Abstract
Bacterial efflux pumps have traditionally been studied as low-level drug resistance determinants. Recent insights have suggested that efflux systems are often involved with fundamental cellular physiological processes, suggesting that drug extrusion may be a secondary function. In Mycobacterium tuberculosis, little is known about the physiological or drug resistance roles of efflux pumps. Using Mycobacterium bovis BCG as a model system, we showed that deletion of the Rv1410c gene encoding the P55 efflux pump made the strain more susceptible to a range of toxic compounds, including rifampin (rifampicin) and clofazimine, which are first- and second-line antituberculosis drugs. The efflux pump inhibitors carbonyl cyanide m-chlorophenylhydrazone (CCCP) and valinomycin inhibited the P55-determined drug resistance, suggesting the active export of the compounds by use of the transmembrane proton and electrochemical gradients as sources of energy. In addition, the P55 efflux pump mutant was more susceptible to redox compounds and displayed increased intracellular redox potential, suggesting an essential role of the efflux pump in detoxification processes coupled to oxidative balance within the cell. Finally, cells that lacked the p55 gene displayed smaller colony sizes and had a growth defect in liquid culture. This, together with an increased susceptibility to the cell wall-targeting compounds bacitracin and vancomycin, suggested that P55 is needed for proper cell wall assembly and normal growth in vitro. Thus, P55 plays a fundamental role in oxidative stress responses and in vitro cell growth, in addition to contributing to intrinsic antibiotic resistance. Inhibitors of the P55 efflux pump could help to improve current treatments for tuberculosis.
Tuberculosis (TB) continues to be one of the main causes of morbidity and mortality worldwide. The most recent WHO report estimates that there are 9.2 million new cases of TB and 1.7 million deaths per year (41), significant numbers of which occur among human immunodeficiency virus-positive patients. Along with human immunodeficiency virus coinfection, multidrug-resistant TB (MDR-TB) and extensively drug-resistant TB (XDR-TB) pose major threats that challenge TB control, especially in those regions of the world with the highest burden of TB.
Efflux pumps are membrane proteins that export substrates from bacterial and eukaryotic cells. They confer resistance to anticancer drugs in tumor cells and to antibiotics in bacteria, often providing low levels of intrinsic multidrug resistance (25). Their activities allow better tolerance of drugs and thus may potentiate the acquisition of chromosomal mutations that provide higher levels of resistance (29, 30). In recent years, it has become evident that efflux pumps have important functions in many other cellular processes, such as physiological homeostasis, resistance to stress conditions, lipid transport, and virulence (24).
While drug resistance in Mycobacterium tuberculosis clinical isolates is often due to the acquisition of mutations in genes encoding drug targets or enzymes activating prodrugs, such mutations are not found in many low-level-drug-resistant isolates, suggesting the contribution of efflux pumps (11). In fact, many M. tuberculosis efflux pumps contribute to drug resistance under laboratory conditions, and there are reports describing increases in the levels of expression of efflux pumps in various drug-resistant M. tuberculosis isolates (11, 15, 17, 32, 33, 38). In addition, the inactivation of certain efflux pumps attenuates M. tuberculosis, indicating that, like other bacterial pathogens, efflux pumps also contribute to virulence (10).
We have previously characterized the contribution of the M. tuberculosis P55 efflux pump to drug resistance in Mycobacterium smegmatis (39). The gene encoding the P55 efflux pump, Rv1410c, forms an operon with Rv1411c (5), which encodes the lipoprotein LprG. Both genes are predicted to support the growth of M. tuberculosis in vivo (6, 37). A recent report has demonstrated that this operon is required for survival in the presence of ethidium bromide and for maintenance of a normal cell surface composition in M. smegmatis (13).
In the study described here, we characterized P55 in the TB vaccine strain, M. bovis BCG. Our results demonstrate that P55 plays a role in at least three important processes: it extrudes and thus provides resistance to several drugs (including rifampin [rifampicin], one of the most important frontline TB drugs), it is part of the oxidative stress response, and it is needed to maintain normal growth characteristics both on solid medium and in liquid medium.
MATERIALS AND METHODS
Gene nomenclature.
M. tuberculosis and M. bovis BCG have a DNA sequence identity of greater than 99.9% (3). The nucleotide sequences of the M. tuberculosis Rv1410c and Rv1411c genes (http://genolist.pasteur.fr/TubercuList/) are identical to those of the BCG1471c and BCG1472c genes, respectively, from M. bovis BCG Pasteur 1173 P2 (http://genolist.pasteur.fr/BCGList/). In this report, both Rv1410c and BCG1471c are referred to as p55, and both Rv1411c and BCG1472c are referred to as p27.
Bacterial strains, growth conditions, and chemicals.
The strains used in this study are listed in Table 1. The sequences of the oligonucleotides used are available upon request. M. bovis BCG was cultivated at 37°C in Middlebrook 7H9 broth (Difco) supplemented with 10% Middlebrook albumin-dextrose-catalase (Difco) and 0.05% (vol/vol) Tween 80 or on Middlebrook 7H10 agar plates (Difco) supplemented with 10% (vol/vol) oleic acid albumin-dextrose-catalase (Difco). Escherichia coli was grown at 37°C in Luria-Bertani (LB) broth or on LB agar plates. For the selection of resistance markers in mycobacteria, hygromycin or kanamycin was added to the cultures at final concentrations of 10 mg/liter and 20 mg/liter, respectively. Plasmids were maintained in E. coli with appropriate antibiotics for selection (100 mg/liter of ampicillin, 20 mg/liter of kanamycin). Acriflavine, amikacin, bacitracin, carbonyl cyanide m-chlorophenylhydrazone (CCCP), cerulenin, d-cycloserine, ethidium bromide, ciprofloxacin, clofazimine, chloramphenicol, chlorpromazine, diamide, dl-dithiothreitol, econazole, ethambutol, ethionamide, fluconazole, gentamicin, hydrogen peroxide, imipenem, isoniazid, l-glutathione (reduced), menadione, monobromobimane (mBBr), novobiocin, ofloxacin, p-aminosalicylic acid, PA-824, reserpine, rifampin, streptomycin, spectinomycin, tetracycline, triclosan, valinomycin, and vancomycin were used in the mycobacterial antimicrobial susceptibility tests.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| M. bovis BCG | ||
| Pasteur 1173 | Wild type | Laboratory collection |
| KOP55 | Pasteur p55::Ωhyg | This study |
| KOP55 PAZ23 | Strain KOP55 containing pPAZ23 | This study |
| PAZ23 | Strain Pasteur 1173 containing pPAZ23 | This study |
| Plasmids | ||
| pGEM-T Easy | E. coli cloning vector, ampicillin resistance | Promega |
| pIJ2925 | Cloning vector, pUC18 derivative with BglII sites flanking MCS,a ampicillin resistance | 16 |
| pSUM series | E. coli-mycobacteria shuttle vectors, nontranscribed MCS, kanamycin resistance | 2 |
| p2NIL | Gene manipulation mycobacterial suicide vector, kanamycin resistance | 27 |
| pGOAL17 | PAg85-lacZ Phsp60-sacB PacI cassette vector, ampicillin resistance | 27 |
| pHP45-Ωhyg | Ωhyg, ampicillin resistance | 31 |
| pSAN6 | p27-p55 cloned into pIJ2925 | This study |
| pPAZ23 | Pp27-p55-p27-p55 cloned into pSUM41 | 39 |
| pILI10 | p27-p55 cloned into p2NIL | This study |
| pILI11 | p27-p55::Ωhyg cloned into p2NIL | This study |
| pILI12 | p27-p55::Ωhyg cloned into p2NIL with pGOAL17 PacI cassette | This study |
MCS, multicloning site.
DNA manipulations.
DNA manipulations were carried out by standard techniques (35). Mycobacterial genomic DNA was isolated as described previously (26). Southern blotting analysis was done with an ECL direct nucleic acid labeling and detection system (Amersham Biosciences), according to the manufacturer's instructions. Both E. coli and the mycobacteria were transformed by electroporation with a Gene Pulser (Bio-Rad Laboratories Inc., Richmond, CA) (26).
Strain construction.
The plasmids used for the overexpression and inactivation of the p55 gene in M. bovis BCG are listed in Table 1.
(i) Overexpression.
The p27-p55 operon, expressed under the control of its own promoter, was cloned into the pSUM41 vector, yielding plasmid pPAZ23 (39). Plasmid pPAZ23 was electroporated into M. bovis BCG, resulting in M. bovis BCG PAZ23 (Table 1).
(ii) Inactivation and complementation.
A suicide delivery plasmid containing the p55 gene interrupted by the insertion of a hygromycin resistance cassette (Ωhyg) was constructed (pILI12). Initially, a 2.1-kb PCR product from M. tuberculosis H37Rv genomic DNA (with an engineered EcoRI site on one side) containing the p27-p55 operon was cloned into the pGEM-T Easy vector. The fragment was then removed by EcoRI digestion and cloned into the EcoRI-digested vector pIJ2925, resulting in plasmid pSAN6. A 2.1-kb BglII-Asp718I fragment from pSAN6 was transferred into BamHI-Asp718I-digested vector p2NIL, resulting in plasmid pILI10. p55 was then interrupted by the insertion of a Ωhyg cassette (derived from BamHI-digested plasmid pHP45-Ωhyg) into its unique Bsp120I restriction site, resulting in plasmid pILI11. A cassette containing the lacZ and sacB genes from pGOAL17 was then cloned into the single PacI site of pILI11 to generate the suicide delivery vector pILI12. pILI12 was used to transform M. bovis BCG. Single-crossover (SXO) and double-crossover (DXO) transformants were selected as described elsewhere (27). Candidates for SXO and DXO were analyzed by PCR with primers specific for the p55 gene flanking the Ωhyg insertion point and primers specific for the kanamycin resistance gene of the vector. Candidates that had the expected PCR patterns were streaked onto plates with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and either kanamycin or hygromycin for phenotypic analysis. Finally, Southern blotting analysis was done with PvuII- and PstI-digested DNA (Fig. 1 and data not shown, respectively) to confirm SXO and DXO. The M. bovis BCG strain with the inactivated p55 gene was named M. bovis BCG KOP55 (Table 1). M. bovis BCG KOP55 was complemented by introducing pPAZ23 containing the p27-p55 operon, resulting in M. bovis BCG KOP55 PAZ23.
FIG. 1.
Southern blot analysis for p55 gene inactivation. Genomic DNA isolated from the M. bovis BCG wild-type (WT), M. bovis BCG single-crossover (SXO), and M. bovis BCG double-crossover (DXO) strains was digested with PvuII and hybridized to a probe corresponding to a 0.9-kb p55 internal region. The DXO strain carrying the inactivated p55 gene lost the wild-type 5-kb band and gained a 7.3-kb band, reflecting the insertion of the 2.3-kb hygromycin resistance cassette (cf. lane WT and lane DXO). The SXO strain showed two bands (9.3 and 7.2 kb) due to the incorporation of the suicide vector in the genome. The diagram showing the expected PvuII digestion fragment is not shown to scale. Ωhyg, cassette containing the hygromycin resistance gene; pGOAL17, cassette containing the lacZ and sacB genes; Km, kanamycin resistance gene from p2NIL vector.
Drug susceptibility assays.
Two methods were used to determine the drug susceptibilities of the M. bovis BCG strains. First, a conventional disc assay was used to test the redox compounds. Briefly, a suspension containing 106 cells/ml was spread on Middlebrook 7H10 agar plates, and discs containing the redox compounds were placed onto the lawn. Halos were recorded after 14 days at 37°C. Second, MICs were determined by using serial twofold dilutions of the compounds in both liquid and solid media. For determination of the MICs in liquid medium, the resazurin assay was carried out essentially as described previously (32), except that the plates were incubated for 8 days at 37°C and for an additional 2 days after the addition of the redox indicator (resazurin). For agar-based determinations, log-phase cultures were diluted to 105 cells/ml and 10 μl was spotted onto Middlebrook 7H10 agar plates containing serial twofold antibiotic dilutions. Visible growth was scored after 21 days of incubation at 37°C. The experiments were carried out in triplicate and were repeated at least three times.
MTT assays.
The tetrazolium salt 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was used to estimate the intracellular redox potential, essentially as described previously (7), with some modifications. Briefly, M. bovis BCG cultures at an optical density (OD) at 600 nm of 0.2 were treated at 37°C with drug or solvent alone for 1 h (100 μl/well in Honeycomb 2 plates in quadruplicate) before the addition of 25 μl of 2 mg/ml MTT and were then incubated for an additional 3 h. The absorbance at 600 nm was recorded after the formazan precipitate was solubilized by the addition of 25 μl of 10% sodium dodecyl sulfate and subsequent shaking for 30 min in a BioScreen C plate reader (Oy Growth Curves Ab Ltd., Helsinki, Finland). All drugs except menadione and diamide were added at a concentration of 10-fold the MIC for the wild-type strain. Menadione was added at eightfold the MIC, and diamide was added at the MIC. The assay was repeated at least three independent times.
Growth kinetics.
Experiments were carried out in a BioScreen C plate reader (Oy Growth Curves Ab Ltd.) at 37°C with continuous shaking. The cells were grown to stationary phase, and quadruplicate Honeycomb 2 plate wells were inoculated with 250 μl of 105 cells/ml. The absorbance at 600 nm was recorded every 12 h for 18 days.
RESULTS
P55 efflux pump drug resistance phenotypes are dependent on genetic background.
A M. bovis BCG Pasteur derivative in which the gene encoding the P55 efflux pump (Rv1410c) was inactivated (strain KOP55) was constructed by the insertion of a cassette containing the hygromycin resistance gene (hyg) and a transcriptional terminator. Previous studies have shown a decrease in susceptibility to tetracycline and aminoglycoside when the p55 gene is overexpressed in M. smegmatis (39) and that deletion of the chromosomal p55 ortholog in M. smegmatis results in increased susceptibility to ethidium bromide (13). However, in M. bovis BCG, the similar p55 overexpression or inactivation constructions did not alter resistance to these drugs (data not shown). The genetic divergence between M. smegmatis (6.98-megabase genome, 67.4% G+C content) and M. bovis BCG (4.37-megabase genome, 65.6% G+C content), which have an average of 68% nucleotide identity between orthologous genes, could explain this difference in their phenotypes. Paralogous transporters in different species may be controlled by different regulatory systems and/or may have different substrate specificities. In addition, comparative genetic studies may be complicated by the presence of transporters with overlapping substrate specificities. These results reinforce the observation that efflux pump substrate specificity and the phenotypes of null mutants or overexpressing strains are determined by the genetic background of the bacterial host (24).
Microarray data analysis reveals possible P55 efflux pump substrates.
Studies of compounds that induce P55 expression might give important insights into its substrates. To investigate this possibility, a microarray database (generated by Boshoff et al. [7]) which recorded the transcriptional responses of M. tuberculosis to a series of 75 different drugs and physiological conditions was analyzed. Eight conditions which increased or decreased p27 and p55 gene expression at least twofold in at least three independent experiments were selected (Table 2). The levels of expression of both genes increased after treatments with chlorpromazine, which is a phenothiazine predicted to affect respiration; novobiocin, which is a gyrase inhibitor; and PA-824, which inhibits the synthesis of protein and cell wall lipid (40). Other compounds induced only either p27 or p55. The level of expression of p55 was increased by treatment with clofazimine (a second-line anti-TB drug), which disrupts the membrane potential; triclosan, which inhibits fatty acid biosynthesis in bacteria; and thioridazine, another phenothiazine which also affects respiration. Menadione, which is both an inhibitor of respiration and a generator of superoxide, increased the expression of only p27. Significant decreases in the levels of expression of these genes were observed only after treatment with valinomycin. This drug, which facilitates the transfer of potassium ions across cell membranes and which thus dissipates the transmembrane potential, decreased the levels of expression of both genes. Together, these data provide general concepts and specific compounds for analysis in the context of the P55 phenotypes.
TABLE 2.
Analysis of Boshoff databasea for p27 and p55 genes from Mycobacterium tuberculosis
| Treatmentb | Fold induction
|
|||
|---|---|---|---|---|
| Up (>2-fold)
|
Down (<0.5-fold)
|
|||
| p27 | p55 | p27 | p55 | |
| Chlorpromazine | x | x | ||
| Clofazimine | x | |||
| Menadione | x | |||
| Triclosan | x | |||
| Thioridazine | x | |||
| Valinomycin | x | x | ||
| Novobiocin | x | x | ||
| PA-824 | x | xc | ||
The database of Boshoff et al. (7).
Treatments for which p27 and p55 gene expression was found to be up- or downregulated at least twofold in three or more independent experiments.
Microarray analyses for which p55 gene expression was upregulated at least twofold only in two independent experiments.
Inactivation of the p55 gene in M. bovis BCG resulted in increased susceptibility to rifampin and other drugs.
Drug susceptibility assays were carried out to determine whether the M. bovis BCG KOP55 mutant had become sensitive to a range of different compounds (Table 3). To explore the possibility that the regulatory responses predict functional properties, compounds in the database of Boshoff et al. (7) that apparently altered the expression of p55 were included in the study. Indeed, the KOP55 mutant was more susceptible to some compounds that alter p55 expression, including clofazimine, novobiocin, PA-824, and valinomycin. However, compound-induced expression was apparently not essential for P55-dependent resistance. While rifampin and econazole treatment did not result in detectable changes in p55 gene expression, the susceptibility to these compounds increased in the mutant strain (for rifampin, eightfold; for econazole, fourfold) (Table 3). On the other hand, susceptibility to chlorpromazine and triclosan, which increased the level of p55 expression according to the information from the database of Boshoff et al. (7), was unchanged. Finally, since the P55 efflux pump may participate in the transport of lipids across the membrane (13), a P55 inhibitor may have an effect on cell envelope biosynthesis, making the bacteria more sensitive to compounds that target the corresponding assembly step. In fact, the KOP55 mutant was more sensitive than the wild type to some cell wall-targeting compounds, such as ethambutol, vancomycin, and bacitracin (Table 3), but not to cerulenin, d-cycloserine, or imipenem (data not shown).
TABLE 3.
Antimicrobial susceptibilities of M. bovis BCG P55 strains
| Compound | MIC (mg/liter) for M. bovis BCG straina:
|
|||
|---|---|---|---|---|
| Wild type | KOP55b | KOP55 PAZ23c | PAZ23d | |
| Bacitracin | >256 | 16 | >256 | NDf |
| Bacitracin + CCCPe | >256 | 16 | >256 | |
| CCCP | 4 | 2 | ND | ND |
| Clofazimine | 0.1 | 0.025 | 0.1 | 0.1 |
| Chlorpromazine | 4 | 4 | ND | ND |
| Econazole | 6.4 | 1.6 | 6.4 | 12.8 |
| Ethambutol | 4 | 2 | ND | ND |
| Novobiocin | 16 | 2 | 16 | ND |
| Novobiocin + CCCPe | 4 | 1 | 4 | |
| Novobiocin + VALe | 8 | 0.5 | 8 | |
| Novobiocin + RPe | 16-8 | 1 | 16-8 | |
| PA-824 | 0.25 | 0.125 | 0.25 | ND |
| Reserpine | 2 | 2 | 2 | ND |
| Rifampin | 0.0032 | 0.0004 | 0.0032 | 0.0032 |
| Rifampin + CCCPe | 0.0008 | 0.0002 | 0.0008 | |
| Rifampin + VALe | 0.0016 | 0.0002 | 0.0016 | |
| Rifampin + RPe | 0.0032-0.0016 | 0.0004 | 0.0032-0.0016 | |
| Valinomycin | 0.4 | 0.2 | 0.4 | ND |
| Vancomycin | 4 | 0.25 | 4 | 8 |
| Vancomycin + CCCPe | 4 | 0.25 | 4 | |
| Triclosan | 8 | 8 | ND | ND |
MICs were assayed over a range of twofold dilutions of antibiotics. The MICs of clofazimine, chlorpromazine, econazole, and triclosan were determined on agar plates. The MICs of bacitracin, CCCP, ethambutol, novobiocin, reserpine, rifampin, valinomycin, and vancomycin were determined by the resazurin assay. No difference in susceptibility between wild-type and KOP55 strains was observed for the following compounds: acriflavine, amikacin, cerulenin, d-cycloserine, ethidium bromide, ciprofloxacin, chloramphenicol, ethionamide, fluconazole, gentamicin, imipenem, isoniazid, ofloxacin, p-aminosalicylic acid, streptomycin, spectinomycin, and tetracycline (data not shown).
KOP55 is M. bovis BCG with the p55 gene inactivated.
KOP55 PAZ23 is complemented M. bovis BCG KOP55 with plasmid pPAZ23 (p27-p55 operon cloned into pSUM41).
PAZ23 is M. bovis BCG containing plasmid pPAZ23 (p27-p55 operon in pSUM36).
CCCP, valinomycin (VAL), and reserpine (RP) were present at concentrations of 1, 0.1, and 0.5 mg/liter, respectively (fourfold the sub-MIC for the M. bovis BCG wild type).
ND, not determined.
Expression of p55 complements the mutant phenotype.
Complementation tests were carried out to confirm that the sensitivity phenotype observed in KOP55 was due to inactivation of the p55 gene rather than suppressor mutations at other locations. Plasmid pPAZ23 was introduced into the KOP55 mutant to express the p27 and p55 genes under the control of the native operon promoter (5). When mutant KOP55 was complemented with the wild-type genes, the levels of resistance to bacitracin, clofazimine, econazole, novobiocin, PA-824, rifampin, valinomycin, and vancomycin returned to the wild-type levels (Table 3). The introduction of pPAZ23 into the parental wild-type strain resulted in a 2-fold increase in the MICs of econazole and vancomycin (whereas inactivation of the p55 gene resulted in 4- and 16-fold decreases in the MICs of these compounds, respectively) and did not alter its susceptibility to clofazimine and rifampin (Table 3). In summary, these results indicate that the P55 efflux pump provides low-level intrinsic resistance to a range of different antimicrobial compounds in M. bovis BCG.
The P55 efflux pump uses the proton and electrochemical gradients across the membrane as sources of energy to extrude compounds from the cell.
To elucidate whether the P55 efflux pump provides intrinsic drug resistance by actively extruding the compounds or by an indirect effect, the drug susceptibilities of the different P55-related strains were tested in the presence of subinhibitory concentrations of standard efflux pump inhibitors. The inhibitors tested included CCCP, a proton uncoupler; valinomycin, a K+-specific transporter that dissipates the electrochemical potential across the membrane; and reserpine, a general inhibitor of bacterial efflux pumps.
Subinhibitory concentrations of CCCP increased the susceptibility of the wild-type strain to novobiocin and rifampin (fourfold). In addition, the novobiocin and rifampin resistance of KOP55 restored by pPAZ23 (expressing p27-p55) was abolished in the presence of CCCP. These results suggest that this proton uncoupler has a direct effect on the function of P55 (Table 3) and reconfirmed that rifampin and novobiocin are substrates of the P55 efflux pump. In contrast, at the same concentration, CCCP did not affect the MICs of bacitracin and vancomycin, suggesting that intrinsic P55-dependent resistance to bacitracin and vancomycin was mediated by a mechanism that is not dependent on active transport. Valinomycin increased the susceptibility of the wild-type strain to rifampin and novobiocin by twofold (Table 3). These data suggest that P55 uses the electrochemical gradient to extrude these antibiotics. Subinhibitory concentrations of reserpine had little or no effect on the sensitivity to rifampin and novobiocin of the M. bovis BCG wild-type, mutant, and complemented strains. Reserpine is apparently not as efficient as CCCP at inhibiting the function of P55 in M. bovis BCG.
Together, these data indicated that rifampin and novobiocin, but probably not bacitracin and vancomycin, are extruded from the cell by P55 and that the efflux pump is using the proton and electrochemical gradients across the membrane as sources of energy.
The P55 efflux pump is involved in oxidative stress-related processes within the cell.
Analysis of the database of Boshoff et al. (7) revealed p27- or p55-inducing compounds that either generate reactive oxygen species or alter redox potential (Table 2). On the basis of these results, the susceptibility of KOP55 to various redox compounds was tested (Table 4). The mutant strain was not altered in its sensitivity to hydrogen peroxide (neither gene was induced by H2O2). However, it was more resistant to menadione, a respiratory inhibitor and superoxide-generating compound (Table 3). Compounds directly involved in the reduction of disulfide bonds were also tested. The mutant strain was more sensitive to the strong reducing agent dl-dithiothreitol but not to glutathione, a weaker thiol reductant. KOP55 was also more susceptible to diamide, a thiol-oxidizing agent. Mycothiol, the primary reducing agent in the mycobacterial cytosol (34), can be specifically alkylated by mBBr; KOP55 was more sensitive to mBBr. These results demonstrate that the inactivation of p55 alters the sensitivity of M. bovis BCG to superoxides and some agents that disrupt systems needed to maintain the thiol redox balance in the cytoplasm.
TABLE 4.
Susceptibilities of M. bovis BCG P55 strains to redox compounds
| Redox compounda | Disc load (μmol) | Inhibition zoneb (mm) for M. bovis BCG strains
|
|
|---|---|---|---|
| Wild type | KOP55c | ||
| Hydrogen peroxide | 80 | 23 | 23 |
| dl-Dithiothreitol | 5 | 0 | 30d |
| l-Glutathione reduced | 10 | 0 | 0 |
| Diamide | 5 | 0 | 30 |
| Menadione | 0.125 | 20 | 16 |
| mBBre | 0.5 | 0.125-0.25 | |
Dimethyl sulfoxide, which was used as the solvent for some of these compounds, had no effect on the growth of either strain (data not shown).
Halos were recorded after 14 days at 37°C.
KOP55 is M. bovis BCG with the p55 gene inactivated.
Delayed growth inside the halo.
Susceptibility to monobromobimane (mBBr) was determined by the Resazurin assay, and the concentrations are shown as the MICs (mM) for M. bovis BCG strains.
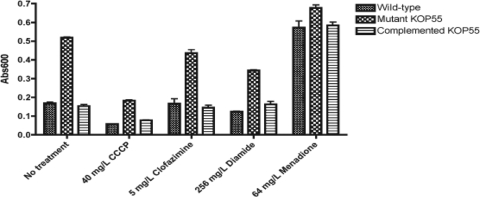
To further investigate the role of the P55 efflux pump in the maintenance of the general redox balance, MTT assays were performed (Fig. 2). The rate of MTT reduction required to generate the colored product formazan is commonly interpreted as a reflection of the levels of NADH, which determines the cytoplasmic redox potential (7). The most dramatic observation was a threefold increase in the MTT reduction activity of KOP55 compared to the activities of the wild type and the complemented mutant strain. MTT could be a P55 efflux pump substrate. To rule out the possibility that a strain lacking the P55 efflux pump might be impaired in its ability to export MTT and thus accumulates more formazan than the wild-type strain, CCCP was used in control experiments. The observed proportional declines in the intracellular redox potentials of all three strains indicated that the assay measured the increased intracellular reductive power of KOP55 and did not quantify the MTT P55-dependent transport activity (Fig. 2). These results suggest an increased intracellular redox potential in KOP55 compared to the redox potentials of the wild-type and complemented mutant strains under normal growth conditions.
FIG. 2.
Intracellular redox potential of M. bovis BCG P55 strains. Cell cultures in quadruplicate wells were treated with drugs for 1 h before the addition of MTT. The reduced formazan product was measured at 600 nm after an additional 3 h of incubation. The dimethyl sulfoxide solvent had no effect compared to no treatment (data not shown). The results shown represent data from one of three independent experiments. Abs600, absorbance at 600 nm.
The effects of drugs on the intracellular redox potentials in the wild type and the KOP55 mutant were compared by the MTT assay (Fig. 2). Rifampin, econazole, and vancomycin treatment induced no significant changes (data not shown). Diamide decreased the abilities of the mutant and wild-type strains to reduce MTT (by approximately 35 and 25%, respectively), while the ability of the complemented strain to reduce MTT remained unchanged. This suggested that the increased expression of the p27-p55 operon cloned into the low-copy-number plasmid (the complemented strain) might protect against the oxidative effects caused by diamide. KOP55 underwent an ∼15% reduction in its intracellular reductive power after treatment with clofazimine, while no effect was observed in either the wild-type or the complemented strain. In contrast, under these conditions, menadione dramatically increased the intracellular redox potential of the wild-type and complemented strains (∼3.5-fold) but had a minimal effect on the mutant (∼1.3-fold), almost abolishing the difference observed among the untreated mutant, complemented, and wild-type strains. In summary, these results suggest that the P55 efflux pump plays a role in the maintenance of the oxidative balance within the cell.
Cell growth is defective in M. bovis BCG KOP55.
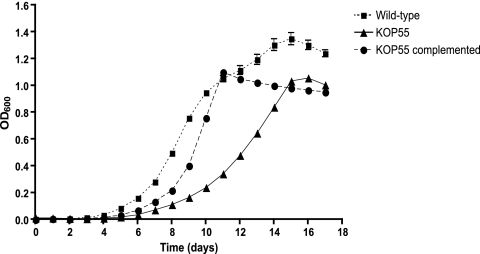
The KOP55 mutant displayed two growth defects in liquid cultures: it had an increased lag phase and reached stationary phase at a lower OD. The wild-type strain reached stationary phase with an OD of 1.3, while the KOP55 mutant was able to reach a stationary-phase OD of only 1.0 (Fig. 3). In addition, strain KOP55 initiated similar growth rates, but only after a 48-h delay, apparently reflecting a prolonged recovery from stationary-phase metabolism. Interestingly, the delayed outgrowth phenotype could be partially complemented by the introduction of the p27-p55 operon. Complementation reduced the lag phase from 48 to 24 h; however, it did not allow the strain to reach the same stationary-phase OD value (OD = 1.0) as the wild type (OD = 1.3). The instability of the complementing plasmid or the reduced level of expression of the cloned gene during this growth phase might explain this incomplete complementation phenotype. In addition, when it was grown in liquid cultures without Tween, KOP55 displayed increased clumping compared with that of either the wild-type or the complemented strains, suggesting an altered cell wall composition. This phenotype was abolished in the presence of Tween (data not shown). The results of these experiments demonstrate that KOP55 has a growth defect in both the early and the final stages of cell growth in liquid culture.
FIG. 3.
Growth kinetics of M. bovis BCG wild-type (squares), KOP55 (triangles), and KOP55 complemented (circles) strains. Growth curves are representative of those from two similar independent experiments done in quadruplicate. Error bars indicate standard deviations. OD600, OD at 600 nm.
To observe bacterial colonial growth, strains were plated on Middlebrook 7H10 agar, and the diameters of 100 colonies were measured. KOP55 mutant colonies were roughly half the size (diameter, 1.52 ± 0.12 mm) of colonies of the wild-type strain (diameter, 2.7 ± 0.15 mm). However, the morphologies of the mutant and wild-type colonies appeared to be similar when they were observed under an inverted microscope (×100 magnification), and no differences in the sizes or the morphologies of single bacilli were observed under a fluorescence microscope after auramine-rhodamine staining (data not shown).
Together, these data suggest that the P55 efflux pump plays an important role in bacterial growth, even though it does not affect the cellular or the colonial morphology, which is probably related to its putative role as a transporter of lipids across the membrane.
DISCUSSION
Efflux pumps have traditionally been linked to their multidrug extrusion capabilities, which confer clinically significant levels of antibiotic resistance (29). However, recent insights suggest that efflux systems are primarily involved with fundamental cellular physiological processes and that drug extrusion is a nonspecific, secondary role (19, 20, 30). Thus, physiological regulatory systems may determine the levels of drug resistance.
The expression database of Boshoff et al. (7) contains information from 430 microarray profiles and has records of the transcriptional regulation of M. tuberculosis genes in response to 75 conditions (including drugs) that elicit defined metabolic or functional changes (7). Our analysis of the results of drug resistance assays in combination with the regulatory data generated by Boshoff et al. (7) allowed us to identify additional P55 efflux pump substrates as well as to elucidate the involvement of P55 in physiological processes. Interestingly, the level of p55 gene expression increased in response to clofazimine, triclosan, and the respiration inhibitors phenothiazines. However, only clofazimine was shown to be a P55 substrate. Boshoff et al. (7) reported a general increased level of expression of potential drug detoxification and efflux mechanisms in response to treatment with the compounds mentioned above, suggesting the presence of generalized, perhaps redundant regulatory systems. In this regard, the P55 efflux pump would play a role in the detoxification systems linked to respiratory processes and maintenance of the redox balance within the cell. This is supported by the increased sensitivity of the P55 mutant to various redox compounds, including clofazimine, dl-dithiothreitol, and diamide, as well as the thiol alkylating agent mBBr. In the cytoplasm, mBBr specifically binds to mycothiol, the thiol reducing agent that is involved in the primary detoxification pathway in mycobacteria. Although the products of this pathway are exported, the nature of the corresponding transport systems is unknown (23). Our data suggest a possible role for the P55 efflux pump in eliminating the toxic products generated by a thiol reductant (including mycothiol) in Mycobacterium.
The MTT assay allowed further analysis of P55's role in general redox processes. Tetrazolium salts are widely used to measure metabolic activity, as reflected by NADH levels. Since NADH is the primary metabolite catalyzing the reduction of MTT to formazan (4), the rate of MTT reduction can be interpreted as a direct reflection of the cellular NADH content. The mutant strain displayed a large (threefold) increase in the level of formazan accumulation, suggesting a major change in the intracellular reductive potential, most likely due to increased NADH levels. This change was fully reversed by genetic complementation by use of the p27-p55 operon (Fig. 2). On the other hand, KOP55 was less susceptible to menadione (Table 4). During 4 h of exposure, menadione caused an overall increase in the amount of MTT reduction products in all strains, probably by increasing the transfer of electrons from NADH to MTT (4). Thus, menadione promotes net NADH synthesis or accumulation, which is reflected in higher levels of MTT reduction. After menadione treatment, the threefold difference in the reductive power of MTT displayed by the KOP55 mutant strain was almost abolished, suggesting that menadione may in some way mimic the effect of the p55 mutation. These data provide additional evidence that the P55 efflux pump serves as a key detoxification system that maintains within the cell an oxidative balance that is probably related to respiratory processes. In fact, the respiratory modulators chlorpromazine and triclosan increased the level of p55 expression (7), even though they are not P55 substrates, as shown by our susceptibility data.
Farrow and Rubin (13) showed that deletion of the p27-p55 operon leads to morphological changes in M. smegmatis and speculated that this was due to an inability to transport cell wall lipids. Thus, any deficiency in lipid transport across the cytoplasmic membrane would affect the integrity of the cell wall, making the mutant more sensitive to drugs targeting the cell envelope and also altering its growth characteristics. In M. bovis, two findings support these speculations. First, the mutant strain has a growth defect. This clearly shows that bacteria lacking P55 have a physiological perturbation that affects their fitness under in vitro growth conditions. Second, the P55 mutant strain is highly sensitive to bacitracin and vancomycin, which both target the peptidoglycan layer of the cell envelope, suggesting the possible lack of an important substrate needed for the integrity of the cell wall. It is tempting to suggest that P55 could actively transport these antibiotics away from the peptidoglycan layer, perhaps in combination with a periplasmic adaptor and TolC-like proteins, as has been described for other efflux pumps (21). However, CCCP was unable to inhibit P55-mediated bacitracin and vancomycin resistance, whereas it could inhibit P55-mediated rifampin and novobiocin resistance. This suggests that P55 contributes to the intrinsic resistance to bacitracin and vancomycin, perhaps by contributing to the impermeability of the cell wall, but it might not be actively transporting both drugs.
In general, the overexpression of an efflux pump decreases the susceptibility of bacteria to substrates of the pump, and susceptibility increases when the gene encoding the efflux pump is inactivated. The introduction of pPAZ23 (which contains the p27 and p55 genes under the control of the operon promoter) into the KOP55 mutant restored drug susceptibility levels, suggesting that p55 was being expressed. However, when additional copies of the operon were introduced (pPAZ23) into the wild-type strain, little or no decrease in drug susceptibility was observed, suggesting the presence of autoregulatory control mechanisms. Indeed, we have obtained preliminary evidence that the p55 gene is also expressed independently under the control of its own promoter. It is noteworthy that the level of p55 transcription from this promoter increased fivefold after exposure to rifampin and vancomycin, both of which are substrates of the efflux pump (S. Ramón-García and J. A. Aínsa, unpublished data). The induction of genes encoding efflux pumps by their own substrates has been described previously (1, 8, 14, 18). In this regard, we propose a model in which p55 can be expressed selectively under its own promoter or under the control of the operon promoter (in combination with p27), depending on the particular conditions that the bacteria confront, such as antibiotics, a need for detoxification, oxidative stress, or other physiological conditions. We are carrying out studies to test this hypothesis.
As efflux pump-mediated drug extrusion in bacteria is of clinical significance (29), determination of the substrate profiles of these transporters could allow the design of new therapies against bacterial infections. In mycobacteria, many efflux pumps may contribute to intrinsic drug resistance (11). However, only a few of them are known to provide resistance to anti-TB drugs and, in particular, to first-line anti-TB drugs. Pasca et al. (28) showed that the resistance-nodulation-division-like transporter MmpL7 extrudes isoniazid when the mmpL7 gene from M. tuberculosis is expressed in M. smegmatis. However, this is not observed in M. tuberculosis, in which the natural substrate of MmpL7, the lipid phthiocerol dimycocerosate, competes with isoniazid. The findings of that study are in agreement with our own data showing that the substrate specificity of the P55 efflux pump is dependent on the genetic background, and it is a clear example of the relevance of the bacterial host when the activities of efflux pumps are analyzed. Other authors have also reported the importance of the iniA gene product for the activity of an efflux pump that confers isoniazid and ethambutol tolerance (9).
To our knowledge, this is the first report of rifampin transport by an efflux pump in mycobacteria. Other studies reported increases in the levels of expression of efflux pumps, including P55, after the treatment of clinical M. tuberculosis strains with rifampin and isoniazid (15, 17, 38). However, those studies did not show a direct correlation between expression of the P55 efflux pump and intrinsic resistance to rifampin in mycobacteria. A decrease in the intracellular levels of rifampin would allow the bacillus to better adapt to the presence of rifampin by acquiring mutations in the rpoB gene. This is of particular importance, since the p55 gene is present in all M. tuberculosis strains whose genomes have been sequenced, such as the laboratory strain M. tuberculosis H37Rv, as well as clinical isolates M. tuberculosis CDC1551, F11, Haarlem, C, KZN 4207, KZN 1435 (an MDR-TB strain), and KZN 605 (an XDR-TB strain) (http://www.broad.mit.edu/). In addition, PCR experiments have detected p55 in 10 of 10 DNA samples from our collection of M. tuberculosis complex clinical isolates, which includes 4 rifampin-resistant strains (the XDR strain MBZ [36] and 3 MDR-TB strains) and 6 rifampin-susceptible isolates of M. tuberculosis from different geographic origins (data not shown).
P55 is a transporter belonging to the drug/H+ antiporter 14-transmembrane-domain (DHA14) family whose members are thought to export cationic molecules by proton motive force (12). We demonstrated that this is the main source of energy used by the P55 efflux pump to extrude drugs. On the other hand, the fact that valinomycin also exerted an effect on drug transport suggests that the electrochemical potential across the membrane can also be used as a source of energy. Surprisingly, reserpine had very little inhibitory effect on the expelling activity of P55 in M. bovis BCG since it inhibits tetracycline efflux by another mycobacterial DHA14 family efflux pump (32), as well as the transport of ethidium bromide by the homologue of P55 in M. smegmatis (13).
In most bacterial species, efflux pump inhibitors can reduce resistance levels both in wild-type strains and in those with acquired target mutations (22). In this regard, an inhibitor of the P55 efflux pump would render the bacillus more susceptible to the first- and second-line drugs rifampin and clofazimine (or other drugs transported by P55), in either susceptible or MDR- and XDR-TB strains. In a scenario in which few or no effective drugs are available for the treatment of patients infected with MDR or XDR M. tuberculosis strains, the possibility that rifampin may be able to be reintroduced is very appealing. In addition, it would also reduce the emergence of rifampin-resistant strains, and since a p55 mutant of M. tuberculosis has been predicted to be attenuated (37), a putative P55 efflux pump inhibitor could also reduce the virulence of the tubercle bacillus. Thus, this inhibitor would help to control the spread of this devastating disease. We currently are performing high-throughput chemical screens to explore this concept.
Acknowledgments
We gratefully acknowledge Helena Boshoff and Clifton E. Barry III for the kind gift of PA-824. We are also grateful to Charles Howes for his help analyzing the database of Boshoff et al. (7) and to Cristina Villellas and Carmen Lafoz for technical assistance with the clinical distribution studies of the p55 gene and Southern blot analysis.
This work was supported by European Union grant QLK2-CT-2000-017610 (to C.M.), Spanish Ministry of Science and Education grants BIO2002-01297 and BIO2005-01810 (to J.A.A.), and Canadian Institute of Health Research grants MOP-82745 and MOP-82855 (to C.J.T.). S.R.-G. held grants from the Spanish Ministry of Science and Education (AP2001-1114) and the Fundación Alfonso Martin Escudero (Spain).
Footnotes
Published ahead of print on 29 June 2009.
REFERENCES
- 1.Ahmed, M., C. M. Borsch, S. S. Taylor, N. Vazquez-Laslop, and A. A. Neyfakh. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506-28513. [PubMed] [Google Scholar]
- 2.Ainsa, J. A., C. Martin, M. Cabeza, F. De la Cruz, and M. V. Mendiola. 1996. Construction of a family of Mycobacterium/Escherichia coli shuttle vectors derived from pAL5000 and pACYC184: their use for cloning an antibiotic-resistance gene from Mycobacterium fortuitum. Gene 176:23-26. [DOI] [PubMed] [Google Scholar]
- 3.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 4.Berridge, M. V., P. M. Herst, and A. S. Tan. 2005. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol. Annu. Rev. 11:127-152. [DOI] [PubMed] [Google Scholar]
- 5.Bigi, F., A. Alito, M. I. Romano, M. Zumarraga, K. Caimi, and A. Cataldi. 2000. The gene encoding P27 lipoprotein and a putative antibiotic-resistance gene form an operon in Mycobacterium tuberculosis and Mycobacterium bovis. Microbiology 146(Pt 4):1011-1018. [DOI] [PubMed] [Google Scholar]
- 6.Bigi, F., A. Gioffre, L. Klepp, M. de la Paz Santangelo, A. Alito, K. Caimi, V. Meikle, M. Zumarraga, O. Taboga, M. I. Romano, and A. Cataldi. 2004. The knockout of the lprG-Rv1410 operon produces strong attenuation of Mycobacterium tuberculosis. Microbes Infect. 6:182-187. [DOI] [PubMed] [Google Scholar]
- 7.Boshoff, H. I., T. G. Myers, B. R. Copp, M. R. McNeil, M. A. Wilson, and C. E. Barry III. 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J. Biol. Chem. 279:40174-40184. [DOI] [PubMed] [Google Scholar]
- 8.Buroni, S., G. Manina, P. Guglierame, M. R. Pasca, G. Riccardi, and E. De Rossi. 2006. LfrR is a repressor that regulates expression of the efflux pump LfrA in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 50:4044-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colangeli, R., D. Helb, S. Sridharan, J. Sun, M. Varma-Basil, M. H. Hazbon, R. Harbacheuski, N. J. Megjugorac, W. R. Jacobs, Jr., A. Holzenburg, J. C. Sacchettini, and D. Alland. 2005. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol. Microbiol. 55:1829-1840. [DOI] [PubMed] [Google Scholar]
- 10.Content, J., M. Braibant, N. D. Connell, and J. A. Ainsa. 2005. Transport processes, p. 379-401. In S. Cole, K. D. Eisenach, D. N. McMurray, and W. R. Jacobs, Jr. (ed.), Tuberculosis and the tubercle bacillus. ASM Press, Washington, DC.
- 11.De Rossi, E., J. A. Ainsa, and G. Riccardi. 2006. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol. Rev. 30:36-52. [DOI] [PubMed] [Google Scholar]
- 12.De Rossi, E., P. Arrigo, M. Bellinzoni, P. A. Silva, C. Martin, J. A. Ainsa, P. Guglierame, and G. Riccardi. 2002. The multidrug transporters belonging to major facilitator superfamily in Mycobacterium tuberculosis. Mol. Med. 8:714-724. [PMC free article] [PubMed] [Google Scholar]
- 13.Farrow, M. F., and E. J. Rubin. 2008. Function of a mycobacterial major facilitator superfamily pump requires a membrane-associated lipoprotein. J. Bacteriol. 190:1783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grkovic, S., M. H. Brown, N. J. Roberts, I. T. Paulsen, and R. A. Skurray. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273:18665-18673. [DOI] [PubMed] [Google Scholar]
- 15.Gupta, A. K., D. S. Chauhan, K. Srivastava, R. Das, S. Batra, M. Mittal, P. Goswami, N. Singhal, V. D. Sharma, K. Venkatesan, S. E. Hasnain, and V. M. Katoch. 2006. Estimation of efflux mediated multi-drug resistance and its correlation with expression levels of two major efflux pumps in mycobacteria. J. Commun. Dis. 38:246-254. [PubMed] [Google Scholar]
- 16.Janssen, G. R., and M. J. Bibb. 1993. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene 124:133-134. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, X., W. Zhang, Y. Zhang, F. Gao, C. Lu, X. Zhang, and H. Wang. 2008. Assessment of efflux pump gene expression in a clinical isolate Mycobacterium tuberculosis by real-time reverse transcription PCR. Microb. Drug Resist. 14:7-11. [DOI] [PubMed] [Google Scholar]
- 18.Kaatz, G. W., and S. M. Seo. 1995. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 39:2650-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krulwich, T. A., O. Lewinson, E. Padan, and E. Bibi. 2005. Do physiological roles foster persistence of drug/multidrug-efflux transporters? A case study. Nat. Rev. Microbiol. 3:566-572. [DOI] [PubMed] [Google Scholar]
- 20.Lewinson, O., E. Padan, and E. Bibi. 2004. Alkali tolerance: a biological function for a multidrug transporter in pH homeostasis. Proc. Natl. Acad. Sci. USA 101:14073-14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 22.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton, G. L., Y. Av-Gay, and R. C. Fahey. 2000. A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry 39:10739-10746. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen, L., and C. J. Thompson. 2006. Foundations of antibiotic resistance in bacterial physiology: the mycobacterial paradigm. Trends Microbiol. 14:304-312. [DOI] [PubMed] [Google Scholar]
- 25.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parish, T., and N. G. Stoker. 1998. Mycobacterial protocols. In T. Parish and N. G. Stoker (ed.), Methods in molecular biology, vol. 101. Humana Press, Totowa, NJ.
- 27.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146(Pt 8):1969-1975. [DOI] [PubMed] [Google Scholar]
- 28.Pasca, M. R., P. Guglierame, E. De Rossi, F. Zara, and G. Riccardi. 2005. mmpL7 gene of Mycobacterium tuberculosis is responsible for isoniazid efflux in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 49:4775-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piddock, L. J. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piddock, L. J. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4:629-636. [DOI] [PubMed] [Google Scholar]
- 31.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 32.Ramon-Garcia, S., C. Martin, J. A. Ainsa, and E. De Rossi. 2005. Characterization of tetracycline resistance mediated by the efflux pump Tap from Mycobacterium fortuitum. J. Antimicrob. Chemother. 57:252-259. [DOI] [PubMed] [Google Scholar]
- 33.Ramon-Garcia, S., C. Martin, E. De Rossi, and J. A. Ainsa. 2007. Contribution of the Rv2333c efflux pump (the Stp protein) from Mycobacterium tuberculosis to intrinsic antibiotic resistance in Mycobacterium bovis BCG. J. Antimicrob. Chemother. 59:544-547. [DOI] [PubMed] [Google Scholar]
- 34.Rawat, M., and Y. Av-Gay. 2007. Mycothiol-dependent proteins in actinomycetes. FEMS Microbiol. Rev. 31:278-292. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Samper, S., and C. Martin. 2007. Spread of extensively drug-resistant tuberculosis. Emerg. Infect. Dis. 13:647-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siddiqi, N., R. Das, N. Pathak, S. Banerjee, N. Ahmed, V. M. Katoch, and S. E. Hasnain. 2004. Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump. Infection 32:109-111. [DOI] [PubMed] [Google Scholar]
- 39.Silva, P. E., F. Bigi, M. de la Paz Santangelo, M. I. Romano, C. Martin, A. Cataldi, and J. A. Ainsa. 2001. Characterization of P55, a multidrug efflux pump in Mycobacterium bovis and Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 45:800-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stover, C. K., P. Warrener, D. R. VanDevanter, D. R. Sherman, T. M. Arain, M. H. Langhorne, S. W. Anderson, J. A. Towell, Y. Yuan, D. N. McMurray, B. N. Kreiswirth, C. E. Barry, and W. R. Baker. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405:962-966. [DOI] [PubMed] [Google Scholar]
- 41.World Health Organization. 2008. Global tuberculosis control: surveillance, planning, financing: WHO report WHO/HTM/TB/2008.393. World Health Organization, Geneva, Switzerland.