Abstract
Six new endomorphin analogues, incorporating constrained amino acids in place of native proline have been synthesized. Residues of (S)-azetidine-2-carboxylic acid (Aze), 3,4-didehydro-(S)-proline (Δ3Pro), azetidine-3-carboxylic acid (3Aze) and didehydro-alanine (ΔAla) have been used to prepare [Δ3Pro2]EM-2 (1), [Aze2]EM-1 (2), [Aze2]EM-2 (3), [3Aze2]EM-1 (4), [3Aze2]EM-2 (5) and [ΔAla2]EM-2 (6). Binding assays and functional bioactivities for μ- and δ-receptors are reported. The highest affinity, bioactivity and selectivity is shown by peptides 2 and 3 containing the Aze residue.
Keywords: azetidine carboxylic acids; 3,4-didehydro-(S)-proline; didehydro-alanine; endomorphins; μ-receptors
The endogenous neuropeptides endomorphin-1 (Tyr-Pro-Trp-Phe-NH2; EM-1) and endomorphin-2 (Tyr-Pro-Phe-Phe-NH2; EM-2), originally isolated from bovine brain1 and subsequently from the human brain cortex,2 continue to attract considerable attention in the field of opioid peptides. In comparison with the other endogenous peptide ligands of opioid receptors, endomorphins (EMs) combine potency and efficacy with high affinity and selectivity towards the μ-opioid receptor,1,3,4 the receptor most involved with acute analgesic effects in the central nervous system.5 Furthermore, the EMs proteolytic stability, although low on a general scale, is the highest among the known endogenous opioid peptides6,7 and their activity is accompanied by reduced cardiorespiratory side effects, as compared with the reference alkaloid ligand morphine.8,9 All of these properties, render EMs the object of continuous investigations and a promising target for the development of new opioid analgesics. However, as found in the case of most short linear natural peptides, native EMs lack critical therapeutic characteristics such as bioavailability, duration of action and oral activity.10 Research efforts are then focused on the design of analogues and peptidomimetics in order to improve EMs properties as potential drugs as well as to better understand the key structural and conformational features on which receptor recognition and binding are based.11
Several chemical modifications performed on EMs have been described previously and many of them were focused on the Pro at position 2.12–20 The presence of Pro, the sole proteinogenic amino acid possessing a cyclic structure and a secondary amino group, represents a crucial factor in determining the structural and conformational properties of proteins and peptides. In the specific context of studies on EMs analogues, the Pro2 is considered a stereochemical spacer, capable of favouring proper spatial orientation of the aromatic side chain groups, a key factor for ligand recognition and interaction with the receptor.21,22
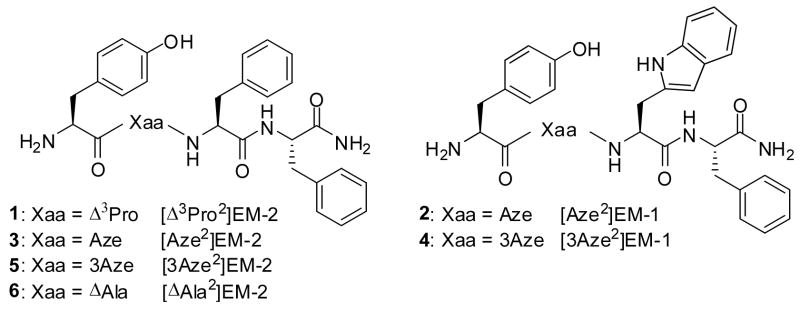
This paper reports on the EM analogues given in Fig. 1, which are obtained by replacing the Pro in position 2 with one of the four amino acid residues depicted in Fig. 2. These amino acids, characterized by different constraints and structural features, can deeply alter the tetrapeptide backbone conformation and thus modulate in new ways the bioactivity of the peptide.
Figure 1.
Molecular structure of the EM analogues 1–6
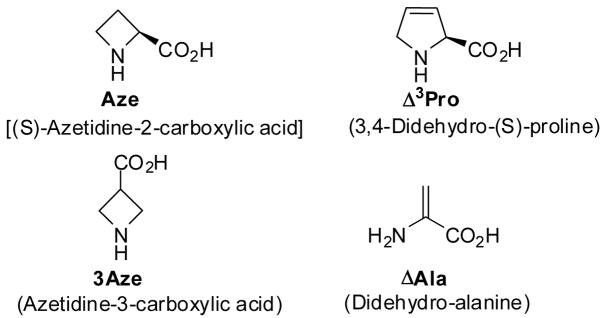
Figure 2.
Molecular structure of the amino acids used in the present paper to replace the native Pro2 residue in endomorphins: (S)-Azetidine-2-carboxylic (Aze); 3,4-Didehydro-(S)-proline (Δ3Pro); Azetidine-3-carboxylic acid (3Aze); Didehydro-alanine (ΔAla).
A first modification, which leads to the EM-2 analogue 1, is based on the introduction of the 3,4-didehydro-(S)-proline (Δ3Pro) residue. This amino acid, as compared with Pro, has an almost planar and less flexible cyclic side chain ring, which does not allow rapid interconversion between ring-puckered forms typical of Pro residues and enhances, at the same time, the population of cis conformers around the CO-N tertiary amide bond.23,24 In addition to this, the presence of the double bond may significantly improve, through π-π interactions, its binding to opioid receptors.25,26 A related Δ3Pro2 containing EM-2 analogue, namely [Dmt1, Δ3Pro2]EM-2, has been previously reported by Toth and Szemenyei27 and utilized for the synthesis of the corresponding tritium labelled [3H2]Pro2 analogue. Two of the other modified ligands here reported analogues contain a cyclic lower homologue of Pro. In particular, in analogues [Aze2]EM-1 (2) and [Aze2]EM-2 (3) the (S)-azetidine-2-carboxylic acid (Aze) has been incorporated. Literature data show that the four-membered ring system of N-acylated (S)-azetidine-2-carboxylic acid (Aze) is either planar or only slightly puckered28–30 and presents, as compared with proline, reduced side chain steric effects, with increased backbone flexibility.31 This latter property generates a larger number of low energy conformers with a population of the cis rotamers around the CO-N amide bond which has been suggested to be higher than that observed in Pro-containing peptides.32 Furthermore, the different ring size may significantly influence the type of folding with β-turns and γ-turns preferentially induced by Pro and Aze, respectively.31
The azetidine ring system also has been incorporated into the backbone of the analogues [3Aze2]EM-1 (4) and [3Aze2]EM-2 (5). Azetidine-3-carboxylic acid (3Aze) is an achiral conformationally restricted analogue of β-alanine which, although not used in opioid chemistry, has been employed for the synthesis of other pharmaceutical compounds.33–35 As compared with Aze and Pro residues, 3Aze maintains the cyclic structure and the secondary amino group in the ring but is devoid of both chirality and restricted rotation around the N-Cα bond. Among 5-membered chiral β-residues related to 3Aze, both β-proline (pyrrolidine-3-carboxylic acid) and β-homoproline (2-pyrrolidineacetic acid) have already been used to replace Pro2 in EM analogues.14,15 Furthermore, EM-2 analogues containing cyclic 6-membered higher homologues of β-proline and 3Aze [viz nipecotic acid (piperidine-3-carboxylic acid) and isonipecotic acid (piperidine-4-carboxylic acid), respectively)] have been recently reported.20
The last analogue under study here is [ΔAla2]EM-2 (6) which contains the rigid skeleton of dehydro-alanine (ΔAla). The local conformational restriction imposed on the backbone by the planar structure of ΔAla moiety, as well as its electronic distribution, significantly influences the properties of this compound. Although a variety of molecular modifications performed on native EMs have been reported, analogues containing α,β-unsaturated amino acid residues are very scarce.36 Thus, the reported data on compound 6 represent, to the best of our knowledge, the first information available in the literature on receptor binding affinity and selectivity of this type of EM analogues.
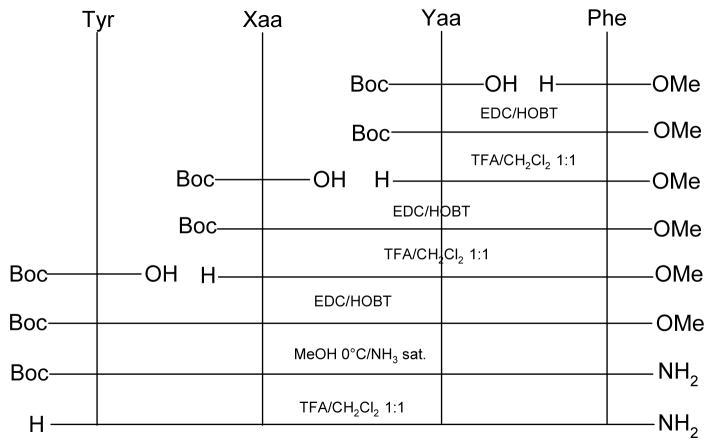
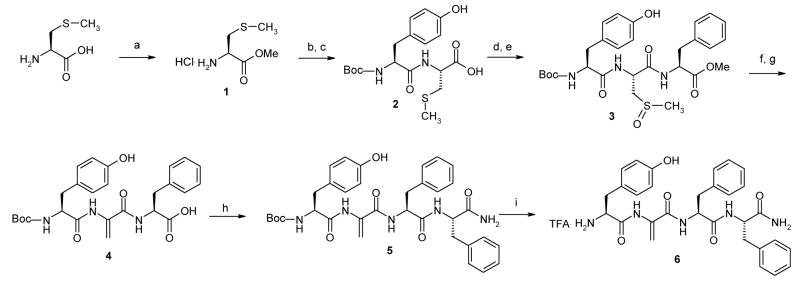
The solution phase synthesis of peptides 1–5 were performed following standard literature methods37 which are given in Scheme 1. The synthesis of the dehydroalanine-containing analogue 6, was accomplished by exploiting the reactivity of the methylcysteine sulfoxide38 as illustrated in Scheme 2. All new compounds were characterized by 1H-NMR data and elemental analysis.39
Scheme 1.
Synthesis of EM analogues 1–5. Xaa = Δ3Pro; Yaa = Phe (1); Xaa = Aze; Yaa = Trp (2); Xaa = Aze; Yaa = Phe (3); Xaa = 3Aze; Yaa = Trp (4); Xaa = 3Aze; Yaa = Phe (5).
Scheme 2.
Reagents and conditions: a) SOCl2, MeOH, 15 min at 0° C then 3h at rt; b) Boc-Tyr-OH, EDC, HOBT, DIPEA, CH2Cl2 30 min at 0° C then 12h at rt; c) 1N NaOH, MeOH 5h at rt; d) HCl·Phe-OMe, EDC, HOBT, DIPEA, CH2Cl2 30 min at 0° C then 12h at rt; e) NaIO4, H2O, Dioxane 1h at 0° C; f) DBU, MeOH, 1.5h at rt; g) 1N NaOH, MeOH 5h at rt; h) HCl·Phe-NH2, EDC, HOBT, DIPEA, CH2Cl2 30 min at 0° C then 12h at rt; i) TFA, CH2Cl21.5h at rt under N2.
Receptor binding affinities to the δ- and μ-opioid receptors were performed using cell membrane preparations from transfected cells that stably express the respective receptors. The radiolabeled ligands used were [3H]DPDPE and [3H]DAMGO for δ- and μ-receptors, respectively (for more details about the procedure see ref. 40). The in vitro tissue bioassays (MVD and GPI/LMMP) were performed as described previously.41 IC50 values represent means of no less than four experiments. IC50 values, relative potency estimates, and their associated standard errors were determined by fitting the data to the Hill equation by a computerized non-linear least-squares method.
As shown in Table 1 all analogues, except [3Aze2]EM-1 (4), which can be considered a μ/δ mixed agonist, show high selectivity for μ-receptors and functional bioactivities in good agreement with the binding values. The highest affinity and selectivity is shown by the Aze-containing peptides 2 and 3. This finding, together with the good activity of the [Δ3Pro2]EM-2 analogue 1, confirms the role of the residue at position 2 as a stereochemical spacer.
Table 1.
Binding affinity and in vitro activity for compounds 1–6.
| Compound | Receptor affinitya,b (nM) |
Selectivity |
Functional Bioactivity |
||
|---|---|---|---|---|---|
| Kiδ | Kiμ | δ/μ | MVDb (IC50) | GPIb (IC50) | |
| 1 [Δ3Pro2]EM-2 | n.c.c | 6.5 ± 2.3 | -- | 70 ±12 | 14±2.1 |
| 2 [Aze2]EM-1 | 3500 ± 360 | 2.3± 0.23 | 1500 | 140 ±21 | 7.9 ± 0.83 |
| 3 [Aze2]EM-2 | 5100 ± 600 | 5.6 ± 1.2 | 920 | 150 ±60 | 6.4 ± 1.2 |
| 4 [3Aze2]EM-1 | 40 ± 8.5 | 34 ± 5.1 | 1.2 | 170 ± 32 | 140 ± 32 |
| 5 [3Aze2]EM-2 | 6900 ± 1200 | 210 ± 51 | 32 | 13 % at 1μM | 1400 ± 280 |
| 6 [ΔAla2]EM-2 | 710 ± 130 | 34 ± 6.3 | 21 | 490 ± 150 | 170 ±23 |
| EM-1d | -- | 10.70 ± 1.5 | -- | 1710 ±110 | 28 ± 3 |
| EM-2 d | -- | 9.56 ± 0.98 | -- | 510 ±35 | 15 ± 2 |
Displacement of [3H]DAMGO (μ-selective) and [3H]DPDPE (δ-selective) from rat brain membrane binding sites.
± S.E.M.
n.c. = data not collected.
Data from ref. 42.
The two analogues containing a β-residue, namely [3Aze2]EM-1 (4) and [3Aze2]EM-2 (5) exhibit highly different activity profiles (Table 1). The much lower activity shown by 5, as compared to the Trp3 containing analogue [3Aze2]EM-1 (4), can be interpreted on the basis of the well recognized role of the aromatic residues on ligand-receptor interactions. Several studies on EM-1 and EM-2 clearly show that aromatic side chains with different size, distinct noncovalent interactions and H-bonding capability, can highly influence conformations43 and binding.21,44,45 Results of ligands 4 and 5 indicate that the beneficial effect on the activity, constantly observed in EM analogues containing the indole ring system at position 3, gives results that are particularly efficient as compared to the Phe3 residue in models containing the achiral 3Aze residue.
The low activity shown by [3Aze2]EM-2 (5) appears, however, is quite unexpected when compared to the high activity shown by EM analogues containing other cyclic β-residues in place of the native Pro2. In particular, among the recently examined EM-2 analogues containing positional isomers of the 6-membered piperidine-carboxylic acid, replacement of Pro2 with the chiral nipecotic acid (viz piperidine-3-carboxylic acid) led, in accordance with the results concerning other related cyclic β-residues,14,15 to a highly active analogue.20 On the contrary, Pro2 replacement with the achiral piperidine-4-carboxylic acid (viz isonipecotic acid) resulted, as in the case of the here reported analogue 5, to a practically inactive ligand.20 It can be argued that the high conformational restriction and the reduced flexibility of the 4-membered achiral 3Aze are detrimental features which prevail over the advantages which, in larger cyclic systems, derive from the presence of a β-positioned carboxylic group.
Peptide (6), containing the rigid skeleton of ΔAla residue, has only modest affinity and bioactivity at the μ-opioid receptor, and this suggests that an efficient constraint at position 2 involves an amino acid residue with a cyclic structure.
Further studies on the synthesis and activity of EM analogues obtained by replacing native amino acid residues with synthetic mimetics are under way in our laboratories and will be reported in due course.
Acknowledgments
This research was supported in part by grants from the U.S. Public Health Service, National Institute of Drug Abuse DA06284 and DA13449 (VJH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Zadina JE, Hackler L, Ge LJ, Kastin AJ. Nature. 1997;386:499. doi: 10.1038/386499a0. [DOI] [PubMed] [Google Scholar]
- 2.Hackler L, Zadina JE, Ge LJ. Peptides. 1997;18:1635. doi: 10.1016/s0196-9781(97)00259-3. [DOI] [PubMed] [Google Scholar]
- 3.Horvath G. Pharmacol Ther. 2000;88:437. doi: 10.1016/s0163-7258(00)00100-5. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg JE, Rossi GC, Letchworth SR, Mathis JP, Ryan-Moro J, Leventhal L, Su W, Emmel D, Bolan EA, Pasternak GW. J Pharmacol Exp Ther. 1998;286:1007. [PubMed] [Google Scholar]
- 5.Matthes HW, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, Befort K, Dierich A, LeMeur M, Dolle P, Tzavara E, Hanoune J, Roques BP, Kieffer BL. Nature. 1996;383:819. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 6.Tömböly Cs, Péter A, Tóth G. Peptides. 2002;23:1573. doi: 10.1016/s0196-9781(02)00100-6. [DOI] [PubMed] [Google Scholar]
- 7.Péter A, Tóth G, Tömböly Cs, Laus G, Tourwé D. J Chromatogr. 1999;846:39. doi: 10.1016/s0021-9673(99)00146-6. [DOI] [PubMed] [Google Scholar]
- 8.Wilson AM, Soignier RD, Zadina JE, Kastin AJ, Nores WL, Olson RD, Olson GA. Peptides. 2000;21:1871. doi: 10.1016/s0196-9781(00)00340-5. [DOI] [PubMed] [Google Scholar]
- 9.Czapla MA, Gozal D, Alea OA, Beckerman RC, Zadina JE. Am J Respir Crit Care Med. 2000;162:994. doi: 10.1164/ajrccm.162.3.9911102. [DOI] [PubMed] [Google Scholar]
- 10.Fichna J, Janecka A, Costentin J, Do Rego J-C. Pharmacol Rev. 2007;59:88. doi: 10.1124/pr.59.1.3. [DOI] [PubMed] [Google Scholar]
- 11.Leitgeb B. Chemistry & Biodiv. 2007;4:2703. doi: 10.1002/cbdv.200790221. [DOI] [PubMed] [Google Scholar]
- 12.Keresztes A, Szűcs M, Borics A, Kövér KE, Forró E, Fülöp F, Tömböly Cs, Péter A, Páhi A, Fábián G, Murányi M, Tóth G. J Med Chem. 2008;51:4270. doi: 10.1021/jm800223t. [DOI] [PubMed] [Google Scholar]
- 13.Fichna J, Do Rego J-C, Chung NN, Lemieux C, Schiller PW, Poels J, Broeck JV, Costentin J, Janecka A. J Med Chem. 2007;50:512. doi: 10.1021/jm060998u. [DOI] [PubMed] [Google Scholar]
- 14.Cardillo G, Gentilucci L, Melchiorre P, Spampinato S. Bioorg Med Chem Lett. 2000;10:2755. doi: 10.1016/s0960-894x(00)00562-x. [DOI] [PubMed] [Google Scholar]
- 15.Cardillo G, Gentilucci L, Qasem AR, Sgarzi F, Spampinato S. J Med Chem. 2002;45:2571. doi: 10.1021/jm011059z. [DOI] [PubMed] [Google Scholar]
- 16.Cardillo G, Gentilucci L, Tolomelli A, Calienni M, Qasem AR, Spampinato S. Org Biomol Chem. 2003;1:1498. doi: 10.1039/b301507f. [DOI] [PubMed] [Google Scholar]
- 17.Doi M, Asano A, Komura E, Ueda Y. Biochem Biophys Res Commun. 2002;297:138. doi: 10.1016/s0006-291x(02)02087-9. [DOI] [PubMed] [Google Scholar]
- 18.Yamada T, Iino M, Matsuoka N, Yanagihara R, Miyazawa T, Shirasu N, Shimohigashi Y. Peptide Sci. 1999;36:441. [Google Scholar]
- 19.Tóth G, Keresztes A, Tömböly Cs, Péter A, Fülöp F, Tourwé D, Navratilova E, Varga É, Roeske WR, Yamamura HI, Szűcs M, Borsodi A. Pure Appl Chem. 2004;76:951. [Google Scholar]
- 20.Staniszewska R, Fichna J, Gach K, Toth G, Poels J, Vanden Broeck J, Janecka A. Chem Biol Drug Des. 2008;72:91. doi: 10.1111/j.1747-0285.2008.00678.x. [DOI] [PubMed] [Google Scholar]
- 21.Leitgeb B, Tóth G. Eur J Med Chem. 2005;40:674. doi: 10.1016/j.ejmech.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Okada Y, Fukumizu A, Takahashi M, Shimizu Y, Tsuda Y, Yokoi T, Bryant SD, Lazarus LH. Biochem Biophys Res Commun. 2000;276:7. doi: 10.1006/bbrc.2000.3416. [DOI] [PubMed] [Google Scholar]
- 23.Benedetti E, Di Blasio B, Pavone V, Pedone C, Felix A, Goodman M. Biopolymers. 1981;20:283. doi: 10.1002/bip.1981.360200204. [DOI] [PubMed] [Google Scholar]
- 24.Flores-Ortega A, Casanovas J, Zanuy D, Nussinov R, Alemán C. J Phys Chem B. 2007;111:5475. doi: 10.1021/jp0712001. [DOI] [PubMed] [Google Scholar]
- 25.Moore S, Felix AM, Meienhofer J. J Med Chem. 1977;20:495. doi: 10.1021/jm00214a007. [DOI] [PubMed] [Google Scholar]
- 26.Werner S, Kasi D, Brummond KM. J Comb Chem. 2007;9:677. doi: 10.1021/cc070011p. [DOI] [PubMed] [Google Scholar]
- 27.Szemenyei E, Toth G. J Label Compd Radiopharm. 2007;50:1148. [Google Scholar]
- 28.Toniolo C. Int J Pept Protein Res. 1990;35:287. doi: 10.1111/j.1399-3011.1990.tb00052.x. [DOI] [PubMed] [Google Scholar]
- 29.Zagari A, Némethy G, Scheraga HA. Biopolymers. 1990;30:951. doi: 10.1002/bip.360300909. [DOI] [PubMed] [Google Scholar]
- 30.Shuman RT, Rothenberger RB, Campbell CS, Smith GF, Gifford-Moore D, Paschal JW, Gesellchen PD. J Med Chem. 1995;38:4446. doi: 10.1021/jm00022a009. [DOI] [PubMed] [Google Scholar]
- 31.Baeza JL, Gerona-Navarro G, Pérez de Vega MJ, García-López MT, González-Muñiz R, Martín-Martínez M. J Org Chem. 2008;73:1704. doi: 10.1021/jo701746w. [DOI] [PubMed] [Google Scholar]
- 32.Kern D, Schutkowski M, Drakenberg T. J Am Chem Soc. 1997;119:8403. [Google Scholar]
- 33.Macdonald SJF, Dowle MD, Harrison LA, Clarke GDE, Inglis GGA, Johnson MR, Shah P, Smith RA, Amour A, Fleetwood G, Humphreys DC, Molloy CR, Dixon M, Godward RE, Wonacott AJ, Singh OM, Hodgson ST, Hardy GW. J Med Chem. 2002;45:3878. doi: 10.1021/jm020881f. [DOI] [PubMed] [Google Scholar]
- 34.Fish P, Barber CG, Brown DG, Butt R, Collis MJ, Dickinson RP, Henry BT, Horne VA, Huggins JP, King E, O’Gara M, McCleverty D, McIntosh F, Phillips C, Webster R. J Med Chem. 2007;50:2341. doi: 10.1021/jm061066t. [DOI] [PubMed] [Google Scholar]
- 35.Klein SI, Czekaj M, Molino BF, Chu V. Bioorg Med Chem Lett. 1997;7:1773. [Google Scholar]
- 36.Brown W, Dimaio J, Schiller P, Martel R. CAN. 1995;124:30439. World Patent WO 9522557. [Google Scholar]
- 37.Bodansky M. Principles of Peptide Synthesis. Springer-Verlag; Berlin: 1984. [Google Scholar]
- 38.Burrage S, Raynham T, Williams G, Essex JW, Allen C, Cardno M, Swali V, Bradley M. Chem Eur J. 2000;6:1455. doi: 10.1002/(sici)1521-3765(20000417)6:8<1455::aid-chem1455>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 39.Spectral data and elemental analysis of compounds 1–6. TFA·H-Tyr-Δ3 Pro-Phe-Phe-NH2 (1): 1H NMR (DMSO- d6)δ: 2.83–3.09 (6H, m, Phe3 β-CH2, Phe4β-CH2, Tyr β-CH2), 3.93–4.61 (5H, m, Phe3 α-CH, Phe4 α-CH, Tyr α-CH and Δ3Pro C5H2), 5.18 (1H, s, Δ3Pro α-CH), 5.72 and 6.01 (2H, 2 br, Δ3Pro CH=CH), 6.09–7.36 (16H, m, aromatics and CONH2), 8.12–8.25 (5H, m, Phe3 NH, Phe4 NH, Tyr NH3+), 9.51 (1H, br, Tyr OH). Anal. Calcd for C34H36F3N5O7: C, 59.73; H, 5.31; N, 10.24. Found: C, 59.55; H, 5.36; N, 10.30. TFA·H-Tyr-Aze-Trp-Phe-NH2 (2): 1H NMR (DMSO-d6)δ: 1.78 and 2.29 (2H, two m, Aze C3H2), 2.60–3.20 (6H, m, Phe β-CH2, Trp β-CH2 and Tyr β-CH2), 3.67 and 4.04 (2H, two m, AzeC4H2), 3.94 (1H, m, Tyr α-CH), 4.41 (1H, m, Trp α-CH), 4.48 (1H, m, Phe α-CH), 4.60 (1H, m, Aze α-CH), 6.55–7.35 (16H, m, aromatics and CONH2), 8–8.25 (5H, m, Phe NH, Trp NH-CO and Tyr NH3+), 9.39 (1H, s, Tyr OH), 10.78 (1H, s, Trp NH). Anal. Calcd for C35H37F3N6O7: C, 59.15; H, 5.25; N, 11.82. Found: C, 59.27; H, 5.26; N, 11.83. TFA·H-Tyr-Aze-Phe-Phe-NH2 (3): 1H NMR (DMSO-d6) δ: 1.78 and 2.31 (2H, two m, Aze C3H2), 2.59–3.22 (6H, m, Phe3 β-CH2, Phe4 β-CH2 and Tyr β-CH2), 3.77 and 4.10 (2H, two m, Aze C4H2), 3.99 (1H, m, Tyr α-CH), 4.45–4.50 (2H, m, Phe3 α-CH and Phe4 α-CH), 4.62 (1H, m, Aze α-CH), 6.60–7.34 (16H, m, aromatic and CONH2), 8.01–8.28 (5H, m, Phe3 NH, Phe4 NH and Tyr NH3+), 9.4 (1H, s, Tyr OH). Anal. Calcd for C33H36F3N5O7: C, 59.01; H, 5.40; N, 10.43. Found: C, 59.24; H, 5.42; N, 10.40. TFA·H-Tyr-3Aze-Trp-Phe-NH2 (4): 1H NMR (DMSO-d6) δ: 2.55–3.90 (11H, m, 3Aze C3H2, 3-Aze C4H2, Tyr β-CH2, Trp β-CH2, Phe β-CH2 and 3Aze α-CH), 3.99 (1H, m, Tyr α-CH), 4.42 (1H, m, Trp α-CH), 4.52 (1H, m, Phe α-CH), 6.58–7.59 (16H, m, aromatic and CONH2), 8.04–8.33 (5H, m, Trp NH-CO, Phe NH and Tyr NH3+), 9.4 (1H, s, Tyr OH), 10.79 (1H, s, Trp NH). Anal. Calcd for C35H37F3N6O7: C, 59.15; H, 5.25; N, 11.82. Found: C, 59.29; H, 5.27; N, 11.78. FA·H-Tyr-3Aze-Phe-Phe-NH2 (5): 1H NMR (DMSO-d6) δ: 2.55–3.90 (11H, m, 3Aze C3H2, 3-Aze C4H2, Tyr β-CH2, Phe3 β-CH2, Phe4 β-CH2 and 3Aze α-CH), 3.99 (1H, m, Tyr α-CH), 4.47–4.53 (2H, m, Phe3 α-CH and Phe4 α-CH), 6.61–7.36 (16H, m, aromatics and CONH2), 8–8.31(5H, m, Phe3 NH, Phe4 NH and Tyr NH3+), 9.41 (1H, s, Tyr OH). Anal. Calcd for C33H36F3N5O7: C, 59.01; H, 5.40; N, 10.43. Found: C, 59.22; H, 5.41; N, 10.45. TFA·H-Tyr-ΔAla-Phe-Phe-NH2 (6): 1H NMR (DMSO- d6) δ: 2.74–3.07 (6H, m, Tyr β-CH2, Phe3 β-CH2, Phe4 β-CH2), 4.24 (1H, m, Tyr α-CH), 4.50-4-58 (2H, m, Phe3 α-CH, Phe4 α-CH), 5.54 and 6.17 (2H, two s, ΔAla CH2), 6.69–7.29 (16H, m, aromatics and CONH2), 7.46 (1H, s, ΔAla NH), 8.09 (3H, br, Tyr NH3+), 8.18 and 8.56 (2H, two d, Phe3 NH, Phe4 NH), 9.69 (1H, s, Tyr OH). Anal. Calcd for C32H34F3N5O7: C, 58.44; H, 5.21; N, 10.65. Found: C, 58.37; H, 5.26; N, 10.61.
- 40.Wang Z, Gardell LR, Ossipov MH, Vanderah TW, Brennan BB, Hochgeschwender U, Hruby VJ, Malan TP, Jr, Lai J, Porreca F. J Neurosci. 2001;21:1779. doi: 10.1523/JNEUROSCI.21-05-01779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kramer TH, Davis P, Hruby VJ, Burks TF, Porreca F. J Pharmacol Exp Ther. 1993;266:577. [PubMed] [Google Scholar]
- 42.Biondi B, Giannini E, Negri L, Melchiorri P, Lattanzi R, Rosso F, Ciocca L, Rocchi R. Int J Pept Res Ther. 2006;2:145. [Google Scholar]
- 43.Gentilucci L, Tolomelli A. Curr Top Med Chem. 2004;4:105. doi: 10.2174/1568026043451627. [DOI] [PubMed] [Google Scholar]
- 44.Yu Y, Shao X, Cui Y, Liu H-m, Wang C-l, Fan Y-z, Liu J, Dong S-l, Cui Y-x, Wang R. Chem Med Chem. 2007;2:309. [Google Scholar]
- 45.Podlogar BL, Paterlini MG, Ferguson DM, Leo GC, Demeter DA, Brown FK, Reitz AB. FEBS Lett. 1998;439:13. doi: 10.1016/s0014-5793(98)01202-2. [DOI] [PubMed] [Google Scholar]