Abstract
We describe the design and characterization of a potent human respiratory syncytial virus (RSV) nucleocapsid gene-specific small interfering RNA (siRNA), ALN-RSV01. In in vitro RSV plaque assays, ALN-RSV01 showed a 50% inhibitory concentration of 0.7 nM. Sequence analysis of primary isolates of RSV showed that the siRNA target site was absolutely conserved in 89/95 isolates, and ALN-RSV01 demonstrated activity against all isolates, including those with single-mismatch mutations. In vivo, intranasal dosing of ALN-RSV01 in a BALB/c mouse model resulted in potent antiviral efficacy, with 2.5- to 3.0-log-unit reductions in RSV lung concentrations being achieved when ALN-RSV01 was administered prophylactically or therapeutically in both single-dose and multidose regimens. The specificity of ALN-RSV01 was demonstrated in vivo by using mismatch controls; and the absence of an immune stimulatory mechanism was demonstrated by showing that nonspecific siRNAs that induce alpha interferon and tumor necrosis factor alpha lack antiviral efficacy, while a chemically modified form of ALN-RSV01 lacking measurable immunostimulatory capacity retained full activity in vivo. Furthermore, an RNA interference mechanism of action was demonstrated by the capture of the site-specific cleavage product of the RSV mRNA via rapid amplification of cDNA ends both in vitro and in vivo. These studies lay a solid foundation for the further investigation of ALN-RSV01 as a novel therapeutic antiviral agent for clinical use by humans.
Human respiratory syncytial virus (RSV) is an ubiquitous virus and the most common cause of serious lower respiratory tract infections in infants and young children worldwide, as well as an important pathogen in elderly individuals and immunocompromised patients (5, 10, 11, 18-21, 62, 64). The worldwide disease burden associated with RSV infection is considerable. RSV is the leading cause of hospitalization for infants (44), with infection rates approaching 70% in the first year of life (25). Approximately 30% of RSV-infected children develop lower respiratory tract infections. RSV results in the hospitalization of approximately 3% of previously healthy infants within their first year of life and a substantially greater percentage of infants and children with underlying diseases (8). RSV is a common cause of childhood bronchiolitis and has been implicated in the development and exacerbation of asthma and reactive airway disease in childhood (39, 50, 51, 54).
Despite nearly four decades of research, no RSV vaccine approach has been successful at conferring protection at a level that exceeds the incomplete protection afforded by natural infection. Currently, the only antiviral approved for use for the treatment of RSV infection is ribavirin; but due to its teratogenicity, limited efficacy, and poorly understood mechanism of action, it has very limited use (43, 73). Prophylactic therapies include the use of the approved humanized monoclonal antibody palivizumab (Synagis), which targets the fusion protein of RSV (2, 27, 36). While this antibody is effective, it is used only for the treatment of high-risk patient populations, including premature infants (3, 48, 66), and as an inhibitor of viral fusion, it may be of limited benefit for the treatment of an established RSV infection. Thus, there is a clear need for an alternative approach to the development of a novel anti-RSV therapeutic agent.
RNA interference (RNAi) is a posttranscriptional mechanism of gene silencing first described as an innate response to viral infections in plants and subsequently in all higher-order eukaryotes (7, 30). RNAi involves the target-specific degradation of RNA transcripts following the incorporation of small double-stranded RNA into the RNA-induced silencing complex. A major advance in the field of RNAi was the demonstration that synthetic double-stranded, small interfering RNAs (siRNAs) were functionally active against target mRNA transcripts in mammalian cells (17). These findings have led to the emergence of a new field of drug discovery with RNAi therapeutics that target a wide variety of human diseases, ranging from cancer to metabolic diseases and viral infections (13). Recent studies have demonstrated the efficacy of siRNAs in inhibiting several viruses, in vitro and in vivo, including hepatitis C virus (9, 59, 75), hepatitis B virus (4, 24, 69), West Nile virus (38, 47, 65), the severe acute respiratory syndrome-associated coronavirus (31, 76, 77, 81), influenza virus (23, 70), and RSV (6, 82), among others. For RSV, Bitko et al. (6) and Zhang et al. (82) have demonstrated the in vitro and in vivo inhibition of RSV by targeting the phosphoprotein (P protein) and nonstructural (NS1) protein siRNAs, respectively, confirming the feasibility of using a strategy that targets siRNA to achieve activity against this virus. However, the P protein siRNA is limited by its specificity to one particular strain of RSV, while the inhibition of the NS1 protein siRNA of RSV may be attributed to immune modulation, which results in the more robust clearance of the virus by the host rather than the direct targeting of the viral RNA. Furthermore, in both cases, definitive proof of an RNAi-mediated mechanism of antiviral activity remains to be established. Here we describe the pharmacology of a highly potent and specific human RSV siRNA with a broad spectrum of activity that demonstrates antiviral activity in vitro and in vivo via an RNAi mechanism of action.
MATERIALS AND METHODS
Animals.
Six- to 8-week old, pathogen-free female BALB/c mice were purchased from Harlan Sprague-Dawley Laboratories (Indianapolis, IN). The mice were housed in microisolator cages and were fed sterilized water and food ad libitum.
Virus preparation, cell lines, and viral titer determination.
Vero E6 cells were maintained in tissue culture medium consisting of Dulbecco's modified Eagle medium (DMEM; Gibco Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT). RSV A2 and RSV B1 were prepared in Vero E6 cells. Briefly, confluent Vero E6 cells (American Type Culture Collection, Manassas, VA) in serum-free DMEM were infected with RSV at a multiplicity of infection of 0.1. The virus was allowed to adsorb for 1 h at 37°C, after which tissue culture medium was added. Infected cells were incubated for 72 to 96 h at 37°C until a cytopathic effect of >90% was observed by light microscopy. Infected cells were harvested by removal of the medium and replacement with a minimal volume of serum-free DMEM, followed by three freeze-thaw cycles at −70°C and 4°C. The contents were collected and centrifuged at 4,000 × g for 20 min at 4°C to remove the cell debris, and the titer was determined by an immunostaining plaque assay, as described previously (72). Briefly, Vero E6 cells were infected with serial dilutions of stock RSV, the virus was allowed to adsorb for 1 h at 37°C, and then the cells were overlaid with 2% methylcellulose medium (DMEM supplemented with 2% fetal bovine serum, 1% antibiotic-antimycotic solution, and 2% methylcellulose). After 5 days at 37°C in 5% CO2, the plates were removed and the cells were fixed with ice-cold acetone-methanol (60:40). The cells were blocked with a universal blocking reagent (Powerblock; Biogenix, San Ramon, CA) and incubated with anti-RSV fusion (F)-protein monoclonal antibody (clone 131-2A; Millipore-Chemicon, Temecula, CA) diluted 1:200, followed by the addition of goat anti-mouse immunoglobulin G whole-molecule alkaline phosphatase secondary antibody. The reaction was developed with an alkaline phosphatase substrate kit (Vector Black; Vector Laboratories, Burlingame, CA), and plaques were visualized and counted under a light microscope. For RSV primary isolate cultures, samples were obtained from John DeVincenzo from the University of Tennessee, Memphis. RSV isolates were obtained from RSV-infected children diagnosed either by a conventional direct fluorescent-antibody method or by a rapid antigen detection method in the Le Bonheur Children's Medical Center Virology Laboratory in Memphis, TN. Nasal secretions were collected by aspiration, and isolates from the secretions were grown and passaged in HEp-2 cells and harvested when the cytopathic effect was 90%. Individual aliquots of the supernatant containing RSV were then subjected to nucleic acid extraction with a QIAmp viral RNA minikit, according to the manufacturer's protocol (Qiagen, Valencia, CA). RSV isolates were also obtained from Mark Van Ranst from the University of Leuven, Leuven, Belgium, and Larry Anderson from the Centers for Disease Control and Prevention, Atlanta, GA.
RSV-specific siRNA selection.
An analysis of the sequences in one of the National Center for Biotechnology Information databases was performed with the Basic Local Alignment Search Tool (BLAST). In that analysis, a sequence comparison algorithm was used to search the sequence databases for optimal local alignments to a query (1). In this case, the query was the 19-nucleotide (nt) sequence comprising the sense or antisense strand of ALN-RSV01, excluding the dTdT overhang. The Reference Sequence (RefSeq) database provides a comprehensive, integrated, nonredundant set of sequences, including genomic DNA, transcript (RNA), and protein products, and is updated weekly (53). Only siRNAs that showed no significant homology with any sequence from the RefSeq database were selected for synthesis and further study.
In vitro RSV inhibition assay.
Vero cells were grown in 24-well plates in a 5% CO2 humidified incubator at 37°C in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 g/ml streptomycin (BioChrom, Cambridge, United Kingdom) to 80% confluence. siRNAs were diluted to the indicated concentrations in 50 μl Opti-MEM reduced serum medium (Invitrogen). Separately, 3 μl Lipofectamine 2000 (Invitrogen) was diluted in 50 μl Opti-MEM, and the componenets were mixed and incubated for 5 min at room temperature. The siRNA and Lipofectamine mixtures were combined, incubated for 20 to 25 min at room temperature, and then added to the cells; and the cells were incubated at 37°C overnight. The mixture was then removed from the cells, and 200 to 400 PFU of RSV A2 (ATCC CC-81) was incubated with the cells for 1 h at 37°C. The infected cells were covered with methylcellulose medium and incubated for 5 days at 37°C, and plaques were visualized by the immunostaining plaque assay in Vero cells, as described previously.
In vivo screening of RSV-specific siRNAs.
For the prophylaxis model, BALB/c mice were anesthetized by the intraperitoneal administration of 2,2,2-tribromoethanol (Avertin), and siRNA in a total volume of 50 μl of phosphate-buffered saline (PBS) was instilled intranasally (i.n.). At 4 h after siRNA instillation, the mice were anesthetized and infected i.n. with 106 PFU of RSV A2 in 50 μl. Prior to removal of the lungs at day 4 postinfection, the anesthetized mice were exsanguinated by severing the right caudal artery. Lung tissue was collected in 1 ml ice-cold PBS (Gibco Invitrogen). The RSV titers in the lung tissue were measured by an immunostaining plaque assay. The lungs were homogenized with a handheld Tissumiser homogenizer (Fisher Scientific, Pittsburg, PA), and the lung tissue homogenates were placed on ice for 5 to 10 min to allow the debris to settle. The clarified lung tissue lysates were serially diluted 10-fold in serum-free DMEM and added to 95% confluent Vero E6 cells cultured in DMEM in 24-well plates (Falcon; Becton Dickinson, San Jose, CA), and plaque assays were performed as described above. For the treatment model, BALB/c mice were anesthetized as described above, and 106 PFU of RSV A2 in 50 μl was instilled i.n. At 1, 2, 3, or 4 days after viral infection, the mice were reanesthetized, siRNA in 50 μl was instilled i.n., and then the viral concentrations in the lungs were measured on day 5 postinfection, as described above.
siRNA generation.
RNA oligonucleotides were synthesized by standard solid-phase oligonucleotide synthesis protocols with commercially available 5′-O-(4,4′-dimethoxytrityl)′-3′O-(2-cyanoethyl-N,N-diisopropyl) phosphoramidite monomers of uridine, 4-N-benzyoylcytidine, 6-N-benzoyladenosine, and 2-N-isobutyrlguanosine with 2′-O-tert-butyldimethylsilyl-protected phosphoramidites. After cleavage and deprotection, the RNA oligonucleotides were purified by anion-exchange high-performance liquid chromatography and characterized by electrospray ionization mass spectrometry. To generate siRNAs from RNA single strands, equimolar amounts of complementary sense and antisense strands were mixed and annealed, and the siRNAs were further characterized by capillary gel electrophoresis.
PBMC assay.
To examine the ability of siRNAs to stimulate alpha interferon (IFN-α) or tumor necrosis factor alpha (TNF-α), human peripheral blood mononuclear cells (PBMCs) were isolated from concentrated fractions of leukocytes (buffy coats) obtained from the Blood Bank Suhl, Institute for Transfusion Medicine, Germany. The buffy coats were diluted 1:1 in PBS and added to a tube of Histopaque (Sigma, St. Louis, MO), and the tubes were centrifuged at 2,200 rpm for 20 min to allow fractionation. White blood cells were collected, washed in PBS, and then centrifuged. The cells (concentration, 1 × 106 cells/ml) were resuspended in RPMI 1640 culture medium (Invitrogen) supplemented with 10% fetal calf serum, interleukin-3 (10 ng/ml; Sigma), and phytohemagglutinin-P (5 μg/ml; Sigma) for the IFN-α assay or with no additive for the TNF-α assay and seeded onto 96-well plates and incubated at 37°C in a 5% CO2 atmosphere. The control oligonucleotides were siRNA AL-DP-5048 duplex (5′-GUCAUCACACUGAAUACCAAU-3′ and 3′-CACAGUAGUGUGACUUAUGGUUA-5′), siRNA AL-DP-7296 duplex (5′-CUACACAAAUCAGCGAUUUCCAUGU-3′ and 3′-GAUGUGUUUAGUCGCUAAAGGUACA-5′), siRNA AL-DP-1730 duplex (5′-CGAUUAUAUUACAGGAUGAdTsdT-3′ and 3′-dTsdTGCUAAUAUAAUGUCCUACU-5′), and siRNA AL-DP-2153 duplex (5′-GGCUCUAAGCUAACUGAAGdTdT-3′ and 3′-dTdTCCGAGAUUCGAUUGACUUC-5′). Cells in culture were combined with either 500 nM oligonucleotide prediluted in Opti-MEM (Invitrogen) or 133 nM oligonucleotide prediluted in Opti-MEM and the Geneporter (GP2) transfection reagent (Genlantis, San Diego, CA) for the IFN-α assay or were combined with N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (Roche, Switzerland) for the TNF-α assay. The cells were then incubated at 37°C for 24 h. IFN-α and TNF-α levels were measured with a Bender MedSystems (Vienna, Austria) instant enzyme-linked immunosorbent assay (ELISA) kit, according to the manufacturer's instruction.
In vitro and in vivo rapid amplification of cDNA ends (RACE).
Total RNA was purified either from in vitro-transfected Vero E6 cells or from lungs harvested at day 5 postinfection, as described above, by using Trizol (Invitrogen), followed by DNase treatment and final processing with RNeasy, according to the manufacturer's instructions (Qiagen). Five to 10 μl of the RNA preparation from pooled samples was ligated to a GeneRacer adaptor (5′-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′) without prior treatment. The ligated RNA was reverse transcribed by use of a gene-specific primer (cDNA primer [5′-CTCAAAGCTCTACATCATTATC-3′]). To detect RNAi-specific cleavage products, two rounds of consecutive PCR were performed with primers complementary to the RNA adaptor and RSV A2 nucleocapsid (N) gene mRNA (primers GR 5′ [5′-CGACTGGAGCACGAGGACACTGA-3′] and Rev Primer [5′-CCACTCCATTTGCTTTTACATGATATCC-3′]) for the first round, followed by a second round of nested PCR with primers GRN (5′-GGACACTGACATGGACTGAAGGAGTA-3′) and Rev N Primer (5′-GCTTTTACATGATATCCCGCATCTCTGAG-3′). The amplified products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. Specific cleavage products migrating at the correct size were excised, cloned into a sequencing vector, and sequenced by standard methods.
Sequence analysis of clinical isolates for ALN-RSV01 target site conservation.
Amplification of the RSV N-gene fragment containing the ALN-RSV01 recognition site was performed by a two-step reverse transcription-PCR. Briefly, RNA was reverse transcribed with random hexamers and Superscript III reverse transcriptase (Invitrogen) at 42°C for 1 h to generate a cDNA library. A 1,200-nt gene-specific fragment was amplified by PCR with Platinum Taq polymerase (Invitrogen) and the RSV N forward primer (5′-AGAAAACTTGATGAAAGACA-3′) and the RSV N reverse primer (5′-ACCATAGGCATTCATAAA-3′) for 35 cycles at 55°C for 30 s, followed by 68°C for 1 min,. The PCR products were analyzed by 1% agarose gel electrophoresis. As a control, a laboratory strain of RSV A Long was subjected to identical procedures for analysis. The PCR products were purified with a QIAquick PCR purification kit (Qiagen), according to the manufacturer's protocol, and sequenced by standard protocols (Agencourt Bioscience, Beverly, MA). For each amplicon, the forward and the reverse sequences were obtained. The sequences were analyzed and aligned by use of the Clustal W program and ContigExpress with Vector NTI software (Invitrogen).
RSV genotyping.
The genotyping of all 21 isolates from Belgium was performed as described previously (85). Genotyping of the remaining 78 isolates (which were of U.S. origin) was performed by Jeffrey Kahn's laboratory, Department of Pediatrics, Yale University, New Haven, CT. Analysis of the RSV attachment (G) protein gene was performed by first generating cDNA with random hexamers and Moloney murine leukemia virus reverse transcriptase (New England Biolabs, Beverly, MA) at 37°C for 1 h. This was followed by PCR amplification with HotStar Taq DNA polymerase (Qiagen) and G-protein-gene-specific primer GTmF (5′-CCGCGGGTTCTGGCAATGATAATCTCAAC-3′) and subgroup-specific G-protein-gene-specific primer RSV A-GAR2 (5′-GCCGCGTGTATAATTCATAAACCTTGGTAG-3′) or RSV B-GBR (5′-GGGGCCCCGCGGCCGCGCATTAATAGCAAGAGTTAGGAAG-3′) by denaturation at 95°C for 15 min, followed by 40 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min and then a single 10-min extension at 72°C. The PCR products were analyzed by 2% agarose gel electrophoresis. If a PCR product of the appropriate size was identified (for RSV serotype A, 1,200 nt; for RSV serotype B, 900 nt), the product was purified by using a QIAquick extraction kit (Qiagen), according to the manufacturer's protocol. The purified PCR products were analyzed by agarose gel electrophoresis and were sequenced on a 3730 XL DNA analyzer (Applied Biosystems, Foster City, CA).
The nucleotide sequences were aligned manually, and the alignment was confirmed by using the Clustal W program. RSV serotype A and RSV serotype B isolates were distinguished by comparing the G-gene nucleotide sequences and laboratory standards. Phylogenetic analysis of the RSV serotype A isolates was performed with an aligned 417-nt segment of the G-protein gene corresponding to nucleotide positions 5010 to 5426 (GenBank accession no. M74568). Phylogenetic analysis of RSV serotype B isolates was performed with an aligned 288-nt segment of the G-protein gene corresponding to nucleotide positions 5036 to 5323 (GenBank accession no. AF013254). Data sets containing 1,000 aligned permuted nucleotide sequence sets for bootstrap analysis were produced by using the SEQBOOT program (PHYLIP package, version 3.65). Maximum-likelihood phylogenetic trees were obtained by using the DNAML program (PHYLIP package, version 3.65) with default settings and the data sets used for bootstrap analysis. The CONSENSE program (PHYLIP package, version 3.65) was used to produce an extended-majority-rule consensus tree, and the trees were produced with the MEGA (version 4) program. Bootstrap values, isolate clustering, and previous genotype assignments (45, 85, 86) were used to determine the RSV genotypes.
RESULTS
Bioinformatic analysis of RSV genome and selection of ALN-RSV01.
RSV is a member of the Pneumovirinae subfamily in the Pneumovirus genus. There are two major groups of human RSV, serotypes A and B, and both serotypes cocirculate in the community. RSV has a negative single-strand RNA genome containing two nonstructural genes (NS2, NS1), followed by eight other genes, namely, the genes for the N, P, matrix (M), small hydrophobic (SH), G, F, second matrix (M2), and RNA-dependent RNA polymerase (L) proteins, in the order 3′-NS1-NS2-N-P-M-SH-G-F-M2-L-5′. The N, P, and L proteins are contained within the nucleocapsid of the virion and are required for various steps within the replication cycle. Consistent with their absence from the outer virus surface, the RNAs encoding the N, P, and L proteins are among the most highly conserved regions of the RSV genome (67, 68). We therefore reasoned that the screening of siRNAs targeting these mRNAs would result in the selection of the most potent and broad-spectrum inhibitors of viral replication.
To select appropriate siRNAs, sequences with GenBank accession numbers AF035006 (RSV A2), AF013255 (RSV B1), AY911262 (RSV A Long), and D00736 (RSV 18537) were aligned by using the Clustal W algorithm to identify conserved 19-mers among all RSV sequences analyzed. To determine the uniqueness of each 19-mer across the human genome, an analysis of the sequences with those in the RefSeq database was performed with BLAST. Only siRNAs with homology to any gene in the human genome of 16 nt or fewer were selected for further analysis.
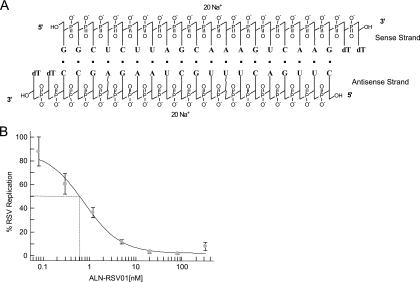
Seventy siRNAs targeting the RSV N, P, and L genes were analyzed in an in vitro RSV plaque inhibition assay; and 19 inhibited plaque formation by >80% compared with that achieved with a PBS control at siRNA concentrations of 20 nM (data not shown). Of these 19 siRNAs, the siRNA designated ALN-RSV01 (Fig. 1A), which targets the N-protein gene, consistently demonstrated the highest antiviral activity. Indeed, ALN-RSV01 showed a 50% inhibitory concentration (IC50) of 0.7 nM in the RSV plaque inhibition assay (Fig. 1B). The in vitro activity of ALN-RSV01 was further evaluated with human lung epithelial cells (A549 cells), with which it demonstrated >97% inhibition and an IC50 of 0.6 nM (data not shown).
FIG. 1.
Structure of ALN-RSV01 duplex and IC50 of ALN-RSV01 in vitro. (A) ALN-RSV01 is a synthetic double-stranded RNA oligonucleotide formed by hybridization of two partially complementary single-stranded RNAs in which the 3′ ends are capped with two thymidine units. (B) Vero cells in 24-well plates were transfected with decreasing concentrations of ALN-RSV01, followed by infection with 200 to 300 PFU of RSV A2. At 5 days postinfection, the cells were fixed, immunostained, and counted. The activity of ALN-RSV01 was plotted as a percentage of that of PBS, and the IC50 was measured with XLFit software. Error bars represent standard deviations.
ALN-RSV01 inhibition of RSV primary isolates.
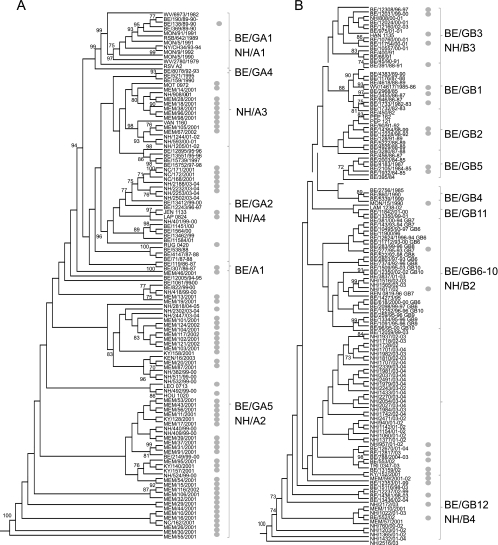
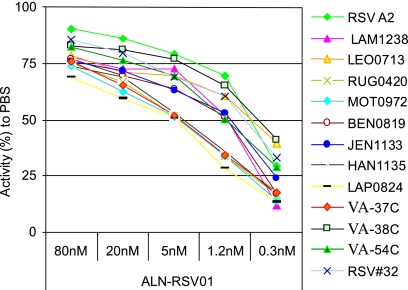
In order to evaluate the ability of ALN-RSV01 to provide broad-spectrum activity across RSV strains, the ALN-RSV01 recognition elements from a series of primary low-passage isolates (genotype analysis; Fig. 2A and B) taken from the nasal washes of children with confirmed RSV disease were sequenced. Of the RSV primary isolates sequenced, 94% (89/95) showed absolute sequence conservation (19/19 nt) across the ALN-RSV01 target site. The six isolates that were not 100% conserved each had a single-base alteration within the ALN-RSV01 target site. Four had C-to-U mutations at position 4 with respect to the 5′ end of the antisense strand of RSV01, one had an A-to-G mutation at position 7, and one had a G-to-A mutation at position 1. A subset of these 95 isolates was tested by the in vitro viral inhibition assay, including one isolate with a mismatch at position 4 and another with a mismatch at position 7 (Table 1). Of these, 12/12 (100%) exhibited ∼70% inhibition with 80 nM ALN-RSV01 compared to the level of inhibition achieved with the PBS control, and all had similar dose-response curves for inhibition by ALN-RSV01 (Fig. 3).
FIG. 2.
Genotype analysis of RSV primary isolates. Maximum-likelihood phylogenetic trees for RSV serotype A (A) and RSV serotype B (B) strains on the basis of analysis of the RSV G-protein gene. Bootstrap values of ≥70% (1,000 replicates) are shown at the corresponding nodes. Circles, isolates whose ALN-RSV01 target sites were analyzed.
TABLE 1.
ALN-RSV01 target site within the RSV N gene of primary clinical isolates and the RSV A2 laboratory strain
| Isolate name | Sequence at target sitea | Subtype | IC50 (nM) |
|---|---|---|---|
| RSV A2 | GGCTCTTAGCAAAGTCAAG | A | 1 |
| LAP 0824 | GGCTCTTAGCAAGGTCAAG | A | 4.9 |
| HAN 1135 | GGCTCTTAGCAAAGTTAAG | B | 4.7 |
| LAM 1238 | GGCTCTTAGCAAAGTCAAG | B | 1.2 |
| LEO 0713 | GGCTCTTAGCAAAGTCAAG | A | 1 |
| RUG 0420 | GGCTCTTAGCAAAGTCAAG | A | 4.8 |
| MOT 0972 | GGCTCTTAGCAAAGTCAAG | A | 4.9 |
| BEN 0819 | GGCTCTTAGCAAAGTCAAG | B | 1.2 |
| JEN 1133 | GGCTCTTAGCAAAGTCAAG | A | 1.1 |
| VA 37C | GGCTCTTAGCAAAGTCAAG | A | 4.8 |
| VA-38C | GGCTCTTAGCAAAGTCAAG | A | 1 |
| VA-54C | GGCTCTTAGCAAAGTCAAG | A | 1.2 |
| RSV 32 | GGCTCTTAGCAAAGTCAAG | A | 1 |
Nucleotides not conserved in the target region are indicated in boldface.
FIG. 3.
In vitro inhibition of primary RSV isolates by ALN-RSV01. Vero cells in 24-well plates were transfected with decreasing concentrations of ALN-RSV01, followed by infection with 200 to 300 PFU of RSV primary isolates. At day 5 postinfection, the cells were fixed, immunostained, and counted. The activity of ALN-RSV01 was plotted as a percentage of that of PBS.
In vivo studies of ALN-RSV01.
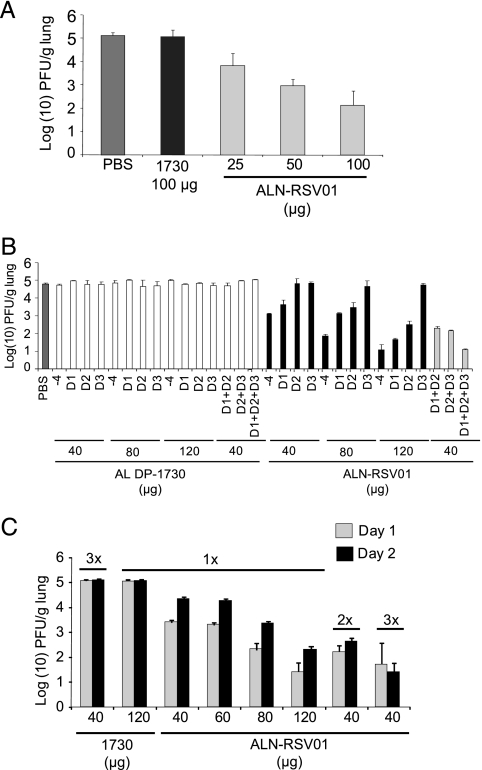
The BALB/c mouse is a well-established model for RSV infection and was thus chosen as the in vivo system to be used for evaluation of the antiviral effect of ALN-RSV01. Studies were initially performed with a prophylaxis model; siRNA was administered i.n. to mice 4 h prior to infection with 106 PFU of RSV A2. There was a dose-dependent inhibition of RSV A2 replication in the lungs of mice, with a 100-μg dose of ALN-RSV01 reducing the titers by between 2.5 and 3.0 log10 PFU/g lung compared to the titers for mice treated with either PBS or a nonspecific siRNA (Fig. 4A). Fifty- and 25-μg doses yielded reductions of approximately 2.0 and 1.25 log10 PFU/g, respectively (Fig. 4A).
FIG. 4.
In vivo activity of ALN-RSV01 following administration of single and multiple doses in BALB/c mice. In each panel, each group represents five animals, error bars represent standard deviations, and the threshold of detection of the assay is 1.7 log10 PFU/g. (A) ALN-RSV01 in vivo dose-response curve. BALB/c mice were treated i.n. with ALN-RSV01 at increasing concentrations (25 μg, 50 μg, or 100 μg), negative control siRNA AL-DP-1730 (1730; 100 μg), or PBS 4 h prior to infection with 1 × 106 PFU of RSV A2. The lungs were harvested, and virus was quantified by a standard immunostaining plaque assay and plotted as log10 PFU/g lung. (B) ALN-RSV01 multidose study. BALB/c mice were treated i.n. with ALN-RSV01 or mismatch siRNA (AL-DP-1730) at either 40 μg, 80 μg, or 120 μg (single-dose treatment) or 40 μg (multiple-dose, daily treatment). The lungs were harvested, and virus was quantified by an immunostaining plaque assay on day 5. −4, 4 h prior to infection; D1, day 1 postinfection; D2, day 2 postinfection; D3, day 3 postinfection. (C) ALN-RSV01 same-day multidose study. BALB/c mice were treated i.n. with ALN-RSV01 or mismatch siRNA (1730) at either 40 μg, 60 μg, 80 μg, or 120 μg for the single-dose groups at day 1 or 2 after RSV infection or 40 μg two or three times daily for the multidose groups at day 1 or 2 after RSV infection. The lungs were harvested, and virus was quantified by an immunostaining plaque assay.
To evaluate the efficiency of viral inhibition in a treatment paradigm, ALN-RSV01 was delivered i.n. in single or multiple daily doses at 1, 2, and/or 3 days postinfection; and the results were compared to those achieved with prophylactic dosing. When ALN-RSV01 was delivered as a single dose, the most efficacious antiviral effect was seen when it was given as a prophylactic (−4 h) dose, and this occurred in a dose-dependent fashion. Compared to the mismatch control (AL-DP-1730), the administration of 120 μg of ALN-RSV01 as a single prophylactic dose resulted in maximal viral inhibition, attenuating the lung concentrations of RSV to background levels in this assay. When ALN-RSV01 was administered in a treatment regimen as a single dose following viral inoculation, the dose-dependent antiviral efficacy was found to decrease as a function of the time of dosing after viral infection (Fig. 4B). Indeed, by day 3 postinfection, single doses as high as 120 μg did not yield any significant viral inhibition. However, when multiple 40-μg doses of ALN-RSV01 were delivered daily on days 1, 2, and 3, potent antiviral activity was maintained and the viral titers were again reduced to background levels (Fig. 4B).
To further explore alternative dosing paradigms that could be employed in future clinical studies, additional multidose regimens were evaluated. To this end, RSV-infected mice were treated with ALN-RSV01 either two times per day or three times per day. Interestingly, the multiple-daily-dose regimen of the RSV-specific siRNA (40 μg three times a day) was found to be as efficacious as a single 120-μg dose (Fig. 4C). In the aggregate, these data show that an ALN-RSV01 multidose treatment regimen can provide a maximal antiviral effect in a fashion readily applicable to human clinical studies.
ALN-RSV01 and cytokine induction.
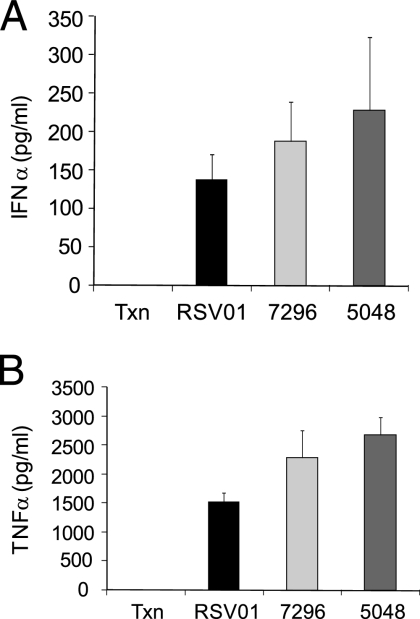
Many nucleic acids, including double-stranded RNAs, single-stranded RNAs, and siRNAs, have been shown to stimulate innate immune responses through a variety of RNA binding receptors (58, 60). This stimulation can be monitored in vitro in a PBMC assay (32, 37, 63). While the immunostimulatory property of an siRNA could act in combination with an RNAi-mediated mechanism to produce a more robust anti-RSV effect, such a feature might also confound the interpretation of the results related to an siRNA treatment strategy. Accordingly, ALN-RSV01 was evaluated for its ability to stimulate IFN-α and TNF-α in vitro by incubating ALN-RSV01 with freshly purified PBMCs, as described above. High concentrations of ALN-RSV01 (133 nM) that exceeded the IC50 for an antiviral effect by over 100-fold were used in these assays. After 24 h, only modest levels of both IFN-α and TNF-α were detected by ELISA, with an average of approximately 147 pg/ml of IFN-α (Fig. 5A) and 1,500 pg/ml of TNF-α (Fig. 5B) being induced, whereas no IFN-α or TNF-α was induced in the controls treated with medium alone.
FIG. 5.
ALN-RSV01 is a modest stimulator of IFN-α and TNF-α in vitro. Transfection reagent alone (Txn) or siRNAs (ALN-RSV01 [RSV01] or mismatch-positive controls siRNA AL-DP-7296 [7296] and siRNA AL-DP-5048 [5048]) were transfected into PBMCs and assayed by ELISA for cytokine induction at 24 h posttransfection. (A) IFN-α induction; (B) TNF-α induction.
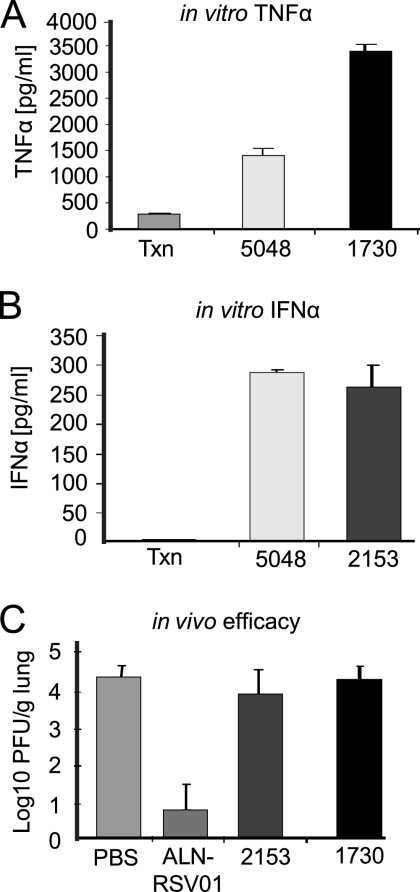
To verify that the antiviral activity of ALN-RSV01 was not influenced by this modest induction of cytokines, two non-RSV-specific siRNAs shown to induce either IFN-α or TNF-α more significantly than ALN-RSV01 were assayed in the in vivo BALB/c mouse model. Neither AL-DP-1730, a TNF-α inducer (Fig. 6A), nor AL-DP-2153, an IFN-α inducer (Fig. 6B), inhibited RSV A2 when administered to the mice i.n. (100 μg), whereas strong inhibition was observed when ALN-RSV01 was administered (Fig. 6C). Importantly, even 10-fold higher doses of AL-DP-1730 had no effect on the viral load when it was delivered prophylactically 4 h prior to infection (data not shown).
FIG. 6.
TNF-α- and IFN-α-stimulatory mismatched siRNAs do not modulate RSV in vivo. (A) Transfection reagent alone (Txn) or siRNAs (AL-DP-5048 [5048] and AL-DP-1730 [1730]) transfected into PBMCs were assayed for TNF-α stimulation at 24 h posttransfection. (B) siRNAs (AL-DP-5048 [5048] and AL-DP-2153 [2153]) transfected into PBMCs were assayed for IFN-α stimulation at 24 h posttransfection. (C) Lung virus concentrations from mice dosed i.n. with RSV at day 0 and with RSV-specific siRNAs (ALN-RSV01) or mismatch control immunostimulatory siRNAs (AL-DP-2153 [2153] and AL-DP-1730 [1730]) at 4 h before inoculation. Lung RSV concentrations were measured by an immunostaining plaque assay at day 5 postinfection.
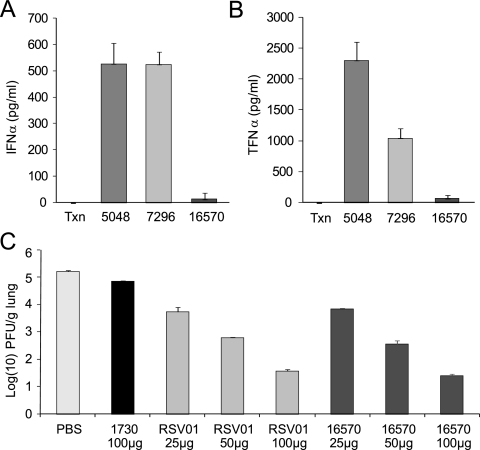
To further evaluate the role of immune activation on the antiviral efficacy of ALN-RSV01, an immune-silent form of this siRNA, AL-DP-16570, containing 2′-methoxy modifications, was synthesized. The sense strand is 5′-GGu uCU UAG cAa AGu cAa GdTdT-HP-3′, and the antisense strand is 5′-Cuu GAC UUU GCu AAG AGC cdTdT-HP-3′. The lowercase letters indicate the 2′-methoxy modifications, and HP indicates hydroxyproline. AL-DP 16570 showed potent antiviral activity in vitro and had an IC50 of ∼1 nM (data not shown), which is comparable to that measured for ALN-RSV01. Furthermore, in PBMC assays, the use of high concentrations of AL-DP-16570 (133 nM) resulted in no significant induction of either IFN-α or TNF-α (Fig. 7A and B, respectively). AL-DP-16570 was then tested for its in vivo antiviral efficacy in the mouse model. Compared with the activity of a nonspecific siRNA control, AL-DP-16570 showed potent antiviral activity that was comparable to that of the parent sequence ALN-RSV01 (Fig. 7C). These data support the conclusion that the antiviral effects of ALN-RSV01 are mediated by an RNAi mechanism and not via the induction of innate immunity.
FIG. 7.
Chemically modified ALN-RSV01 (AL-DP-16571) is immunologically silent, while it maintains antiviral activity. (A) Transfection reagent alone (Txn), AL-DP-16570 (16570), or positive control siRNAs AL-DP-5048 (5048) and AL-DP-7296 (7296) were transfected into PBMCs and assayed for IFN-α stimulation at 24 h posttransfection. (B) AL-DP-16570 or positive control siRNAs AL-DP-5048 and AL-DP-7296 were transfected into PBMCs and assayed for TNF-α stimulation at 24 h postinfection. (C) Lung virus concentrations from mice dosed i.n. with RSV at day 0 and with ALN-RSV01 (RSV01), AL-DP-16570 (16570), or a mismatched control at the indicated concentrations at 4 h before inoculation. Lung RSV concentrations were measured by an immunostaining plaque assay at day 5 postinfection.
In vitro and in vivo RACE analysis of ALN-RSV01 cleavage product.
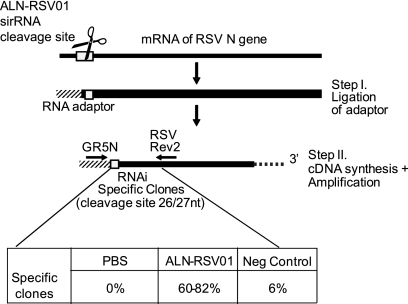
The RNA-induced silencing complex-mediated cleavage of a specific mRNA transcript occurs exactly 10 nt from the 5′ end of the siRNA antisense strand. To definitively confirm an RNAi-mediated mechanism of action for ALN-RSV01, a 5′ RACE assay was used. This assay allows the capture and sequence analysis of the specific RNAi cleavage product mRNA intermediate following ALN-RSV01 treatment both in vitro and in vivo. Following siRNA transfection into Vero cells and subsequent infection with RSV A2, a specific cleavage fragment could be detected only in the samples treated with ALN-RSV01 and not in samples treated with either PBS or a mismatch control siRNA (AL-DP-2153) (data not shown). In these experiments, 92% of the sequenced clones resulted from site-specific cleavage (between positions 26 and 27 of RSV A2 N mRNA) (data not shown). When they were analyzed in vivo, 60 to 82% of the clones isolated from the lung tissue of ALN-RSV01 treated, RSV-infected mice demonstrated site-specific cleavage of the N-gene transcript between positions 26 and 27 (Fig. 8). Only animals treated with ALN-RSV01, in contradistinction to those treated with PBS or the mismatch control siRNA, yielded significant numbers of clones whose sequence was confirmed to be the result of cleavage at the predicted site (Fig. 8).
FIG. 8.
ALN-RSV01 viral inhibition is mediated by RNAi in vivo. Shown is a schematic representation of the 5′ RACE assay used to demonstrate the generation of the site-specific cleavage product. Boxed are the results of sequence analysis of individual clones from PCR amplification of cDNAs generated from linker-adapted RSV N-gene mRNA isolated from an in vivo viral inhibition assay in which mice were inoculated with RSV at day 0 and treated with ALN-RSV01, AL-DP-2153 (Neg Control), or PBS at day 3, followed by lung homogenization and evaluation by RACE at day 5 postinfection.
DISCUSSION
While a large number of anti-RSV therapeutic strategies have been evaluated, including strategies that use antisense oligonucleotides (12, 52, 71), deoxyribozymes (79, 83, 84), small-molecule inhibitors (15, 16, 55), fusion protein inhibitors (22, 26, 56), and morpholino compounds (42), no approach has led to any new positive clinical data or FDA-approved products. At the same time, the level of unmet need for safe and effective therapies remains high because of the more than 125,000 pediatric hospitalizations (61) and 170,000 adult hospitalizations (19) in the United States annually. In this report, we describe the generation of a novel RNAi therapeutic agent for the treatment of RSV infection. This pan-RSV siRNA, ALN-RSV01, demonstrates antiviral activity both in vitro and in vivo in a number of dosing paradigms relevant to clinical use and shows an RNAi-specific antiviral mechanism of action.
ALN-RSV01 is a double-stranded siRNA duplex with 19 paired nucleotides complementary to a highly conserved region of the RSV N-protein gene and 2-nt dT overhangs at the 3′ end of each strand; this structure represents the canonical Tuschl design structure that is most validated in the literature (17). The molecule was purposefully designed to be stable as an agent for local delivery to the lung and unstable in systemic circulation to minimize any potential systemic exposure. Indeed, in serum, ALN-RSV01 has a half-life of ∼30 min, while in respiratory secretions from RSV-positive children, the half life is >5 h (data not shown). Thus, ALN-RSV01 would be rapidly degraded after distribution from the lung, preventing any prolonged systemic exposure.
A major consideration when an RSV therapeutic is developed is the requirement that the molecule be broadly active across the spectrum of viral genotypes. RSV can be classified into two subgroups, serotypes A and B, and the sequences of all 11 genes identified differ between the two serotypes. Genotypes can vary from year to year, with certain serotypes dominating in some years, but the cocirculation of multiple RSV strains is commonly observed (28, 41). Although RSV serotype A may produce greater viral loads in infants (29, 46, 49, 74) and RSV serotype A may produce disease slightly more severe than that produced by RSV serotype B, both RSV serotype A and RSV serotype B produce a similar and clinically indistinguishable disease spectrum. In the case of an RNAi therapeutic, broad-spectrum activity is achieved by selecting an siRNA molecule whose corresponding mRNA target site is conserved across the various circulating RSV isolates. This is achieved for ALN-RSV01 by targeting the relatively conserved viral N protein, which is less prone to mutation than surface viral proteins, such as the G or the F protein. Indeed, within RSV, the N-protein gene is among the most conserved, with approximately 86% identity at the nucleic acid level and 96% identity at the amino acid level. Sequence differences within the target site were observed at a low frequency in clinical specimens and largely created a G-U wobble between the antisense strand of the siRNA and the target RSV mRNA. As expected, all clinical isolates tested were effectively inhibited in vitro by ALN-RSV01 (Fig. 4). Although the IC50 for the two isolates with G-U wobbles was 5 nM (the top of the range), it should be noted that other isolates, namely, isolates RUG 0420 and MOT 0472, both of which show 100% sequence conservation at the ALN-RSV01 target site, also exhibited IC50s of 5 nM (Table 1). This suggests that the target site divergence in the isolates with nonconserved sequences does not account for the slightly higher IC50. It is likely that the growth and replication properties of these isolates account for their slightly higher IC50s. These primary isolates spanned genotypes A and B, indicating that ALN-RSV01 is likely active against all currently circulating strains.
The mechanism of RNAi is such that the specificity is determined by sequence complementarity, and thus, ALN-RSV01 would not be effective against other viruses, even closely related viruses within the subfamily Pneumoviridae (i.e., human metapneumovirus). In fact, the ALN-RSV01 target region within the N-protein gene of human metapneumovirus diverges from ALN-RSV01 target region within the N-protein gene of RSV almost completely, and we therefore predict that ALN-RSV01 would not inhibit human metapneumovirus. In addition, Bitko et al. (6) have also demonstrated the specificity of RNAi-mediated inhibition of RSV by showing that their RSV-specific siRNA did not inhibit parainfluenza virus type 3. Although we have not attempted to generate recombinant viruses via reverse genetics to identify the positions which would make viruses resistant to ALN-RSV01, our large genetic data set of circulating primary RSV isolates shows a high degree of conservation of the sequence within this target region in both subtype A and subtype B viruses (Fig. 2A and B). This conservation likely predicts an important function for this region and suggests an evolutionary bias against sequence changes there.
The importance of target sequence conservation was also explored in a study evaluating an siRNA targeting the RSV P gene (data not shown). Specifically, this siRNA was identical to the sequence in laboratory strain RSVA2 but had a single-base variation compared with the sequence of laboratory strain RSV A Long. Interestingly, while complete inhibition of RSV A2 was achieved, no inhibition of RSV A Long was observed in vitro. This result was expected on the basis of the specific position of the mismatch (position 7 with respect to the antisense strand of the siRNA), as this position in the siRNA seed is known to be important for target recognition (34).
In a large series of in vivo studies, ALN-RSV01 was found to be a potent antiviral in both prophylaxis and treatment paradigms, achieving up to 3-log-unit viral load reductions compared to the reductions achieved with either PBS or nonspecific siRNA controls. Bitko et al. (6) and Zhang et al. (82) have also previously demonstrated siRNA inhibition of RSV in BALB/c mice. They targeted either the P-protein mRNA (6) or the NS1-protein mRNA (82) and achieved reductions similar to those described here. These RNAi-mediated antiviral activities are similar to those previously reported for ribavirin and palivizumab (33, 35, 78). While these other siRNAs were effective inhibitors of viral replication in vivo, neither is ideally suited for RNAi therapeutic development. The P-protein gene targeting the siRNA of Bitko et al. (6) is directed against a region of the virus that is not conserved across RSV serotypes A and B. Furthermore, the NS1 protein-targeting siRNA of Zhang et al. (82) is believed to function (at least in part) as an immune modulator by inhibiting the production of the RSV protein that inhibits the induction of IFN.
Applications of ALN-RSV01 in clinical settings will require optimization of the dosing strategy in terms of dose level and frequency. In the current study, we evaluated the effects of multidosing by comparing a single-dose regimen with the administration of either multiple daily doses or multiple doses on only a single day. Multidosing with low doses of siRNA was equivalent or slightly more effective than the equivalent total dose delivered in a single administration, suggesting that multidosing may be the optimal strategy for the delivery of ALN-RSV01.
Recent studies have suggested that some siRNAs may function not by the intended mechanism of RNAi but by nonspecifically activating the innate immune system (37, 40, 57, 80). Such a nonspecific effect might confound the interpretation of a direct antiviral mechanism, as immune stimulation could itself provide an antiviral response. Numerous experiments were performed to confirm that the mechanism of viral inhibition by ALN-RSV01 is due to RNAi. First, as a control for the antiviral efficacy of ALN-RSV01, nonspecific siRNAs were used. Second, non-RSV-targeting siRNAs with robust IFN-α (AL-DP-2153) or TNF-α (AL-DP-1730) stimulatory activity were found to be ineffective in our in vivo model. Third, a derivative of ALN-RSV01 (AL-DP-16570) that was chemically modified to reduce immune stimulatory activity maintained essentially indistinguishable antiviral activity in vitro and in vivo. AL-DP-16570 is one of a series of molecules currently under evaluation in our in vivo RSV model. Fourth, and most importantly, 5′ RACE was employed to capture the specific cleavage products from both the in vitro and the in vivo degradation of the target N-gene mRNA. By this technique, isolated clones from ALN-RSV01-treated mice demonstrated site-specific cleavage of the N-gene mRNA at a position corresponding to that exactly 10 nt from the 5′ end of the siRNA antisense strand (Fig. 8). The small percentage of clones resulting from nonspecific cleavage at other sites in the RSV N gene reflects the inherent instability of the 5′ end of the RSV N gene mRNA. In aggregate, these data demonstrate that the inhibition of RSV replication by ALN-RSV01 is via an RNAi mechanism and not via immune modulation.
RNAi represents a promising strategy for the discovery and development of antiviral medicines. Our studies of ALN-RSV01 thus provide a road map for considerations in the advancement of this approach in general, including the selection of conserved target sequences, in vivo pharmacology, and proof of the mechanism of action. Furthermore, these studies lay a foundation for the further study of ALN-RSV01 and other virus-targeting siRNAs in human clinical trials (14).
Footnotes
Published ahead of print on 8 June 2009.
REFERENCES
- 1.Altschul, S. F., and E. V. Koonin. 1998. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci. 23:444-447. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1998. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 102:26-28. [PubMed] [Google Scholar]
- 3.Anonymous. 2003. Revised indications for the use of palivizumab and respiratory syncytial virus immune globulin intravenous for the prevention of respiratory sycytial virus infections. Pediatrics 112:1442-1446. [PubMed] [Google Scholar]
- 4.Arbuthnot, P., S. Carmona, and A. Ely. 2005. Exploiting the RNA interference pathway to counter hepatitis B virus replication. Liver Int. 25:9-15. [DOI] [PubMed] [Google Scholar]
- 5.Billings, J. L., M. I. Hertz, and C. H. Wendt. 2001. Community respiratory virus infections following lung transplantation. Transpl. Infect. Dis. 3:138-148. [DOI] [PubMed] [Google Scholar]
- 6.Bitko, V., A. Musiyenko, O. Shulyayeva, and S. Barik. 2005. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 11:50-55. [DOI] [PubMed] [Google Scholar]
- 7.Bosher, J. M., and M. Labouesse. 2000. RNA interference: genetic wand and genetic watchdog. Nat. Cell Biol. 2:E31-E36. [DOI] [PubMed] [Google Scholar]
- 8.Boyce, T. G., B. G. Mellen, E. F. Mitchel, Jr., P. F. Wright, and M. R. Griffin. 2000. Rates of hospitalization for respiratory syncytial virus infection among children in Medicaid. J. Pediatr. 137:865-870. [DOI] [PubMed] [Google Scholar]
- 9.Carmichael, G. G. 2002. Medicine: silencing viruses with RNA. Nature 418:379-380. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2002. Respiratory syncytial virus activity-United States, 2000-01 season. MMWR Morb. Mortal. Wkly. Rep. 51:26-28. [PubMed] [Google Scholar]
- 11.Couch, R. B., J. A. Englund, and E. Whimbey. 1997. Respiratory viral infections in immunocompetent and immunocompromised persons. Am. J. Med. 102:2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cramer, H. 2005. Antisense approaches for inhibiting respiratory syncytial virus. Expert Opin. Biol. Ther. 5:207-220. [DOI] [PubMed] [Google Scholar]
- 13.de Fougerolles, A., H. P. Vornlocher, J. Maraganore, and J. Lieberman. 2007. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 6:443-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeVincenzo, J., J. E. Cehelsky, R. Alvarez, S. Elbashir, J. Harborth, I. Toudjarska, L. Nechev, V. Murugaiah, A. Van Vliet, A. K. Vaishnaw, and R. Meyers. 2008. Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV). Antivir. Res. 77:225-231. [DOI] [PubMed] [Google Scholar]
- 15.Douglas, J. L., M. L. Panis, E. Ho, K. Y. Lin, S. H. Krawczyk, D. M. Grant, R. Cai, S. Swaminathan, X. Chen, and T. Cihlar. 2005. Small molecules VP-14637 and JNJ-2408068 inhibit respiratory syncytial virus fusion by similar mechanisms. Antimicrob. Agents Chemother. 49:2460-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglas, J. L., M. L. Panis, E. Ho, K. Y. Lin, S. H. Krawczyk, D. M. Grant, R. Cai, S. Swaminathan, and T. Cihlar. 2003. Inhibition of respiratory syncytial virus fusion by the small molecule VP-14637 via specific interactions with F protein. J. Virol. 77:5054-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 18.Falsey, A. R. 1998. Respiratory syncytial virus infection in older persons. Vaccine 16:1775-1778. [DOI] [PubMed] [Google Scholar]
- 19.Falsey, A. R., P. A. Hennessey, M. A. Formica, C. Cox, and E. E. Walsh. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 352:1749-1759. [DOI] [PubMed] [Google Scholar]
- 20.Falsey, A. R., and E. E. Walsh. 2000. Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 13:371-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fixler, D. E. 1996. Respiratory syncytial virus infection in children with congenital heart disease: a review. Pediatr. Cardiol. 17:163-168. [DOI] [PubMed] [Google Scholar]
- 22.Gazumyan, A., B. Mitsner, and G. A. Ellestad. 2000. Novel anti-RSV dianionic dendrimer-like compounds: design, synthesis and biological evaluation. Curr. Pharm. Des. 6:525-546. [DOI] [PubMed] [Google Scholar]
- 23.Ge, Q., M. T. McManus, T. Nguyen, C. H. Shen, P. A. Sharp, H. N. Eisen, and J. Chen. 2003. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. USA 100:2718-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giladi, H., M. Ketzinel-Gilad, L. Rivkin, Y. Felig, O. Nussbaum, and E. Galun. 2003. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol. Ther. 8:769-776. [DOI] [PubMed] [Google Scholar]
- 25.Glezen, P., and F. W. Denny. 1973. Epidemiology of acute lower respiratory disease in children. N. Engl. J. Med. 288:498-505. [DOI] [PubMed] [Google Scholar]
- 26.Gower, T. L., and B. S. Graham. 2001. Antiviral activity of lovastatin against respiratory syncytial virus in vivo and in vitro. Antimicrob. Agents Chemother. 45:1231-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groothuis, J. R., E. A. Simoes, M. J. Levin, C. B. Hall, C. E. Long, W. J. Rodriguez, J. Arrobio, H. C. Meissner, D. R. Fulton, R. C. Welliver, et al. 1993. Prophylactic administration of respiratory syncytial virus immune globulin to high-risk infants and young children. N. Engl. J. Med. 329:1524-1530. [DOI] [PubMed] [Google Scholar]
- 28.Guney, C., A. Kubar, M. Yapar, A. B. Besirbellioglu, and L. Doganci. 2004. An outbreak of respiratory infection due to respiratory syncytial virus subgroup B in Ankara, Turkey. Jpn. J. Infect. Dis. 57:178-180. [PubMed] [Google Scholar]
- 29.Hall, C. B., E. E. Walsh, K. C. Schnabel, C. E. Long, K. M. McConnochie, S. W. Hildreth, and L. J. Anderson. 1990. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J. Infect. Dis. 162:1283-1290. [DOI] [PubMed] [Google Scholar]
- 30.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 31.He, M. L., B. Zheng, Y. Peng, J. S. Peiris, L. L. Poon, K. Y. Yuen, M. C. Lin, H. F. Kung, and Y. Guan. 2003. Inhibition of SARS-associated coronavirus infection and replication by RNA interference. JAMA 290:2665-2666. [DOI] [PubMed] [Google Scholar]
- 32.Hornung, V., M. Guenthner-Biller, C. Bourquin, A. Ablasser, M. Schlee, S. Uematsu, A. Noronha, M. Manoharan, S. Akira, A. de Fougerolles, S. Endres, and G. Hartmann. 2005. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 11:263-270. [DOI] [PubMed] [Google Scholar]
- 33.Hruska, J. F., P. E. Morrow, S. C. Suffin, and R. G. Douglas, Jr. 1982. In vivo inhibition of respiratory syncytial virus by ribavirin. Antimicrob. Agents Chemother. 21:125-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson, A. L., J. Burchard, J. Schelter, B. N. Chau, M. Cleary, L. Lim, and P. S. Linsley. 2006. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12:1179-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, S., S. D. Griego, D. S. Pfarr, M. L. Doyle, R. Woods, D. Carlin, G. A. Prince, S. Koenig, J. F. Young, and S. B. Dillon. 1999. A direct comparison of the activities of two humanized respiratory syncytial virus monoclonal antibodies: MEDI-493 and RSHZl9. J. Infect. Dis. 180:35-40. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, S., C. Oliver, G. A. Prince, V. G. Hemming, D. S. Pfarr, S. C. Wang, M. Dormitzer, J. O'Grady, S. Koenig, J. K. Tamura, R. Woods, G. Bansal, D. Couchenour, E. Tsao, W. C. Hall, and J. F. Young. 1997. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 176:1215-1224. [DOI] [PubMed] [Google Scholar]
- 37.Judge, A. D., V. Sood, J. R. Shaw, D. Fang, K. McClintock, and I. MacLachlan. 2005. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 23:457-462. [DOI] [PubMed] [Google Scholar]
- 38.Kachko, A. V., A. V. Ivanova, E. V. Protopopova, V. Netesov, and V. B. Loktev. 2006. Inhibition of West Nile virus replication by short interfering RNAs. Dokl. Biochem. Biophys. 410:260-262. [DOI] [PubMed] [Google Scholar]
- 39.Kalina, W. V., and L. J. Gershwin. 2004. Progress in defining the role of RSV in allergy and asthma: from clinical observations to animal models. Clin. Dev. Immunol. 11:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinman, M. E., K. Yamada, A. Takeda, V. Chandrasekaran, M. Nozaki, J. Z. Baffi, R. J. Albuquerque, S. Yamasaki, M. Itaya, Y. Pan, B. Appukuttan, D. Gibbs, Z. Yang, K. Kariko, B. K. Ambati, T. A. Wilgus, L. A. DiPietro, E. Sakurai, K. Zhang, J. R. Smith, E. W. Taylor, and J. Ambati. 2008. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature 452:591-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuroiwa, Y., K. Nagai, L. Okita, and H. Tsutsumi. 2004. Genetic variability and molecular epidemiology of respiratory syncytial virus subgroup A strains in Japan determined by heteroduplex mobility assay. J. Clin. Microbiol. 42:2048-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai, S. H., D. A. Stein, A. Guerrero-Plata, S. L. Liao, T. Ivanciuc, C. Hong, P. L. Iversen, A. Casola, and R. P. Garofalo. 2008. Inhibition of respiratory syncytial virus infections with morpholino oligomers in cell cultures and in mice. Mol. Ther. 16:1120-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Law, B. J., E. E. Wang, N. MacDonald, J. McDonald, S. Dobson, F. Boucher, J. Langley, J. Robinson, I. Mitchell, and D. Stephens. 1997. Does ribavirin impact on the hospital course of children with respiratory syncytial virus (RSV) infection? An analysis using the Pediatric Investigators Collaborative Network on Infections in Canada (PICNIC) RSV database. Pediatrics 99:E7. [DOI] [PubMed] [Google Scholar]
- 44.Leader, S., and K. Kohlhase. 2002. Respiratory syncytial virus-coded pediatric hospitalizations, 1997 to 1999. Pediatr. Infect. Dis. J. 21:629-632. [DOI] [PubMed] [Google Scholar]
- 45.Martinello, R. A., M. D. Chen, C. Weibel, and J. S. Kahn. 2002. Correlation between respiratory syncytial virus genotype and severity of illness. J. Infect. Dis. 186:839-842. [DOI] [PubMed] [Google Scholar]
- 46.McConnochie, K. M., C. B. Hall, E. E. Walsh, and K. J. Roghmann. 1990. Variation in severity of respiratory syncytial virus infections with subtype. J. Pediatr. 117:52-62. [DOI] [PubMed] [Google Scholar]
- 47.McCown, M., M. S. Diamond, and A. Pekosz. 2003. The utility of siRNA transcripts produced by RNA polymerase i in down regulating viral gene expression and replication of negative- and positive-strand RNA viruses. Virology 313:514-524. [DOI] [PubMed] [Google Scholar]
- 48.Meissner, H. C., M. B. Rennels, S. S. Long, and L. K. Pickering. 2004. Immunoprophylaxis with RespiGam. Pediatrics 113:629. [DOI] [PubMed] [Google Scholar]
- 49.Mufson, M. A., B. Akerlind-Stopner, C. Orvell, R. B. Belshe, and E. Norrby. 1991. A single-season epidemic with respiratory syncytial virus subgroup B2 during 10 epidemic years, 1978 to 1988. J. Clin. Microbiol. 29:162-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogra, P. L. 2004. Respiratory syncytial virus: the virus, the disease and the immune response. Paediatr. Respir. Rev. 5(Suppl. A):S119-S126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peebles, R. S., Jr. 2004. Viral infections, atopy, and asthma: is there a causal relationship? J. Allergy Clin. Immunol. 113:S15-S18. [DOI] [PubMed] [Google Scholar]
- 52.Player, M. R., D. L. Barnard, and P. F. Torrence. 1998. Potent inhibition of respiratory syncytial virus replication using a 2-5A-antisense chimera targeted to signals within the virus genomic RNA. Proc. Natl. Acad. Sci. USA 95:8874-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pruitt, K. D., T. Tatusova, and D. R. Maglott. 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33:D501-D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Psarras, S., N. G. Papadopoulos, and S. L. Johnston. 2004. Pathogenesis of respiratory syncytial virus bronchiolitis-related wheezing. Paediatr. Respir. Rev. 5(Suppl. A):S179-S184. [DOI] [PubMed] [Google Scholar]
- 55.Razinkov, V., A. Gazumyan, A. Nikitenko, G. Ellestad, and G. Krishnamurthy. 2001. RFI-641 inhibits entry of respiratory syncytial virus via interactions with fusion protein. Chem. Biol. 8:645-659. [DOI] [PubMed] [Google Scholar]
- 56.Razinkov, V., C. Huntley, G. Ellestad, and G. Krishnamurthy. 2002. RSV entry inhibitors block F-protein mediated fusion with model membranes. Antivir. Res. 55:189-200. [DOI] [PubMed] [Google Scholar]
- 57.Robbins, M., A. Judge, E. Ambegia, C. Choi, E. Yaworski, L. Palmer, K. McClintock, and I. MacLachlan. 2008. Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation. Hum. Gene Ther. 19:991-999. [DOI] [PubMed] [Google Scholar]
- 58.Robbins, M., A. Judge, L. Liang, K. McClintock, E. Yaworski, and I. MacLachlan. 2007. 2′-O-Methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 15:1663-1669. [DOI] [PubMed] [Google Scholar]
- 59.Saksela, K. 2003. Human viruses under attack by small inhibitory RNA. Trends Microbiol. 11:345-347. [DOI] [PubMed] [Google Scholar]
- 60.Schlee, M., V. Hornung, and G. Hartmann. 2006. siRNA and isRNA: two edges of one sword. Mol. Ther. 14:463-470. [DOI] [PubMed] [Google Scholar]
- 61.Shay, D. K., R. C. Holman, R. D. Newman, L. L. Liu, J. W. Stout, and L. J. Anderson. 1999. Bronchiolitis-associated hospitalizations among US children, 1980-1996. JAMA 282:1440-1446. [DOI] [PubMed] [Google Scholar]
- 62.Simoes, E. A. 1999. Respiratory syncytial virus infection. Lancet 354:847-852. [DOI] [PubMed] [Google Scholar]
- 63.Sioud, M. 2005. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J. Mol. Biol. 348:1079-1090. [DOI] [PubMed] [Google Scholar]
- 64.Staat, M. A. 2002. Respiratory syncytial virus infections in children. Semin. Respir. Infect. 17:15-20. [DOI] [PubMed] [Google Scholar]
- 65.Stein, D. A., and P. Y. Shi. 2008. Nucleic acid-based inhibition of flavivirus infections. Front Biosci. 13:1385-1395. [DOI] [PubMed] [Google Scholar]
- 66.Stevens, T. P., and C. B. Hall. 2004. Controversies in palivizumab use. Pediatr. Infect. Dis. J. 23:1051-1052. [DOI] [PubMed] [Google Scholar]
- 67.Sullender, W. M. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin. Microbiol. Rev. 13:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sullender, W. M., L. Sun, and L. J. Anderson. 1993. Analysis of respiratory syncytial virus genetic variability with amplified cDNAs. J. Clin. Microbiol. 31:1224-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor, J. A., and N. V. Naoumov. 2005. The potential of RNA interference as a tool in the management of viral hepatitis. J. Hepatol. 42:139-144. [DOI] [PubMed] [Google Scholar]
- 70.Tompkins, S. M., C. Y. Lo, T. M. Tumpey, and S. L. Epstein. 2004. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc. Natl. Acad. Sci. USA 101:8682-8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torrence, P. F. 1999. 2-5A-antisense chimeras: inhibitors of respiratory syncytial virus infection. Curr. Opin. Mol. Ther. 1:307-315. [PubMed] [Google Scholar]
- 72.Tripp, R. A., D. Moore, L. Jones, W. Sullender, J. Winter, and L. J. Anderson. 1999. Respiratory syncytial virus G and/or SH protein alters Th1 cytokines, natural killer cells, and neutrophils responding to pulmonary infection in BALB/c mice. J. Virol. 73:7099-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ventre, K., and A. Randolph. 2004. Ribavirin for respiratory syncytial virus infection of the lower respiratory tract in infants and young children. Cochrane Database Syst. Rev. CD000181. [DOI] [PubMed]
- 74.Walsh, E. E., K. M. McConnochie, C. E. Long, and C. B. Hall. 1997. Severity of respiratory syncytial virus infection is related to virus strain. J. Infect. Dis. 175:814-820. [DOI] [PubMed] [Google Scholar]
- 75.Wang, Q. C., Q. H. Nie, and Z. H. Feng. 2003. RNA interference: antiviral weapon and beyond. World J. Gastroenterol. 9:1657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, Z., L. Ren, X. Zhao, T. Hung, A. Meng, J. Wang, and Y. G. Chen. 2004. Inhibition of severe acute respiratory syndrome virus replication by small interfering RNAs in mammalian cells. J. Virol. 78:7523-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu, C. J., H. W. Huang, C. Y. Liu, C. F. Hong, and Y. L. Chan. 2005. Inhibition of SARS-CoV replication by siRNA. Antivir. Res. 65:45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wyde, P. R., D. K. Moore, T. Hepburn, C. L. Silverman, T. G. Porter, M. Gross, G. Taylor, S. G. Demuth, and S. B. Dillon. 1995. Evaluation of the protective efficacy of reshaped human monoclonal antibody RSHZ19 against respiratory syncytial virus in cotton rats. Pediatr. Res. 38:543-550. [DOI] [PubMed] [Google Scholar]
- 79.Xie, Y. Y., X. D. Zhao, L. P. Jiang, H. L. Liu, L. J. Wang, P. Fang, K. L. Shen, Z. D. Xie, Y. P. Wu, and X. Q. Yang. 2006. Inhibition of respiratory syncytial virus in cultured cells by nucleocapsid gene targeted deoxyribozyme (DNAzyme). Antivir. Res. 71:31-41. [DOI] [PubMed] [Google Scholar]
- 80.Zamanian-Daryoush, M., J. T. Marques, M. P. Gantier, M. A. Behlke, M. John, P. Rayman, J. Finke, and B. R. Williams. 2008. Determinants of cytokine induction by small interfering RNA in human peripheral blood mononuclear cells. J. Interferon Cytokine Res. 28:221-233. [DOI] [PubMed] [Google Scholar]
- 81.Zhang, R., Z. Guo, J. Lu, J. Meng, C. Zhou, X. Zhan, B. Huang, X. Yu, M. Huang, X. Pan, W. Ling, X. Chen, Z. Wan, H. Zheng, X. Yan, Y. Wang, Y. Ran, X. Liu, J. Ma, C. Wang, and B. Zhang. 2003. Inhibiting severe acute respiratory syndrome-associated coronavirus by small interfering RNA. Chin Med. J. (English Ed.) 116:1262-1264. [PubMed] [Google Scholar]
- 82.Zhang, W., H. Yang, X. Kong, S. Mohapatra, H. San Juan-Vergara, G. Hellermann, S. Behera, R. Singam, R. F. Lockey, and S. S. Mohapatra. 2005. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat. Med. 11:56-62. [DOI] [PubMed] [Google Scholar]
- 83.Zhao, C. A., X. D. Zhao, H. G. Yu, Y. P. Wu, and X. Q. Yang. 2003. Inhibition of respiratory syncytial virus replication in cultured cells by RNA-cleaving DNAzyme. Zhonghua Er Ke Za Zhi 41:594-597. (In Chinese.) [PubMed] [Google Scholar]
- 84.Zhou, J., X. Q. Yang, Y. Y. Xie, X. D. Zhao, L. P. Jiang, L. J. Wang, and Y. X. Cui. 2007. Inhibition of respiratory syncytial virus of subgroups A and B using deoxyribozyme DZ1133 in mice. Virus Res. 130:241-248. [DOI] [PubMed] [Google Scholar]
- 85.Zlateva, K. T., P. Lemey, E. Moes, A. M. Vandamme, and M. Van Ranst. 2005. Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J. Virol. 79:9157-9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zlateva, K. T., P. Lemey, A. M. Vandamme, and M. Van Ranst. 2004. Molecular evolution and circulation patterns of human respiratory syncytial virus subgroup A: positively selected sites in the attachment G glycoprotein. J. Virol. 78:4675-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]