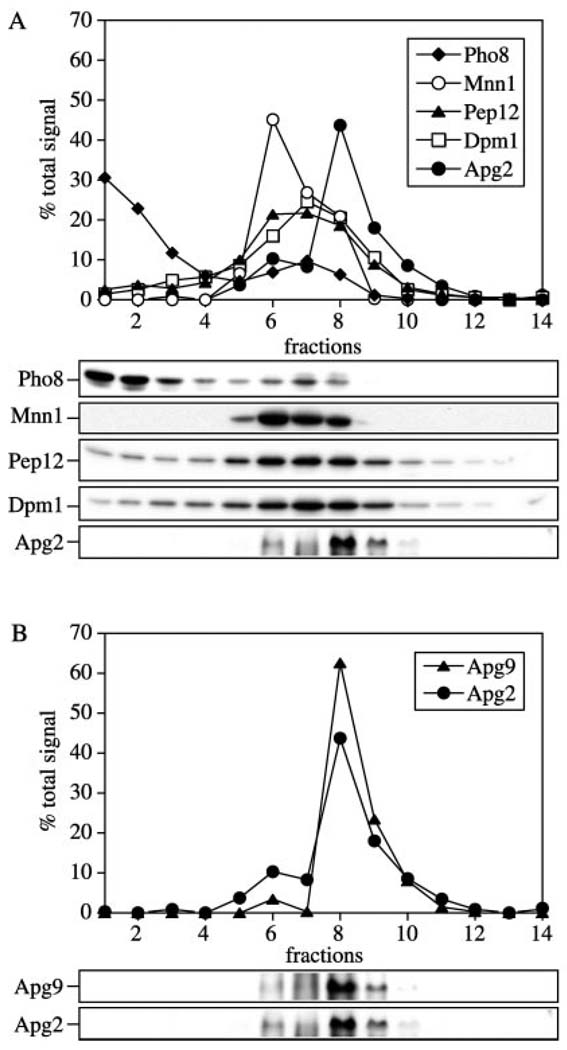
The GFP analysis suggested that Apg2 was localized to the same membrane compartment as Apg9. To extend our analysis of Apg2 localization, we resolved the Apg2 compartment by a density gradient analysis. Spheroplasts from wild type cells transformed with a 2µ APG9 plasmid were osmotically lysed and separated into supernatant and total membrane fractions by centrifugation at 100,000 × g. The membrane pellet was then resuspended and resolved on a linear 0–66% OptiPrep gradient. After centrifugation at 100,000 × g for 16 h, 14 fractions were collected from the top to the bottom and analyzed by immunoblot. The distribution of Apg2 was detected at fractions 6 through 9 with the peak concentration of Apg2 detected in fraction 8, while the vacuole membrane marker Pho8 localized at fractions 1 and 2 (Fig. 6A). We also examined the endomembrane markers Dpm1 (endoplasmic reticulum), Mnn1 (Golgi), and Pep12 (endosome). These proteins displayed fractionation patterns that were distinct from Apg2 (Fig. 6A). The unique density gradient profile of Apg2 was reminiscent of the fractionation pattern observed for Apg9. Apg9 was also characterized as a protein that localized to a membrane compartment that did not co-fractionate with known endomembrane markers (23). We compared the distribution of Apg2 and Apg9 on the OptiPrep gradients and found that they co-localized to the same peak fractions, suggesting residence in the same membrane compartment (Fig. 6B).
FIG. 6. Apg2 co-localized with the Apg9 compartment.
The wild type (SEY6210) strain transformed with the APG9 2µ plasmid was grown in SMD to A600 = 1.0, converted to spheroplasts, and osmotically lysed. Crude cell lysates were pre-cleared, and centrifuged to obtain a total membrane pellet by centrifugation at 100,000 × g for 20 min. Pellets were resuspended in lysis buffer and loaded on an OptiPrep density gradient ranging from 0 to 66%, and centrifuged for 16 h at 100,000 × g as described under “Experimental Procedures.” A total of 14 fractions were collected from the top of the gradient and examined by immunoblot with antibodies or antiserum against A, Pho8 (vacuole), Dpm1 (endoplasmic reticulum), Mnn1 (Golgi), Pep12 (endosome), and Apg2; and B, Apg2 and Apg9.
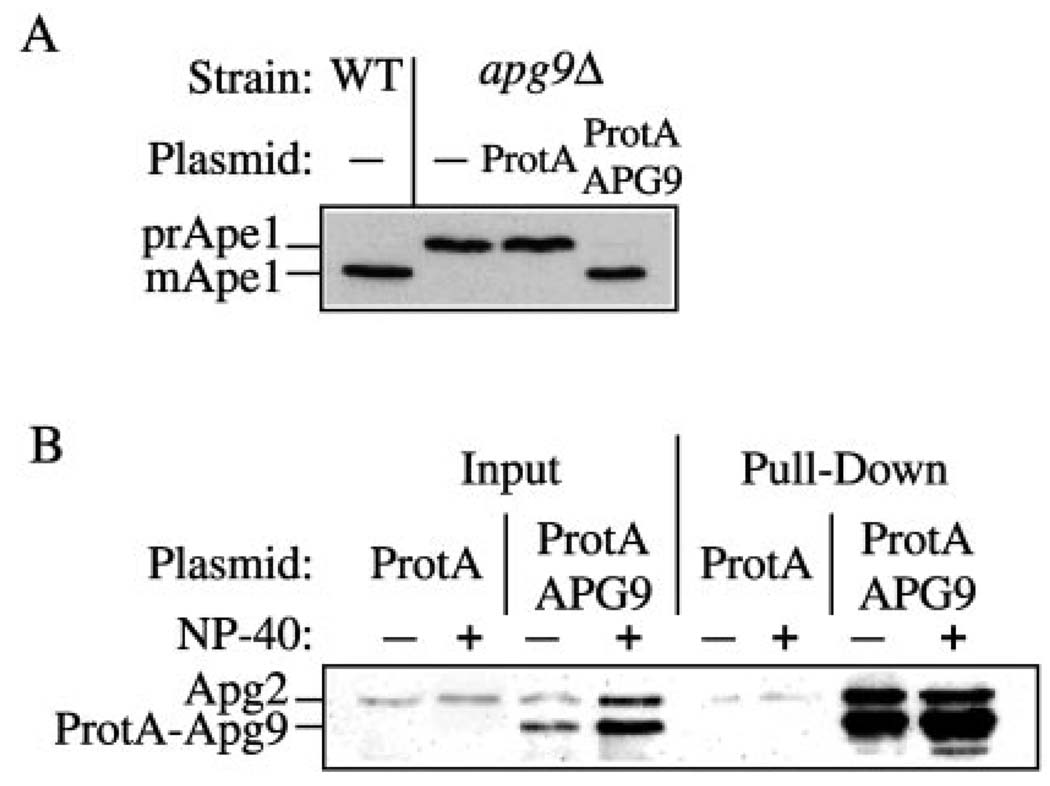
The Apg9-dependent localization of Apg2 suggested that Apg2 and Apg9 might interact. To determine whether Apg2 associates with Apg9, we carried out a protein pull-down assay. Protein A was fused to the NH2 terminus of Apg9. The protein A-Apg9 fusion protein rescued the prApe1 transport defect of an apg9Δ strain, indicating that it was functional (Fig. 7A). For the pull-down assay, spheroplasts expressing protein A-Apg9 or protein A alone were lysed, and the protein A constructs was recovered by incubating with Dynabeads coupled to IgG in the absence or presence of detergent. Interacting proteins were detected by Western blot. The Apg2 protein was recovered along with protein A-Apg9 whether or not detergent was present, although the recovery was ~5-fold higher in the absence of detergent (Fig. 7B). Apg2 was not recovered from cells expressing the protein A vector alone, indicating that the interaction was specific to Apg9 and not the protein A domain. These results further suggest that Apg2 is localized to the Apg9 compartment and that the two proteins interact directly or indirectly.
FIG. 7. The Apg2 protein interacts with Apg9.
A, the apg9Δ (JKY007) strain was transformed with the ProtA or ProtA-APG9 plasmids as indicated. Wild type (SEY6210) or apg9Δ cells were grown in SMD, and protein extracts were prepared and analyzed by immunoblot with antiserum against Ape1. The ProtA-Apg9 fusion protein complements the prApe1 accumulation defect. B, cell lysates were prepared from wild type (SEY6210) cells overexpressing Apg2 and protein A (ProtA) or protein A-Apg9 (ProtA-APG9) in the absence or presence of Nonidet P-40 detergent as described under “Experimental Procedures.” Lysates were incubated with Dynabeads M-500 cross-linked to human IgG. Protein A-Apg9 and associated proteins were isolated by collecting the Dynabeads from the crude cell lysates by magnetic separation (Pull-Down). Proteins were resolved by SDS-polyacrylamide gel electrophoresis and detected by immunoblot with antiserum against Apg2. Protein A-Apg9 was detected in the blot due to the affinity between the protein A moiety and rabbit IgG. The positions of Apg2 and protein A-Apg9 are indicated. Input lanes corresponded to 2.5% of cell lysates used for each pull-down reaction. Apg2 is pulled down by the ProtA-Apg9 fusion but not by protein A alone.
Apg2 Is Required for the Peroxisome Degradation Pathway
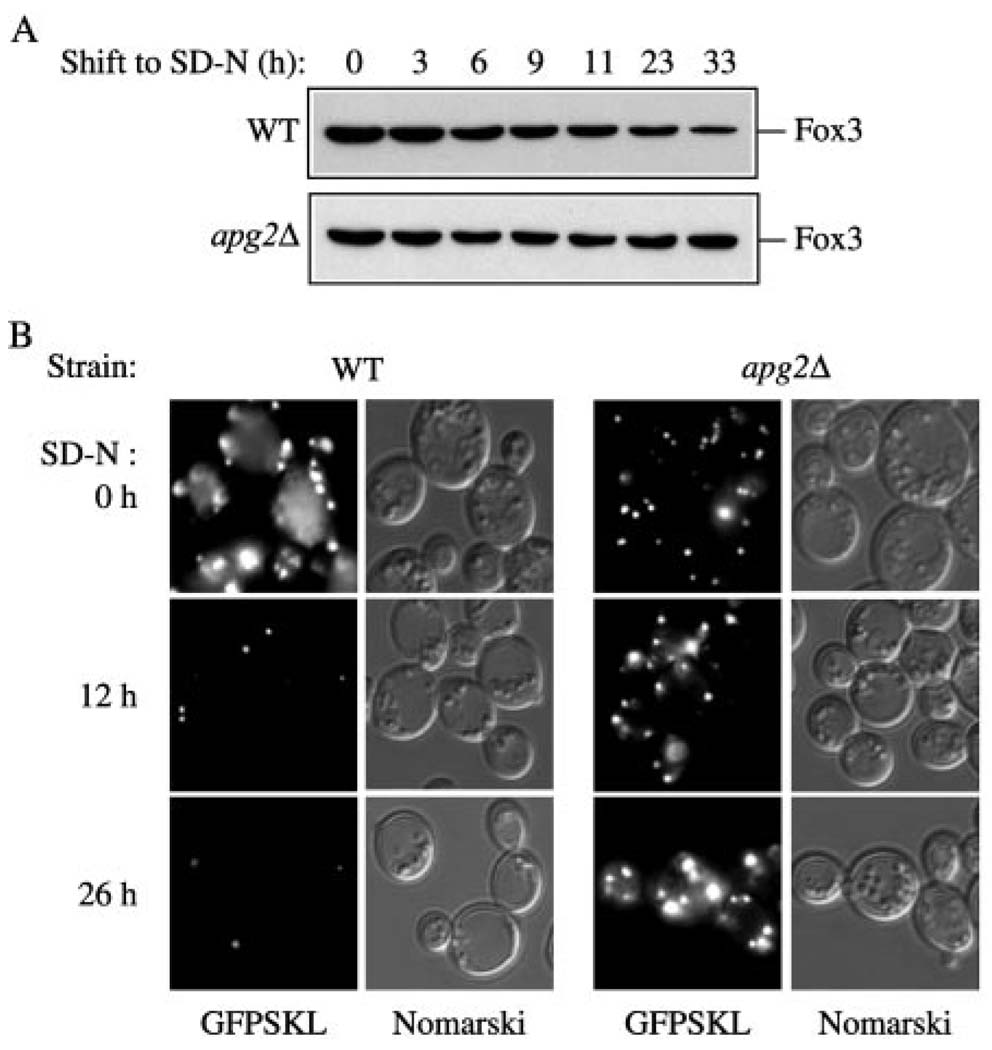
The selective degradation of excess peroxisomes is a critical feature of cellular adaptation when the yeast Saccharomyces cerevisiae are shifted from oleic acid to glucose medium. Previously, we have demonstrated an overlap among some of the components required for the Cvt, autophagy, and pexophagy pathways in S. cerevisiae (11). To investigate if Apg2 is a component required for pexophagy, we monitored the fate of Fox3. Fox3 is the thiolase enzyme that localizes to the peroxisome matrix. Fox3 is induced when yeast are grown in medium (YTO) with oleic acid as the sole carbon source. The degradation of Fox3 when cells are shifted to glucose serves as a convenient means of monitoring peroxisome levels. In particular, the prolonged accumulation of this protein following a shift from oleic acid to glucose represents a defect in peroxisome degradation. Both wild type and apg2Δ cells were incubated in YTO medium to induce peroxisomes as described under “Experimental Procedures,” and then shifted to SD-N medium. Crude cell lysates from both strains were collected at the indicated time points and examined by immunoblot. In wild type cells, Fox3 levels decreased as expected (Fig. 8A). In contrast, the apg2Δ strain accumulated Fox3 indicating it was defective in peroxisome degradation.
FIG. 8. The apg2Δ mutant is unable to degrade peroxisomes.
A, The apg2Δ mutant is defective in the ability to degrade Fox3. Wild type (WCGa) and apg2Δ (CWY2) strains were grown in YPD to mid-log phase, shifted to YTO medium for peroxisome induction, and then shifted to degradation medium (SD-N) as described under “Experimental Procedures.” Samples were taken at the indicated times after shift to SD-N, trichloroacetic acid precipitated, resolved by SDS-polyacrylamide gel electrophoresis, and probed by immunoblot with Fox3 antiserum. B, a GFP-SKL peroxisome-targeted fusion protein is maintained in the apg2Δ mutant following a shift to pexophagy conditions. Wild type (WCGa) and apg2Δ (CWY2) strains harboring the pCuGFPSKL(416) plasmid were grown in SMD-Ura to an A600 = 1.0 and then shifted to peroxisome-inducing conditions as described in “A.” Following a shift to SD-N, samples were removed at the indicated times and examined by fluorescence microscopy as described under “Experimental Procedures.” Fluorescent (GFP) panels are shown on the left for each strain and Nomarski panels are shown on the right.
We also monitored peroxisome levels in vivo by following a fluorescent peroxisome reporter protein. Addition of the amino acids serine-lysine-leucine (SKL) to the COOH terminus of GFP causes the fusion protein to be targeted to the peroxisome lumen. The plasmid expressing GFPSKL was transformed into wild type and apg2Δ strains. Following induction of peroxisomes in YTO medium, cells expressing GFPSKL were shifted to starvation conditions. The cultures were examined at various time points and the level of peroxisomes determined by fluorescence microscopy as described under “Experimental Procedures.” In wild type cells, the GFPSKL signal was dramatically reduced by 12 h in SD-N and almost completely eliminated by 26 h (Fig. 8B). In contrast, in apg2Δ cells there was essentially no change in peroxisome levels again indicating a defect in peroxisome degradation (Fig. 8B). Together, these data suggest that Apg2 in S. cerevisiae plays a similar role in peroxisome degradation to the Gsa11 protein4 of P. pastoris.
DISCUSSION
Apg2 Is Required for the Formation/Completion of Cvt Vesicles and Autophagosomes
Autophagy is the specialized degradation process that cells use to transport bulk cytoplasm to the compartmentalized hydrolase-containing organelle, the vacuole/lysosome. From studies over the past few years, the topology of autophagy has been described in detail and a few of the molecular mechanisms have been uncovered (1, 2). The membrane trafficking machinery used in autophagy is similar to that of the Cvt pathway that is used for the biosynthetic delivery of prApe1 to the vacuole in S. cerevisiae (1–4). Both processes require the formation of a sequestering vesicle in the cytosol. At present, the origin of the sequestering membrane is still a mystery. The identified mechanism required for the completion of the sequestering vesicle includes a novel ubiquitin-like conjugation machinery composed of Apg5, -7, -10, -12, and -16, and a regulatory multiprotein complex(es) that includes Apg1, -13, and -17 as well as Cvt9 and Vac8 (Refs. 13 and 14; reviewed in Ref. 2). However, our knowledge about the exact sequence of events required for the completion of the Cvt vesicle/autophagosome is still limited. Therefore, the identification of additional molecular components involved in these pathways will further our understanding about the molecular details of the vesicle formation process. In this study, we characterized Apg2 as a new protein component in the Cvt and autophagy pathways. Taking advantage of the established model for the Cvt pathway, we assessed the role of Apg2 through biochemical analyses. Examining the apg2Δ strain, we found that prApe1 accumulated in a protease-accessible form, suggesting that the sequestration step for the Cvt complex was not completed (Fig. 2A). Furthermore, a membrane-associated form of prApe1 was detected in the float fraction through a Ficoll gradient (Fig. 2B). Taken together, these findings implicate a direct role for Apg2 in the completion of the Cvt vesicle/autophagosome.
To survive starvation conditions, the vacuole/lysosome mediates the turnover and recycling of non-essential intracellular material for re-use in critical biosynthetic reactions through the autophagy pathway. Accordingly, the level of vacuolar proteolysis increases dramatically under these conditions and accounts for more than 85% of the degradative capacity of the cell (31). The degree of viability of apg mutants during starvation is not equal, suggesting that there may be differing requirements for various proteins in the progression of autophagy (32). For example, the apg1Δ strain shows a greater loss of viability than the aut7Δ strain during starvation. Thus it has been suggested that proteins involved at the nucleation/signaling step show a complete loss of viability within a few days in starvation conditions, because they could not bypass the induction of autophagy. In contrast, mutants that have defects in genes whose products are required for the autophagosome expansion step, such as aut7Δ, retain partial viability (32). The apg2Δ strain exhibited a complete loss of viability quickly after a shift to starvation conditions (Fig. 1B). This phenotype indicated an absolutely essential role for Apg2 in autophagy.
Apg2 Is a Peripheral Membrane Protein That Localizes to the Apg9 Compartment
To investigate the function of Apg2 in the Cvt and autophagy pathways, we raised specific antiserum against this protein. In this study, we have demonstrated that Apg2 accumulated in both a soluble and pelletable fraction (Fig. 4B). Further examination of the pelletable pool of Apg2, suggested that it behaved as a peripheral membrane protein (Fig. 4C). We constructed a GFPApg2 fusion protein to carry out an in vivo analysis of Apg2. Interestingly, GFPAg2 showed better complementation of the prApe1 accumulation phenotype of the apg2Δ strain under starvation conditions than in nutrient-rich conditions even though the apg2Δ mutant alone did not show any change in complementation in SMD versus SD-N (data not shown). This result suggests that the induction of other components during starvation may compensate for the partial function of GFPApg2 seen in nutrient-rich growth conditions. Microscopy analysis of GFPApg2 indicated that the fusion protein had both a cytosolic and perivacuolar, punctuate distribution (Fig. 5). The solubility of Apg2 and GFPApg2 during subcellular fractionation was salt-dependent. In physiological salt buffers containing MgCl2, the majority of Apg2 was found in a pelletable fraction. We think that the dispersed in vivo staining pattern corresponding to cytosolic GFPApg2 may in part present membrane-associated protein that is not localized to the perivacuolar dot. This is in agreement with observations in the accompanying paper from Shintani et al. (38) based on analysis of the mutant protein GFPApg2pG83E. This protein is still partially pelletable following subcellular fractionation even though it shows a dispersed distribution in vivo. The punctate staining pattern seen with GFPApg2 was more intense under starvation conditions (Fig. 5), but this was not accompanied by a substantial increase in the level of the Apg2 protein. Instead, we think this reflects a higher level of recruitment of GFPApg2 to the perivacuolar dot structure during autophagy.
Because the GFPApg2 visual pattern was similar to other proteins including the integral membrane protein Apg9, we decided to extend our analysis by examining the fluorescent staining pattern in an apg9Δ strain. We found that GFPApg2 displayed a completely cytosolic distribution in the apg9Δ mutant (Fig. 5). The loss of the punctate dot was not due to degradation of GFPApg2, suggesting that the protein relocated from, or could not be recruited to, the perivacuolar structure. These data suggest that in wild type cells, Apg2 is localized to a compartment that contains Apg9. To examine this more carefully, we looked at the localization of native Apg2 using a density gradient approach. By examining the comparative localization of Apg2 with several endomembrane markers, including the vacuole, Golgi, endoplasmic reticulum, and endosome, we conclude that Apg2 localized at a compartment largely distinct from these organelles. Our gradient data suggest that Apg2 is present at the membrane compartment that contains the essential autophagy component Apg9 (Fig. 6). Furthermore, a protein affinity analysis with protein A-tagged Apg9 pulled down Apg2, suggesting a direct or indirect physical interaction between the two proteins (Fig. 7B). Our results concerning the localization of Apg2 are in agreement with those of Shintani et al. (38) who carried out an independent analysis of this protein. They found that the punctate distribution of Apg2 was blocked in the apg9Δ strain as well as in strains having mutations in APG1, APG6, or APG14.
As mentioned previously, the membrane sequestration process requires conjugation machinery that covalently attaches Apg12 to Apg5 (17, 20). The Apg12-Apg5 conjugates were able to form in the apg2Δ strain. The conjugation reaction as well as post-translational processing and lipidation are required for the membrane binding of Aut7 (21, 22). The apg2Δ strain was not defective in the membrane association of Aut7 (data not shown). Therefore, we suggest that Apg2 does not have a role in the conjugation of Apg12 to Apg5 or in other reactions that are required for membrane recruitment of Aut7. The question of whether Apg2 belongs to part of the regulatory complex that comprises Apg1, Apg13, Cvt9, Vac8, and Apg17 remains to be elucidated.
The apg2Δ Strain Is Defective in Peroxisome Degradation
The monitoring of Fox3 degradation provides a useful tool to identify molecular components that are required for peroxisome degradation. In this study, we explored the role of Apg2 in pexophagy by following the stability of Fox3 following a shift from peroxisome-inducing to peroxisome-degrading conditions. We found that the apg2Δ strain was defective in peroxisome degradation as measured by the inability to degrade Fox3 (Fig. 8A). Similarly, we monitored peroxisome levels in vivo with a peroxisome-targeted GFPSKL fusion protein. As with the Fox3 analysis, we found that loss of the GFPSKL signal was blocked in the apg2Δ strain (Fig. 8B). Together, these results suggest that Apg2 is the functional homolog of Gsa11 in P. pastoris.4 Although it has been shown previously that some components shared by the Cvt pathway and autophagy are also involved in pexophagy (11), little is known about how the sequestration of peroxisomes is achieved in S. cerevisiae. In the macropexophagy model as seen during ethanol adaptation in P. pastoris (1), peroxisomes are sequestered within cytosolic vesicles, which is a similar process to that of the Cvt and autophagy pathways. Alternatively, the engulfment of peroxisomes might occur at the vacuole surface by micropexophagy as is seen in P. pastoris during glucose adaptation (33). In either case, in S. cerevisiae we speculate that Apg2 has a general role in the completion stage for these pathways, however, it will be interesting to determine which mechanism is utilized in this yeast. APG2 also has homologs in humans (FLJ10242, 24% identity) and S. pombe (Spbc31e1.01cp, 22% identity), although in both cases nothing is known about the protein’s function. There are now several examples of homologs to the yeast APG genes that are required for autophagy in animal cells (34). Hence, we can predict that in future studies Apg2 will also be shown to be involved in this process in higher eukaryotes.
Characterization of proteins involved in the Cvt and autophagy pathways has been under way for several years. We know that the majority of Cvt/Apg gene products are involved in the sequestration step, yet the exact mechanism that results in the formation of double-membrane vesicles is still unknown. In this study, we identify Apg2 as a novel protein in the sequestration step for the Cvt, autophagy, and pexophagy pathways. The donor membrane source for the Cvt vesicles and autophagosomes remains unclear. One candidate protein for a marker of the source membrane is the transmembrane protein Apg9 that is required for both the Cvt pathway and autophagy. The present study is the first report of a protein that co-localizes at the putative donor membrane source for the Cvt vesicle/autophagosome that is marked by Apg9. Future studies will be aimed at an analysis of the Apg9 compartment as well as the mechanism by which Apg2 directly participates in the vesicle sequestration stage for the Cvt and autophagy pathways.
Acknowledgments
We thank Drs. Scott Emr, Todd Graham, and Jeremy Thorner for supplying antiserum, Dr. Sidney Scott for critical reading of the manuscript, and members of the Klionsky lab for helpful discussions and advice.
Footnotes
This work was supported by National Institutes of Health Public Health Service Grant GM53396 (to D. J. K.), the Lewis E. and Elaine Prince Wehmeyer Trust (to C.-W. W.), a grant from The Norwegian Cancer Society (to P. E. S.), and National Science Foundation Grant MCB-9817002 (to W. A. D.).
The abbreviations used are: Ape1, aminopeptidase I; Apg, autophagy; Cvt, cytoplasm to vacuole targeting; GFP, green fluorescent protein; Mnn1, α1,3-mannosyltransferase; ORF, open reading frame; Pgk1, phosphoglycerate kinase; Pho8, alkaline phosphatase; SMD, synthetic minimal medium with glucose and nitrogen; SD-N, synthetic minimal medium lacking nitrogen; YNB, yeast nitrogen base; PCR, polymerase chain reaction; PIPES, 1,4-piperazinediethanesulfonic acid.
J. Guan, P. E. Stromhaug, M. D. George, P. Habibzadegah-Tari, A. Bevan, W. A. Dunn, Jr., and D. J. Klionsky, submitted for publication.
J. Kim, W.-P. Huang, P. E. Stromhaug, and D. J. Klionsky, manuscript in preparation.
P. E. Stromhaug and W. A. Dunn, Jr., manuscript in preparation.
REFERENCES
- 1.Klionsky DJ, Ohsumi Y. Annu. Rev. Cell Dev. Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Klionsky DJ. Annu. Rev. Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- 3.Klionsky DJ. J. Biol. Chem. 1998;273:10807–10810. doi: 10.1074/jbc.273.18.10807. [DOI] [PubMed] [Google Scholar]
- 4.Klionsky DJ. J. Membr. Biol. 1997;157:105–115. doi: 10.1007/s002329900220. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Scott SV, Oda M, Klionsky DJ. J. Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott SV, Baba M, Ohsumi Y, Klionsky DJ. J. Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harding TM, Morano KA, Scott SV, Klionsky DJ. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. J. Biol. Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 11.Hutchins MU, Veenhuis M, Klionsky DJ. J. Cell Sci. 1999;112:4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- 12.Noda T, Ohsumi Y. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 13.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr, Klionsky DJ. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang WP, Scott SV, Kim J, Klionsky DJ. J. Biol. Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 16.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 18.Mizushima N, Noda T, Ohsumi Y. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ. Mol. Biol. Cell. 2000;11:969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Huang W-P, Klionsky DJ. J. Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. J. Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow E, Lane D. In: Using Antibodies: A Laboratory Manual. Marlow E, Lane D, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1999. pp. 61–99. [Google Scholar]
- 25.Klionsky DJ, Cueva R, Yaver DS. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Mol. Biol. Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan W, Stromhaug PE, Dunn WA., Jr Mol. Biol. Cell. 1999;10:1353–1366. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukada M, Ohsumi Y. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 29.Saiz JE, de Los Angeles Santos M, Vázquez de Aldana CR, Revuelta JL. Yeast. 1999;15:155–164. doi: 10.1002/(SICI)1097-0061(19990130)15:2<155::AID-YEA342>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 31.Teichert U, Mechler B, Muller H, Wolf DH. J. Biol. Chem. 1989;264:16037–16045. [PubMed] [Google Scholar]
- 32.Abeliovich H, Dunn WA, Kim J, Klionsky DJ. J. Cell Biol. 2000;151:1025–1033. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuttle DL, Dunn WA., Jr J. Cell Sci. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- 34.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. J. Biol. Chem. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 35.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinemeyer W, Gruhler A, Möhrle V, Mahé Y, Wolf DH. J. Biol. Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- 37.Wurmser AE, Emr SD. EMBO J. 1998;17:4930–4942. doi: 10.1093/emboj/17.17.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shintani T, Suzuki K, Kamada Y, Noda T, Ohsumi Y. J. Biol. Chem. 2001;276:30452–30460. doi: 10.1074/jbc.M102346200. [DOI] [PubMed] [Google Scholar]