SUMMARY
TRIM25 mediates Lys 63-linked ubiquitination of the N-terminal CARDs of the viral RNA sensor RIG-I, leading to type I interferon (IFN) production. Here, we report that the influenza A virus non-structural protein 1 (NS1) specifically inhibits TRIM25-mediated RIG-I CARD ubiquitination, thereby suppressing RIG-I signal transduction. A novel domain in NS1 comprising E96/E97 residues mediates its interaction with the coiled-coil domain of TRIM25, thus blocking TRIM25 multimerization and RIG-I CARD ubiquitination. Furthermore, a recombinant influenza A virus expressing an E96A/E97A NS1 mutant is defective in blocking TRIM25-mediated anti-viral IFN response and loses virulence in mice. Our findings reveal a novel mechanism of influenza virus to inhibit host IFN response and also emphasize the vital role of TRIM25 in modulating viral infections.
INTRODUCTION
An integral component of innate immunity is the production of type I interferons (IFNs), a family of anti-viral cytokines that functions to prevent completion of the virus lifecycle as well as virus dissemination in vivo (Garcia-Sastre et al., 1998a). To elicit IFN responses, mammalian hosts have evolved a variety of cellular pattern recognition receptors (PRRs) including Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs), which sense the presence of viral nucleic acids or other conserved molecular components of invading microbes (Akira et al., 2006; Meylan et al., 2006; Yoneyama and Fujita, 2007). With respect to the respiratory pathogens influenza and respiratory syncytial viruses, analysis of infected RIG-I−/− or MDA5−/− cells illustrated the essential role of RIG-I in initiating anti-viral responses against these RNA viruses (Loo et al., 2008). Upon viral infection, the cytosolic receptor RIG-I recognizes viral RNA in a 5’-triphosphate-dependent manner and initiates an anti-viral signaling cascade by interacting with the downstream partner MAVS/VISA/IPS-1/Cardif (Cui S, et al. (2008); (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). For MAVS binding and downstream signaling, the N-terminal Caspase recruitment domains (CARDs) of RIG-I are responsible.
In addition, tripartite motif (TRIM) proteins, containing a N-terminal RING domain with potential ubiquitin E3 ligase activity, represent a new class of anti-viral molecules involved in innate immunity (Nisole et al., 2005; Ozato et al., 2008). TRIM5α has been extensively studied as a host restriction factor for HIV-1 infection (Stremlau et al., 2004). TRIM25 has recently been shown to induce Lys 63-linked ubiquitination of the N-terminal CARDs of RIG-I, which is crucial for the cytosolic RIG-I signaling pathway to elicit host anti-viral innate immunity (Gack et al., 2007). Specifically, TRIM25 interacts with the 1st CARD of RIG-I, and this interaction effectively delivers the Lys 63-linked ubiquitin moiety to the 2nd CARD of RIG-I, leading to efficient interaction with MAVS/VISA/IPS-1/Cardif. Additionally, a splice variant of RIG-I which carries a deletion (amino acids 36–80) within the 1st CARD, loses TRIM25 binding, CARD ubiquitination, and downstream signaling ability, demonstrating the critical role of TRIM25-mediated ubiquitination for RIG-I anti-viral activity (Gack et al., 2008).
On the other hand, viruses have evolved sophisticated mechanisms to evade or counteract the host IFN system. Specifically, virus-encoded IFN antagonists inhibit host innate anti-viral responses by targeting IFN gene expression or IFN-induced host effector molecules (Garcia-Sastre and Biron, 2006; Haller et al., 2006). Indeed, the NS1 is the main IFN antagonist encoded by influenza A viruses (Garcia-Sastre et al., 1998b), and was shown to antagonize the assembly of the IFN-β enhanceosome by inhibiting activation of IRF-3 (Talon et al., 2000), NFκB (Wang et al., 2000), and ATF-2/c-Jun (Ludwig et al., 2002) transcription factors. The ability of NS1 to prevent the nucleation of the IFN-β enhanceosome appears to be due at least in part to its binding to double-stranded RNA (dsRNA), likely resulting in the sequestration of this viral mediator from cellular sensors including RIG-I. Indeed, an influenza virus expressing an NS1 mutant defective in RNA-binding activity induces high levels of IFN in vitro and is attenuated in vivo (Donelan et al., 2003). In addition, it has recently been shown that NS1 interacts with RIG-I and efficiently suppresses its signal transducing activity (Guo et al., 2007; Mibayashi et al., 2007; Opitz et al., 2006; Pichlmair et al., 2006). However, the precise details of the molecular mechanism by which the NS1 protein antagonizes RIG-I function remains unknown. Specifically, it is not known whether the interaction of NS1 with RIG-I directly antagonizes RIG-I function (i.e. sensing viral RNA), or whether the NS1-RIG-I interaction precludes RIG-I interactions with and/or modifications by additional cellular proteins, such as TRIM25. In this study, we describe the long-sought mechanism by how the influenza A virus NS1 achieves this inhibition of the host IFN system. Surprisingly, this involves direct inhibition of the ubiquitin ligase activity of TRIM25, a novel mechanism not yet described for any other viral protein.
RESULTS
Influenza A virus NS1 suppresses RIG-I CARD ubiquitination
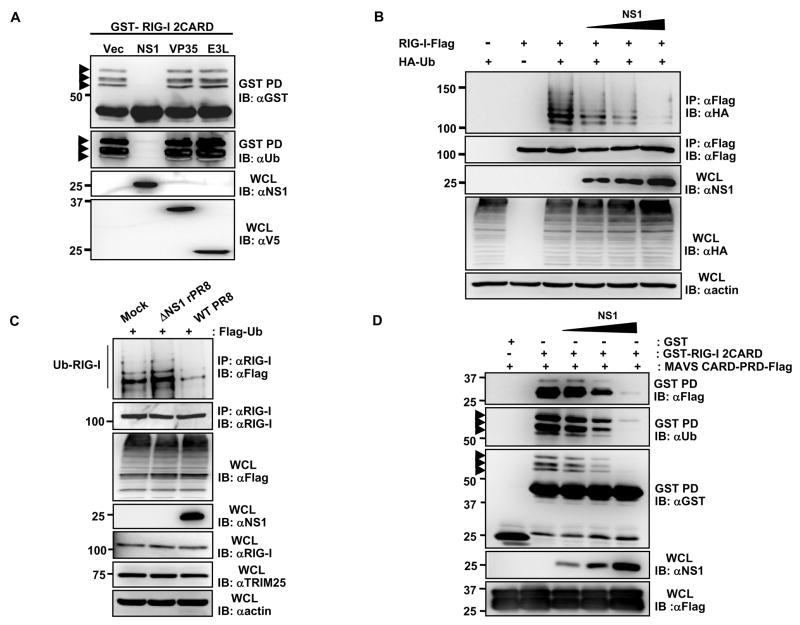
Given that TRIM25-induced ubiquitination of the N-terminal CARDs is essential for RIG-I to elicit anti-viral signal transduction (Gack et al., 2007), we postulated that virus-encoded IFN antagonists might specifically inhibit this step to prevent RIG-I activation. To test this, we examined the effect of expressing various viral proteins known to prevent IFN-β promoter activation on the ubiquitination of GST-RIG-I 2CARD in HEK293T cells. Among the three viral IFN antagonist proteins tested, influenza A virus NS1 was unique in that it potently inhibited the RIG-I 2CARD ubiquitination; whereas the other IFN antagonists Ebola virus VP35 (Cardenas et al., 2006) and vaccinia virus E3L (Smith et al., 2001) showed no effect under the same conditions (Fig. 1A). Consistently, influenza A NS1 effectively suppressed the ubiquitination of full-length RIG-I in a dose-dependent manner (Fig. 1B). Furthermore, the ubiquitination levels of endogenous RIG-I markedly decreased in cells infected with wild type (WT) influenza A/PR/8/34 virus compared to mock-infected cells (Fig. 1C). In contrast, cells infected with influenza A/PR/8/34 virus containing a deletion of the NS1 gene (ΔNS1 virus) exhibited a slight increase in RIG-I ubiquitination compared to mock-infected cells (Fig. 1C). Since the Lys 63-linked ubiquitination of RIG-I CARDs is crucial for efficient MAVS interaction, we further tested the effect of increasing amounts of influenza A NS1 on the CARD-dependent binding of RIG-I to MAVS (Fig. 1D). Consistent with its ability to suppress the RIG-I CARD ubiquitination, NS1 inhibited the RIG-I-MAVS interaction in a dose-dependent manner. These results indicate that influenza A virus NS1 specifically inhibits the RIG-I CARD ubiquitination, resulting in the suppression of RIG-I-MAVS complex formation.
Fig. 1. Influenza A virus NS1 inhibits the TRIM25-mediated RIG-I CARD ubiquitination.
(A)Whole cell lysates (WCLs) of HEK293T cells transfected with GST-RIG-I 2CARD together with vector, NS1, V5-VP35 or V5-E3L were subjected to GST-pulldown (GST-PD) followed by immunoblotting (IB) with α-GST or α-Ub. Arrows: ubiquitinated bands. (B) After transfection with RIG-I-Flag, HA-ubiquitin, and increasing amount of NS1, HEK293T were infected with SeV (50 HA units/ml) for 10 h. WCLs were subjected to immunoprecipitation (IP) with α-Flag, followed by IB with α-HA or α-Flag. (C) At 36 h post-transfection with Flag-ubiquitin, HEK293T were either mock-infected or infected with influenza A/PR/8/34 WT or ΔNS1 virus at MOI 2 for 11 h. WCLs were subjected to IP with α-RIG-I followed by IB with α-Flag or α-RIG-I. (D) WCLs of HEK293T transfected with MAVS-CARD-proline-rich domain (PRD)-Flag and GST or GST-RIG-I 2CARD together with increasing amounts of NS1 were subjected to GST-PD, followed by IB with α-Flag, α-Ub or α-GST. Arrows: ubiquitinated bands.
NS1 interacts with TRIM25
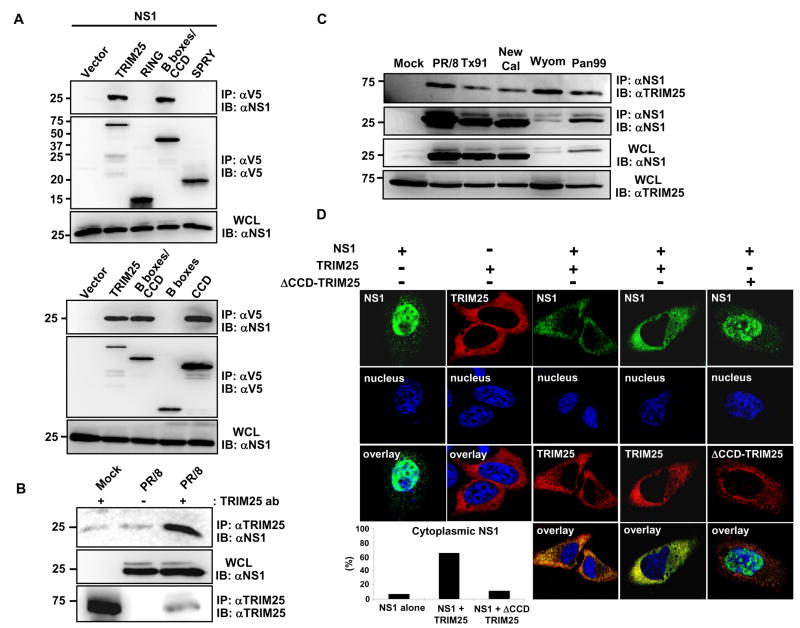
In order to elucidate the mechanism by which influenza A virus NS1 inhibits RIG-I ubiquitination, we tested the potential interaction between NS1 and TRIM25 in HEK293T cells. Co-immunoprecipitation (Co-IP) studies in transfected cells showed an interaction between NS1 and TRIM25 (Fig. 2A). TRIM25 is composed of an N-terminal RING domain, two B-boxes, a central coiled-coil domain (CCD), and a C-terminal SPRY domain (Meroni and Diez-Roux, 2005). The RING and the SPRY domains of TRIM25 have been shown to interact with E2 ubiquitin-conjugating enzymes and with the N-terminal CARDs of RIG-I, respectively (Gack et al., 2007; Horie-Inoue and Inoue, 2006; Zou et al., 2007). TRIM25 polypeptides corresponding to RING, B-boxes/CCD, B-boxes, CCD, and SPRY domains were examined for NS1 binding. This showed that NS1 specifically interacted with the central CCD (aa 180–450) of TRIM25 (Fig. 2A). Accordingly, a TRIM25 mutant, in which the CCD was deleted (TRIM25 ΔCCD), was incapable of binding NS1 (Supplementary Fig. 1A). HEK293T cells infected with various human influenza A virus strains including A/PR/8/34, and human virus isolates A/Texas/36/91, A/New Caledonia/20/99, A/Wyoming/3/2003, and A/Panama/2007/99 also showed an interaction between NS1 and endogenous TRIM25 (Fig. 2B, C). Furthermore, the NS1 protein of several avian and swine influenza A virus strains and of the 1918 pandemic strain of influenza A virus readily bound TRIM25 (Supplementary Fig. 1B). Confocal microscopy confirmed an interaction between TRIM25 and NS1 (Fig. 2D). While expression of NS1 in HeLa cells resulted in nuclear localization with a minor cytoplasmic component, TRIM25 over-expression led to a marked increase of NS1 cytoplasmic localization (Fig. 2D). However, expression of a TRIM25 ΔCCD mutant that no longer interacted with NS1 showed little or no effect on the subcellular localization of NS1 (Fig. 2D). In addition, we observed an in vitro interaction between bacterially purified GST-PR8 NS1 and MBP-TRIM25-Flag (Supplementary Fig. 2A), suggesting the direct binding of influenza A NS1 to TRIM25.
Fig. 2. The NS1 proteins of various influenza A virus strains interact with TRIM25.
(A) HEK293T were transfected with NS1 together with vector, V5-tagged TRIM25, RING, B-boxes/CCD, SPRY (upper panels), TRIM25, B-boxes/CCD, B-boxes or CCD (lower panels). WCLs were used for IP with α-V5 followed by IB with α-NS1 or α-V5. (B) HEK293T were either mock-infected or infected with influenza A/PR/8/34 at MOI 2. At 18 h post-infection, WCLs were used for IP with α-TRIM25 or without antibody followed by IB with α-NS1 or α-TRIM25. (C) WCLs of HEK293T either mock-infected or infected with the indicated influenza A virus strains (MOI 2 for 12 h) were used for IP with α-NS1 followed by IB with α-TRIM25 or α-NS1. (D) At 20 h post-transfection with NS1 alone, TRIM25-V5 alone, NS1 together with TRIM25-V5, or NS1 together with ΔCCD-TRIM25-V5, HeLa cells were stained with α-NS1 (green), α-V5 (red) and Hoechst 33256 (nucleus, blue). More than 100 cells with expression of NS1 alone or NS1 together with TRIM25 or ΔCCD-TRIM25, respectively, were counted and percentage of the cells with cytoplasmic localization of NS1 is shown (lower left).
WT NS1 but not R38A/K41A and E96A/E97A NS1 mutants inhibit TRIM25-mediated RIG-I ubiquitination and signal transduction
The structure of the N-terminal RNA binding domain (RBD; aa 1–73) and the C-terminal effector domain (ED; aa 74–230) of the influenza A virus NS1 has been obtained (Bornholdt and Prasad, 2006; Chien et al., 1997; Chien et al., 2004; Hale et al., 2008a; Liu et al., 1997). The NS1 RBD likely sequesters viral dsRNA from cytoplasmic RNA sensors, such as PKR, OAS, and RIG-I, while the ED mediates interactions with several cellular factors, including CPSF (binding cleavage and polyadenylation specificity factor), PAB II (inhibiting poly-(A)-binding protein) and eIF4G-I to regulate viral and host gene expression (Garcia-Sastre, 2001). An initial mapping study showed that both domains were required to bind TRIM25 (Supplementary Fig. 1C). We then mutated conserved amino acids within both NS1 domains to analyze their impact on TRIM25 binding. The R38A/K41A NS1 mutant is known to be deficient in dsRNA binding activity and in IFN antagonism (Donelan et al., 2003; Talon et al., 2000). The E96A/E97A mutation is located in a highly conserved putative protein-protein interacting motif (S/T-x-E-E), identified by a computational analysis of the NS1 sequence using the Eukaryotic Linear Motif resource (ELM) (Puntervoll et al., 2003). R38A/K41A and E96A/E97A NS1 mutants showed a complete loss of TRIM25 binding ability in Co-IP assays (Fig. 3A). Furthermore, both E96A/E97A and R38A/K41A NS1 mutants did not co-localize with over-expressed TRIM25 in HeLa cells (Supplementary Fig. 3). In agreement with these results, the R38A/K41A and E96A/E97A NS1 mutants, in contrast to NS1 WT, did not interact with full-length RIG-I (Supplementary Fig. 4).
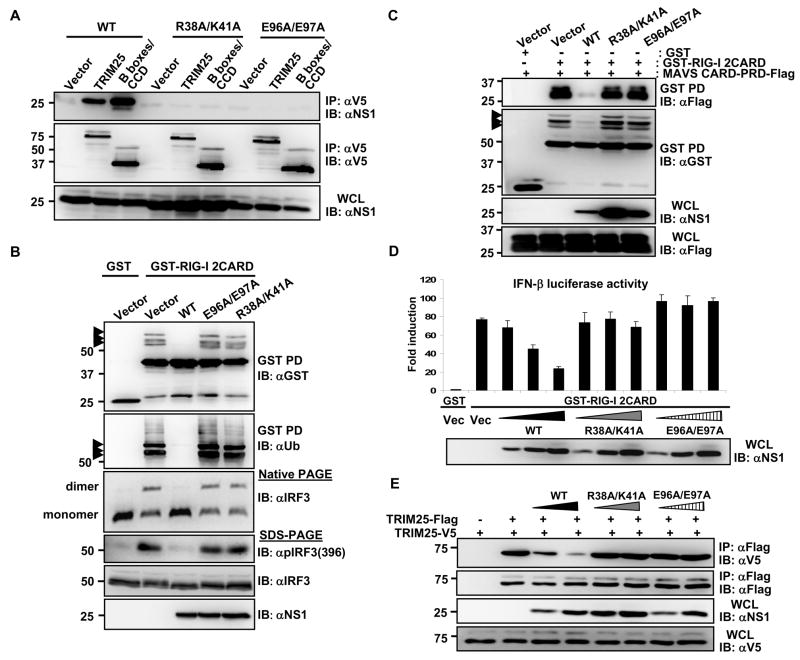
Fig. 3. Inhibition of TRIM25-mediated RIG-I signaling by WT NS1 but not E96A/E97A and R38A/K41A NS1 mutants.
(A)At 48 h post-transfection of HEK293T with vector, V5-TRIM25 or TRIM25-B-boxes/CCD together with NS1 WT, R38A/K41A, or E96A/E97A, WCLs were subjected to IP with α-V5 followed by IB with α-NS1 or α-V5. (B) After transfection with GST or GST-RIG-I 2CARD together with vector, NS1 WT, R38A/K41A, or E96A/E97A, WCLs were used for GST-PD followed by IB with α-GST or α-Ub (upper two panels). WCLs were either subjected to native PAGE followed by IB with α-IRF3 (middle panel), or used for SDS-PAGE and immunoblotted with α-Phospho-IRF3 (Ser396), α-IRF3, or α-NS1 antibodies (lower panels). Arrows: ubiquitinated bands. (C) WCLs of HEK293T transfected with MAVS-CARD-PRD-Flag and GST or GST-RIG-I 2CARD together with NS1 WT or mutants were subjected to GST-PD, followed by IB with α-Flag or α-GST. Arrows: ubiquitinated bands. (D) HEK293T were transfected with GST or GST-RIG-I 2CARD together with vector or increasing amount of NS1 WT, R38A/K41A, or E96A/E97A as well as IFN-β-luciferase and pGK-β-gal. Luciferase and β-galactosidase values were determined as previously described (Gack et al., 2007). Data represent the mean + SD (n=3). (E) At 48 h post-transfection with vector, TRIM25-Flag, TRIM25-V5 and increasing amount NS1 WT, R38A/K41A, or E96A/E97A, HEK293T WCLs were subjected to IP with α-Flag followed by IB with α-V5 or α-Flag.
We next investigated the ability of NS1 WT and mutants E96A/E97A and R38A/K41A to inhibit the TRIM25-mediated RIG-I ubiquitination and downstream signaling (Fig. 3B, C and D). NS1 WT potently suppressed the RIG-I CARD ubiquitination, whereas the E96A/E97A and R38A/K41A NS1 mutants had no effect on the ubiquitination level of RIG-I CARDs (Fig. 3B, upper panels). In line with this, NS1 WT, but not the E96A/E97A and R38A/K41A NS1 mutants, potently suppressed the ubiquitination-dependent CARD interaction between RIG-I and MAVS (Fig. 3C). To further delineate the effect of NS1 WT and mutants on RIG-I anti-viral downstream signaling, we examined the phosphorylation and dimerization of interferon regulatory factor 3 (IRF3) triggered by RIG-I CARDs (Fig. 3B, lower panels). IRF3 has been well characterized to undergo virus-induced phosphorylation at serine 396 and dimerization, which leads to its nuclear translocation and contribution to IFN-β promoter activation (Paz et al., 2006). We found that NS1 potently inhibited the Ser-396 phosphorylation of IRF3 and almost completely blocked IRF3 dimerization (Fig. 3B, lower panels), consistent with the previous published results (Mibayashi et al., 2007). In contrast, the E96A/E97A and R38A/K41A NS1 mutants, which had no inhibitory effect on RIG-I 2CARD ubiquitination, did not interfere with the phosphorylation or dimerization of IRF3 (Fig. 3B, lower panels). Consistently, while NS1 WT effectively inhibited the RIG-I 2CARD-induced IFN-β promoter activation in a dose-dependent manner, the E96A/E97A and R38A/K41A NS1 mutants showed no effect (Fig. 3D).
To provide further evidence whether NS1 directly targets TRIM25 to inhibit RIG-I signal transduction, we addressed if TRIM25 overexpression can overcome the RIG-I inhibition by NS1. While NS1 markedly suppressed the RIG-I 2CARD-induced IFN-β gene expression, increasing amounts of TRIM25 were able to overcome the NS1-mediated inhibition of RIG-I 2CARD (Supplementary Fig. 5). Collectively, these results strongly suggest that TRIM25 ubiquitin E3 ligase is the direct target of NS1 inhibition.
Influenza A virus NS1 inhibits CCD-dependent TRIM25 oligomerization
The CCD of the TRIM family proteins has been characterized to form a hyper-secondary structure with multiple α-helices involved in homo-oligomeric interactions (Meroni and Diez-Roux, 2005). For example, TRIM5α, an intracellular inhibitor of retroviral replication with a structure similar to TRIM25, has been shown to form a trimer in vivo (Mische et al., 2005). To decipher the mechanism by which NS1 interaction with the CCD of TRIM25 leads to the suppression of RIG-I ubiquitination, we addressed whether: i) TRIM25 underwent oligomerization, ii) whether TRIM25 oligomerization was necessary for its ubiquitin E3 ligase activity, and iii) whether NS1 affected TRIM25 oligomerization and thereby suppressed RIG-I ubiquitination. As shown in Supplementary Fig. 6A, V5-tagged TRIM25, TRIM25-B-boxes/CCD and TRIM25-CCD readily formed a complex with Flag-TRIM25; in contrast, the TRIM25 ΔCCD mutant was unable to multimerize. When tested for its ability to ubiquitinate the RIG-I CARDs, the exogenously expressed TRIM25 ΔCCD mutant, in contrast to TRIM25 WT, did not increase the ubiquitination of GST-RIG-I 2CARD (Supplementary Fig. 6B). These results strongly indicate that TRIM25 multimerizes through its central CCD, and that this multimerization appears to be essential for its E3 ligase activity to ubiquitinate the RIG-I CARDs. We then tested whether influenza A virus NS1 interfered with the TRIM25 multimerization. Indeed, NS1 expression effectively inhibited the CCD-mediated TRIM25 multimerization in a dose-dependent manner (Supplementary Fig. 7A). Consistent with their inability to bind TRIM25, the E96A/E97A and R38A/K41A NS1 mutants did not interfere with TRIM25 multimerization (Fig. 3E). Finally, we did not observe any significant alteration of endogenous or exogenous TRIM25 protein levels upon NS1 overexpression or infection with influenza A/PR/8 virus (Fig. 1C and Supplementary Fig. 7B). These results collectively indicate that influenza A virus NS1 does not affect TRIM25 expression level, but its interaction with the central CCD of TRIM25 abolishes TRIM25 multimerization and enzymatic activity, resulting in the inhibition of RIG-I CARD ubiquitination.
Inhibition of TRIM25 is required for suppression of IFN induction and optimal replication by influenza A virus
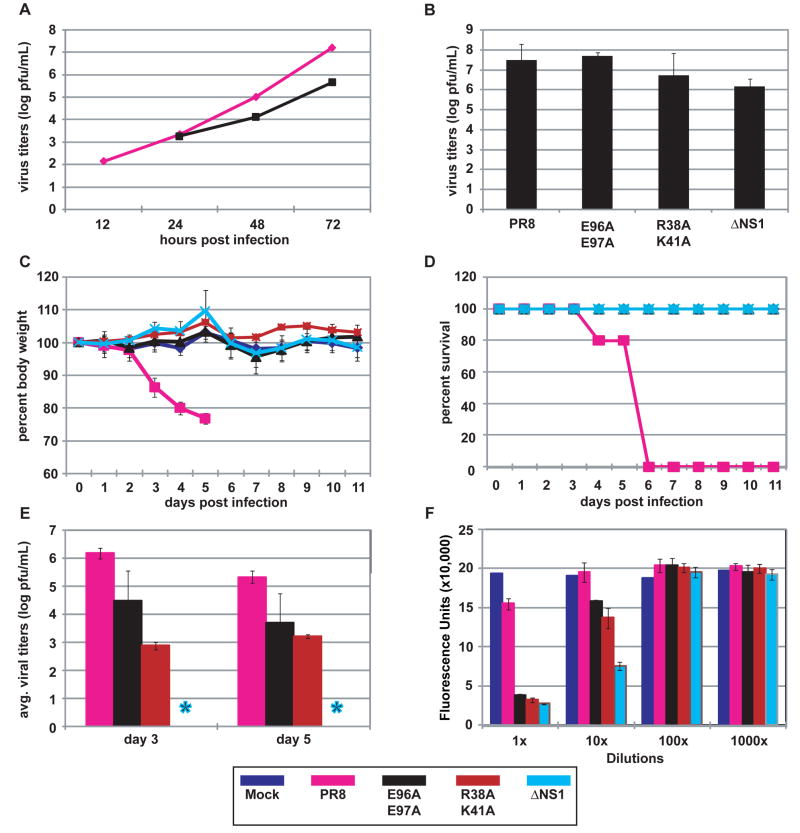
The R38A/K41A NS1 mutant was previously reported to be deficient in binding dsRNA, and a recombinant influenza virus expressing this mutant NS1 protein was defective in IFN antagonism and attenuated in mice (Donelan et al., 2003; Talon et al., 2000). Considering that the R38A/K41A NS1 mutant is defective both in sequestering viral RNA from molecular sensors and in suppressing TRIM25-mediated RIG-I ubiquitination, it is difficult to determine which NS1 function is more critical to its antagonism of the type I interferon system. In contrast to the R38A/K41A NS1 mutant, the E96A/E97A NS1 mutant retains dsRNA-binding activity (Supplementary Fig. 2B), yet it is defective in blocking TRIM25-triggered anti-viral responses (Fig. 3). Thus, the E96A/E97A mutation in the context of a recombinant influenza virus should unveil the contribution of TRIM25 inhibition by NS1 in IFN antagonism and virulence. We therefore generated a recombinant A/PR/8/34 virus expressing NS1 E96A/E97A and tested its replication in A549 cells, a human lung epithelial cell line, in comparison to WT virus. As compared to titers achieved with WT virus, the E96A/E97A virus exhibited an approximately 1.5 log reduction in titer at 72 hours post-infection (Fig. 4A). To determine whether the reduction in titer for the E96A/E97A virus was attributed to induction of IFN, virus production from IFN-deficient, 8-day old embryonated chicken eggs was determined (Fig. 4B). At 24 hours post-infection with 100 pfu per egg, the titers obtained for the E96A/E97A virus were comparable to those of the WT virus. The titers of the R38A/K41A and ΔNS1 viruses were slightly reduced as compared to the WT virus. These results suggest that the reduced titers obtained for the E96A/E97A virus following infection of A549 cells are attributed to IFN production and not to an intrinsic defect in NS1 function resulting in impaired virus replication. To establish whether the loss of TRIM25 regulation correlated with attenuation in pathogenicity, we assessed the replication and virulence of WT and E96A/E97A viruses in mice. For comparative purposes we also included R38A/K41A and ΔNS1 viruses. Intranasal infection of Balb/C mice with WT A/PR/8/34 resulted in lethal infection, in which all mice succumbed by day 6 post-infection (Fig. 4C and D). However, infection with the mutant viruses did not result in any observable morbidity. Consistent with the absence of morbidity, infections with E96A/E97A and R38A/K41A viruses resulted in reduced pulmonary titers at day 3 and 5 post-infection as compared to titers achieved by WT virus (Fig. 4E). The ΔNS1 virus was not detectable by plaque assay at these time points.
Fig. 4. Replication and pathogenicity of recombinant influenza viruses.
(A) Multicycle replication of recombinant influenza viruses in A549 cells infected at an MOI of 0.001 pfu per cell. (B) Virus production from interferon-deficient 8-day old embryonated chicken eggs at 24 h post-infection with 100 pfu of each virus per egg. (C) Percent body weight loss of mice following virus infection. Female Balb/C (n = 5) were infected intranasally with 1x104 pfu of one of the indicated viruses or mock-infected with PBS. Body weights were measured for 12 consecutive days following infection. (D) Kaplan-Meier survival curves of mice infected with the indicated recombinant viruses. (E) Viral pulmonary titers of infected mice determined on day 3 and 5 post-infection (n = 3) by plaque assay. Asterisk indicates ΔNS1 not detected by plaque assay. Bars indicate standard deviation. (F) IFN bioassays measured IFN production by influenza virus-infected A549 cells. Fluorescence units represent the extent of NDV-GFP replication in Vero cells treated with supernatants from virus-infected A549 cells. Bars indicate standard deviation. The color scheme for the indicated viruses is provided at the bottom of the figure.
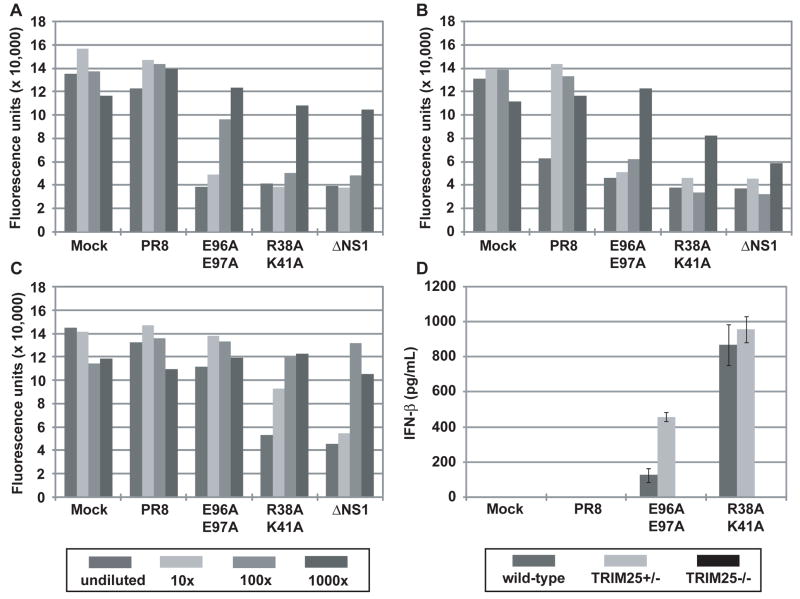
Previous studies have indicated a correlation between the IFN-inducing phenotype in vitro and reduced virulence in mice of influenza viruses expressing mutant NS1 (Donelan et al., 2003; Falcon et al., 2004; Garcia-Sastre et al., 1998b). Therefore, we measured IFN production from infected A549 cells by bioassay (Park et al., 2003; Solorzano et al., 2005). Infection of A549 cells with the E96A/E97A and R38A/K41A mutant viruses resulted in substantial IFN production as compared to infection with WT virus (Fig. 4F and Supplementary Fig. 8). Cells infected with ΔNS1 virus produced the highest levels of IFN. In order to assess the contribution of TRIM25 in IFN induction by the NS1 mutant viruses, interferon bioassays were conducted with mouse embryonic fibroblasts (MEF) derived from either wild-type, heterozygous (TRIM25+/−) or homozygous (TRIM25−/−) mice (Fig. 5). Whereas WT virus resulted in minimal IFN production in all MEF lines tested, the mutant viruses were efficient inducers of IFN. Infection with the E96A/E97A mutant virus resulted in detectable IFN production from wild-type and heterozygous cells (Fig. 5A and B), but not from the homozygous (TRIM25−/−) cells (Fig. 5C). The IFN-inducing phenotypes of the R38A/K41A and ΔNS1 viruses were reduced in at least one order of magnitude but they were still evident in the TRIM25−/− cell line. These data indicate that the loss of TRIM25 inhibition by the E96A/E97A mutant virus is responsible for its IFN inducing properties. In contrast, mutations associated with loss of multiple IFN antagonistic NS1 functions, such as R38A/K41A, result in viruses with lower but still detectable IFN-inducing properties in TRIM25−/− cells. Specifically, while some remaining IFN was detectable by bioassay in ΔNS1 and R38A/K41A virus infected cells in the absence of TRIM25, these levels were approximately 10 to 100 times lower than in the presence of TRIM25, indicating that most of the IFN produced in MEFs is dependent on TRIM25 expression and suggesting that additional NS1 functions, such as dsRNA binding, are needed to fully prevent a suboptimal TRIM25-independent induction of IFN during viral infection. As an alternative approach to measuring IFN-β secretion, ELISA was performed to measure IFN-β production by the MEF cell lines at 18 hours post-infection (Fig. 5D). Although the IFN bioassay appears to be a more sensitive assay for IFN-β production, the ELISA results are in agreement with the bioassay results. While the WT virus did not produce detectable amounts of IFN-β, the E96A/E97A and R38A/K41A viruses induced between 100 to 1000 pg/mL IFN-β from wild-type and heterozygous (TRIM25+/−) MEFs. By contrast, no IFN-β was detected by ELISA from the TRIM25−/− infected cells.
Fig. 5. IFN inducing/suppressing phenotypes of recombinant influenza viruses in MEFs.
IFN bioassays were performed to quantitate IFN production resulting from influenza virus infection of (A) wild-type, (B) heterozygous (TRIM25+/−), or (C) homozygous (TRIM25−/−) MEFs. Fluorescence units represent the extent of VSV-GFP replication in L929 cells treated with supernatants from virus-infected MEF cells. The data are representative of 3 independent experiments. The legend for panels (A), (B), and (C) is provided below panel (C). (D) ELISA was performed to measure IFN-β production from wild-type, heterozygous (TRIM25+/−), or homozygous (TRIM25−/−) MEFs upon influenza virus infection. The results are expressed as means ± s.d. (n = replicate of 3).
DISCUSSION
TRIM proteins, so named due to the presence of RING, B-box, and coiled-coil domains, constitute a family of proteins that are involved in a broad range of biological processes including cell cycle progression and anti-viral responses (Meroni and Diez-Roux, 2005; Nisole et al., 2005; Ozato et al., 2008). Among them, TRIM19 (promyelocytic leukemia protein (PML)) and TRIM5α have been shown to exhibit potent anti-viral activity. Whereas TRIM19, a component of nuclear bodies, inhibits the replication of a wide variety of DNA and RNA viruses, TRIM5α has been demonstrated to interfere with the uncoating of the pre-integration complex of lentiviruses including HIV-1 (Kratovac et al., 2008; Stremlau et al., 2004; Takeuchi and Matano, 2008). The importance of TRIM molecules in host defense against virus infection has also been illustrated by the finding that the RING ubiquitin E3 ligase TRIM25 is crucial for RIG-I-mediated induction of type I IFN (Gack et al., 2007). Due to their anti-viral activities, TRIM molecules might be targeted by viral proteins enabling viruses to evade the host immune response. Although a few viral proteins have been shown to associate with TRIM family members (Ahn et al., 1998; Hoppe et al., 2006; Yondola and Hearing, 2007), the biological consequences of these interactions have not been elucidated. Our study is the first demonstration of how a viral protein directly interferes with the activity of a TRIM molecule to disrupt IFN-mediated innate immunity, which ultimately leads to viral pathogenesis.
For influenza viruses, the main IFN-antagonistic action is encoded by the NS1 gene. Several functions have been documented that may account for NS1 inhibition of type I IFN production, including sequestration of dsRNA, interaction with RIG-I, and inhibition of downstream processes after RIG-I-MAVS interactions such as the processing and trafficking of cellular mRNAs and the activities of IFN-inducible enzymes PKR and OAS (for recent reviews see (Albrecht and Garcia-Sastre, 2009; Hale et al., 2008b)). However, the precise mechanism by which the NS1 avoids recognition of virus infection by the host and prevents IFN production was not elucidated. In this report, we describe that the NS1 protein directly binds TRIM25; specifically, the NS1 targets the TRIM25 CCD, thus interfering with TRIM25 multimerization. TRIM25 multimerization is crucial for ubiquitination of RIG-I CARDs, a modification that was found to be necessary for maximal IFN production in response to virus infection (Gack et al., 2007). Thus, by directly binding to and inhibiting the enzymatic activity of TRIM25, the NS1 suppresses RIG-I signal transduction and ultimately IFN-β production.
Since NS1 and RIG-I bind to non-overlapping domains of TRIM25, the CCD and SPRY domains (Gack et al., 2007), respectively, our protein interaction studies suggest a NS1/TRIM25/RIG-I triple complex in influenza virus-infected cells. In further support of this, we did not observe a competition between RIG-I and NS1 for TRIM25 binding (data not shown). The detailed molecular architecture of the RIG-I/TRIM25/NS1 triple complex is currently being investigated. We have shown that the interaction of NS1 with TRIM25 is dependent on amino acids located both in the NS1 RNA-binding domain (basic residues 38 and 41) and outside of this domain (acidic residues 96 and 97). The recently published crystal structure of full-length NS1 reveals that these four residues are not in close proximity (Bornholdt and Prasad, 2008). Interestingly, the crystal structure could only be determined for an RNA-binding NS1 mutant; R38 and K41 were determined to govern aggregation and stability of the full-length NS1. It is plausible that the R38A/K41A mutations have created conformational changes that have resulted in the unanticipated loss of binding to TRIM25. Further studies are then required to understand the structural contribution of these amino acids to TRIM25 binding. Nevertheless, the use of a recombinant influenza virus bearing the NS1 E96A/E97A mutations and of TRIM25 knockout cells allowed us to conclude that NS1 binding to TRIM25 is needed for optimal inhibition of IFN production in virus-infected cells. This mutant virus showed attenuated replication in human cells and pathogenesis in vivo, and since amino acids 96 and 97 of NS1 have not been implicated in additional NS1 functions, this strongly suggests that TRIM25 binding by NS1 is required for virulence. Interestingly, the NS1 R38A/K41A mutant virus, impaired in both NS1 binding to dsRNA and TRIM25, also shows reduced IFN production in TRIM25−/− cells. The lack of a complete loss of IFN induction indicates that TRIM25 is not completely required for IFN induction by influenza virus infection, and that NS1 functions other than binding to TRIM25, such as binding to dsRNA, also contribute to maximal inhibition of type I IFN synthesis during infection.
It is interesting that despite sequence variations, NS1 proteins encoded by human, avian, and swine influenza viruses interacted with TRIM25 in infected or transfected cells, indicating that targeting TRIM25 is a conserved function of NS1 of various influenza viruses. However, it is conceivable that sequence variations in the NS1 proteins of different virus strains influence the affinity for TRIM25 binding, which may correlate with viral pathogenesis. This concept is important when considering the adaptation of an influenza virus to a new host species since cellular factors targeted by NS1 may have divergent sequences from one host to the other. Indeed, human and avian TRIM25 show a detectable degree of sequence variation, such that the human TRIM25 coiled-coil domain, which is targeted by NS1, exhibits only 33% identity with the corresponding domain of its avian orthologue. Thus, further studies are directed to address the influence of sequence variations in TRIM25 and NS1 on their interactions and on the NS1-mediated IFN antagonizing activity.
Our study demonstrates that the influenza A virus NS1 targets multiple checkpoints of the IFN-mediated signaling pathway by sequestering RNA from cellular sensors like RIG-I and by inhibiting TRIM25 E3 ligase. These NS1 activities, combined with the ability of the NS1 of several viral strains to suppress host gene expression collectively establish a comprehensive suppression of IFN production during viral infection. Our findings also provide the first example of a virus suppressing IFN production by directly antagonizing the enzymatic function of a TRIM family member, in this case the RING-mediated E3 ligase activity of TRIM25. In addition, by identifying NS1 mutants lacking TRIM25 inhibition and by testing their replication in mice, we demonstrate the important role of the NS1-mediated TRIM25 inhibition in influenza A virus virulence. Finally, the crucial role of TRIM25 for maximal type I IFN production in response to influenza A virus was demonstrated by viral infection studies in TRIM25 knock-out cells. Thus, our findings not only describe a new class of viral immune evasion mechanism being crucial for in vivo virulence, but also provide detailed insights into the biological role of TRIM25 in anti-viral host defense. These observations should stimulate the search for additional viral antagonists of innate immune responses that target TRIM proteins and for additional cellular proteins modified by TRIM25 which may play specific roles in anti-viral immunity.
EXPERIMENTAL PROCEDURES
Viruses
The A/Puerto Rico/8/1934 mutant virus with NS1 E96A/E97A mutations was rescued as described previously (Grimm et al., 2007). The rescue plasmid pDZ-PR8 (NS1 E96A/E97A) was cloned by PCR mutagenesis of the A/Puerto Rico/8/1934 NS segment with the primer pair (respective to cRNA): 3′ NC-Ambi G (5′-GATCGCTCTTCTGGGAGCGAAAGCAGG-3′) and E9697A-rev (5′-CGCGTTACCTAACTGACATGACTCTTGCGGCAATGTCAAGGGACTGGTC-3′). The E9697A-rev primer contained two point mutations (A313C and A316C) which resulted in the substitutions Glu96Ala and Glu97Ala. The E9697A-rev primer also contained a native BsrI restriction enzyme site that facilitated ligation of the amplified fragment with the wild-type NS sequence. The viral NS segments from the rescued viruses were amplified by RT-PCR for subsequent sequence confirmation.
The parental A/Puerto Rico/8/1934 virus, the mutant viruses encoding NS1 R38A/K41A and NS1 E96A/E97A, A/Texas/36/1991 (H1N1), A/New Caledonia/20/1999 (H1N1), A/Wyoming/3/2003 (H3N2), A/Panama/2007/1999 (H3N2), and NDV-GFP were propagated in 8-day old, specific pathogen-free embryonated eggs (Charles River Laboratories, North Franklin, CT). The A/Puerto Rico/8/1934 ΔNS1 virus was propagated in NS1-expressing MDCK cells. Influenza viruses titers were determined by plaque assay or by indirect immunofluorescence microscopy with an anti- A/Puerto Rico/8/1934 NP polyclonal antibody (Bourmakina and Garcia-Sastre, 2005) in MDCK cells. VSV-GFP was propagated in Vero cells (Ebert et al., 2003).
GST Pulldown Assay, Immunoprecipitation, and Immunoblot Analysis
HEK293T cells were lysed in NP40 buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1% (v/v) NP40 and protease inhibitor cocktail (Roche)). GST pulldown and Immunoprecipitations were performed as described previously (Gack et al., 2007). For immunoblotting, proteins were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a PVDF membrane (BioRad Laboratories, Hercules, CA). A549 cell lysates were resolved by SDS-PAGE and transferred to nitrocellulose membranes (BioRad Laboratories, Hercules, CA). The following primary antibodies were used: anti-V5 (1:5,000) (Invitrogen), anti-Flag (1:5,000) (Sigma), anti-HA (1:5,000) (Sigma), anti-GST (1:10,000) (Sigma), anti-ubiquitin (P4D1, Santa Cruz), anti-TRIM25 (1:5,000) (BD Biosciences), anti-RIG-I (1:1,000) (Alexis), anti-IRF3 (1:500) (Santa Cruz), anti-phospho-Ser396-IRF3 (1:200) (Upstate), anti-NP (1:2,000), anti-NS1 (1:1000; (Solorzano et al., 2005)), anti-β-actin (Abcam, Cambridge, MA), and anti-M1/M2 monoclonal antibody E10 (Bourmakina and Garcia-Sastre, 2005). Immunoblot analyses of A549 cell lysates were developed with the following secondary antibodies: ECL anti-rabbit IgG horse radish peroxidase conjugated whole antibody from donkey and ECL anti-mouse IgG horse radish peroxidase conjugated whole antibody from sheep (GE Healthcare, Buckinghamshire England). The proteins were visualized by an enhanced chemiluminescence reagent (Pierce) and detected by a phospho imager (Fuji LAS-4000).
Direct Protein Interaction Assay
Recombinant TRIM25 purified from bacteria as an amino-terminal Maltose-binding protein and carboxy-terminal Flag fusion protein (MBP-TRIM25-FLAG (Gack et al., 2007)) was incubated with bacteria-produced recombinant GST-PR8 wt NS1 or GST in 50 mM Tris HCl, pH 7.4, 150 mM NaCl, and 0.1% NP-40 (IGEPAL). TRIM25-NS1 protein complexes were then precipitated with anti-Flag M1 affinity gel (Sigma, St. Louis, MO). Precipitated protein complexes were resolved by SDS-PAGE and visualized by Coomassie staining.
Poly (I)-Poly (C) Pulldown Assay
Total cell lysates were prepared from A549 cells infected with WT, E96A/E97A, or R38A/K41A viruses by lysing cells in 50 mM HEPES, pH 7.5, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mM sodium chloride, and 10% glycerol. NS1 proteins were precipitated from total cell lysates by poly (I)-poly (C) conjugated to sepharose as described previously (Cardenas et al., 2006). Precipitated proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes (BioRad Laboratories). Precipitated NS1 proteins were visualized by immunoblot analysis with the anti-NS1 (1–73) polyclonal antibody (Solorzano et al., 2005) and ECL anti-rabbit IgG horse radish peroxidase conjugated whole antibody from donkey (GE Healthcare).
Quantification of interferon production
Interferon produced from cells infected with the indicated influenza viruses at an MOI of 2 was examined by an interferon bioassay as described elsewhere (Park et al., 2003; Quinlivan et al., 2005; Solorzano et al., 2005). Interferon produced from A549 cells was determined by NDV-GFP infection of Vero cells treated with A549 supernatants. Interferon produced from MEF cells was determined by VSV-GFP infection of L929 cells treated with MEF supernatants. Mouse IFN-β production from MEFs was measured by enzyme-linked immunosorbent assays (ELISA; PBL Biomedical Laboratories) as per manufacturer’s instructions. The limit of detection for the IFN-β ELISA was 15.6 pg/mL.
Mouse Infections
Six week old female Balb/C mice were anesthetized with ketamine/xylazine prior to intranasal infection with 1x104 pfu of virus diluted in phosphate buffered saline (PBS) or mock-infected. Mice were monitored daily for weight loss or other signs of morbidity over a 12-day period. All animal procedures were performed in accordance with guidelines established by the Mount Sinai School of Medicine Institutional Animal Care and Use Committee and National Institutes of Health for the care and use of laboratory animals.
Supplementary Material
Acknowledgments
We wish to thank Richard Cadagan for excellent technical support. We thank Jonathan Yewdell, Savio L.C. Woo and Luis Martínez-Sobrido for kindly providing the monoclonal NS1 antibody (Mibayashi et al., 2007) VSV-GFP (Ebert et al., 2003), and MDCK-NS1 cell line, respectively. This research was supported in part by grants from the NIH, R01 AI46954, U19 AI62623 (Center for Investigating Viral Immunity and Antagonism), U54 AI57158 (North East Biodefense Center) and by CRIP (Center for Research on Influenza Pathogenesis, NIAID contract HHSN266200700010C) to A.G-S, CA082057, CA31363, CA115284, DE019085, and RR00168 to J.U.J, and U19 AI083025 (Center for Immune Mechanisms of Virus Control) to A.G-S and JUJ. This work was further supported by the grant of the Genome Network Project from the MEXT to S.I. and the exchange program between Harvard Medical School and the graduate training program 1071 at the Friedrich-Alexander University Erlangen-Nuremberg, Germany (M.U.G.).
Michaela U. Gack and Randy A. Albrecht performed all aspects of this study and prepared manuscript. Tomohiko Urano and Satoshi Inoue provided recombinant TRIM25, Trim25−/−, and TRIM25 −/+ MEFs. Kyung-Soo Inn assisted in data collection. I-Chueh Huang and Michael Farzan contributed reagents and assisted in data collection. Elena Carnero performed virus rescue transfections. Jae U. Jung and Adolfo García-Sastre organized this study and prepared manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn JH, Brignole EJ, 3rd, Hayward GS. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Albrecht RA, Garcia-Sastre A. Suppression of innate immunity by orthomyxoviruses. In: Brasier AR, Garcia-Sastre A, Lemon SM, editors. Cellular signalling and innate immune responses to RNA virus infections. Washington: ASM Press; 2009. pp. 267–315. [Google Scholar]
- Bornholdt ZA, Prasad BV. X-ray structure of influenza virus NS1 effector domain. Nat Struct Mol Biol. 2006;13:559–560. doi: 10.1038/nsmb1099. [DOI] [PubMed] [Google Scholar]
- Bornholdt ZA, Prasad BV. X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature. 2008 doi: 10.1038/nature07444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourmakina SV, Garcia-Sastre A. The morphology and composition of influenza A virus particles are not affected by low levels of M1 and M2 proteins in infected cells. J Virol. 2005;79:7926–7932. doi: 10.1128/JVI.79.12.7926-7932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas WB, Loo YM, Gale M, Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CY, Tejero R, Huang Y, Zimmerman DE, Rios CB, Krug RM, Montelione GT. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat Struct Biol. 1997;4:891–895. doi: 10.1038/nsb1197-891. [DOI] [PubMed] [Google Scholar]
- Chien CY, Xu Y, Xiao R, Aramini JM, Sahasrabudhe PV, Krug RM, Montelione GT. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry. 2004;43:1950–1962. doi: 10.1021/bi030176o. [DOI] [PubMed] [Google Scholar]
- Donelan NR, Basler CF, Garcia-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert O, Shinozaki K, Huang TG, Savontaus MJ, Garcia-Sastre A, Woo SL. Oncolytic vesicular stomatitis virus for treatment of orthotopic hepatocellular carcinoma in immune-competent rats. Cancer Res. 2003;63:3605–3611. [PubMed] [Google Scholar]
- Falcon AM, Marion RM, Zurcher T, Gomez P, Portela A, Nieto A, Ortin J. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J Virol. 2004;78:3880–3888. doi: 10.1128/JVI.78.8.3880-3888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, Myong S, Ha T, Hopfner KP, Jung JU. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci U S A. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology. 2001;279:375–384. doi: 10.1006/viro.2000.0756. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Durbin RK, Zheng H, Palese P, Gertner R, Levy DE, Durbin JE. The role of interferon in influenza virus tissue tropism. J Virol. 1998a;72:8550–8558. doi: 10.1128/jvi.72.11.8550-8558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998b;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Grimm D, Staeheli P, Hufbauer M, Koerner I, Martinez-Sobrido L, Solorzano A, Garcia-Sastre A, Haller O, Kochs G. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc Natl Acad Sci U S A. 2007;104:6806–6811. doi: 10.1073/pnas.0701849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Chen LM, Zeng H, Gomez JA, Plowden J, Fujita T, Katz JM, Donis RO, Sambhara S. NS1 Protein of Influenza A Virus Inhibits the Function of Intracytoplasmic Pathogen Sensor, RIG-I. Am J Respir Cell Mol Biol. 2007;36:263–269. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- Hale BG, Barclay WS, Randall RE, Russell RJ. Structure of an avian influenza A virus NS1 protein effector domain. Virology. 2008a;378:1–5. doi: 10.1016/j.virol.2008.05.026. [DOI] [PubMed] [Google Scholar]
- Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008b;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- Haller O, Kochs G, Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe A, Beech SJ, Dimmock J, Leppard KN. Interaction of the adenovirus type 5 E4 Orf3 protein with promyelocytic leukemia protein isoform II is required for ND10 disruption. J Virol. 2006;80:3042–3049. doi: 10.1128/JVI.80.6.3042-3049.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie-Inoue K, Inoue S. Epigenetic and proteolytic inactivation of 14-3-3sigma in breast and prostate cancers. Semin Cancer Biol. 2006;16:235–239. doi: 10.1016/j.semcancer.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Kratovac Z, Virgen CA, Bibollet-Ruche F, Hahn BH, Bieniasz PD, Hatziioannou T. Primate lentivirus capsid sensitivity to TRIM5 proteins. J Virol. 2008;82:6772–6777. doi: 10.1128/JVI.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lynch PA, Chien CY, Montelione GT, Krug RM, Berman HM. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol. 1997;4:896–899. doi: 10.1038/nsb1197-896. [DOI] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig S, Wang X, Ehrhardt C, Zheng H, Donelan N, Planz O, Pleschka S, Garcia-Sastre A, Heins G, Wolff T. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J Virol. 2002;76:11166–11171. doi: 10.1128/JVI.76.21.11166-11171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr, Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mische CC, Javanbakht H, Song B, Diaz-Griffero F, Stremlau M, Strack B, Si Z, Sodroski J. Retroviral restriction factor TRIM5alpha is a trimer. J Virol. 2005;79:14446–14450. doi: 10.1128/JVI.79.22.14446-14450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- Opitz B, Rejaibi A, Dauber B, Eckhard J, Vinzing M, Schmeck B, Hippenstiel S, Suttorp N, Wolff T. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2006 doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008 doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, Shaw ML, Munoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, Garcia-Sastre A, Basler CF. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol. 2003;77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz S, Sun Q, Nakhaei P, Romieu-Mourez R, Goubau D, Julkunen I, Lin R, Hiscott J. Induction of IRF-3 and IRF-7 phosphorylation following activation of the RIG-I pathway. Cell Mol Biol (Noisy-le-grand) 2006;52:17–28. [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, et al. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlivan M, Zamarin D, Garcia-Sastre A, Cullinane A, Chambers T, Palese P. Attenuation of equine influenza viruses through truncations of the NS1 protein. J Virol. 2005;79:8431–8439. doi: 10.1128/JVI.79.13.8431-8439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EJ, Marie I, Prakash A, Garcia-Sastre A, Levy DE. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Ikappa B kinase but is blocked by Vaccinia virus E3L protein. J Biol Chem. 2001;276:8951–8957. doi: 10.1074/jbc.M008717200. [DOI] [PubMed] [Google Scholar]
- Solorzano A, Webby RJ, Lager KM, Janke BH, Garcia-Sastre A, Richt JA. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J Virol. 2005;79:7535–7543. doi: 10.1128/JVI.79.12.7535-7543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Matano T. Host factors involved in resistance to retroviral infection. Microbiol Immunol. 2008;52:318–325. doi: 10.1111/j.1348-0421.2008.00040.x. [DOI] [PubMed] [Google Scholar]
- Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, Garcia-Sastre A. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yondola MA, Hearing P. The adenovirus E4 ORF3 protein binds and reorganizes the TRIM family member transcriptional intermediary factor 1 alpha. J Virol. 2007;81:4264–4271. doi: 10.1128/JVI.02629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev. 2007;18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Zou W, Wang J, Zhang DE. Negative regulation of ISG15 E3 ligase EFP through its autoISGylation. Biochem Biophys Res Commun. 2007;354:321–327. doi: 10.1016/j.bbrc.2006.12.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.