Abstract
Nontyphoidal Salmonella enterica strains with a nonclassical quinolone resistance phenotype were isolated from patients returning from Thailand or Malaysia to Finland. A total of 10 isolates of seven serovars were studied in detail, all of which had reduced susceptibility (MIC ≥ 0.125 μg/ml) to ciprofloxacin but were either susceptible or showed only low-level resistance (MIC ≤ 32 μg/ml) to nalidixic acid. Phenotypic characterization included susceptibility testing by the agar dilution method and investigation of efflux activity. Genotypic characterization included the screening of mutations in the quinolone resistance-determining regions (QRDR) of gyrA, gyrB, parC, and parE by PCR and denaturing high-pressure liquid chromatography and the amplification of plasmid-mediated quinolone resistance (PMQR) genes qnrA, qnrB, qnrS, qnrD, aac(6′)-Ib-cr, and qepA by PCR. PMQR was confirmed by plasmid analysis, Southern hybridization, and plasmid transfer. No mutations in the QRDRs of gyrA, gyrB, parC, or parE were detected with the exception of a Thr57-Ser substitution within ParC seen in all but the S. enterica serovar Typhimurium strains. The qnrA and qnrS genes were the only PMQR determinants detected. Plasmids carrying qnr alleles were transferable in vitro, and the resistance phenotype was reproducible in Escherichia coli DH5α transformants. These data demonstrate the emergence of a highly mobile qnr genotype that, in the absence of mutation within topoisomerase genes, confers the nontypical quinolone resistance phenotype in S. enterica isolates. The qnr resistance mechanism enables bacteria to survive elevated quinolone concentrations, and therefore, strains carrying qnr alleles may be able to expand during fluoroquinolone treatment. This is of concern since nonclassical quinolone resistance is plasmid mediated and therefore mobilizable.
Fluoroquinolones are among the most extensively used antimicrobial agents for the treatment of bacterial infections both in human and in veterinary medicine (16, 31). Unfortunately, in many countries, extensive fluoroquinolone use has led to increasing numbers of resistant isolates, including nontyphoidal strains of Salmonella enterica, being recorded (11, 12, 30). In Finland, the fluoroquinolone susceptibility of Salmonella isolates has been monitored in Finnish patients since 1995. Between 1995 and 2002, all Salmonella isolates with reduced fluoroquinolone susceptibility were uniformly resistant to nalidixic acid (9, 10, 17). In 2003, we discovered a novel population of S. enterica strains with reduced fluoroquinolone susceptibility (MIC ≥ 0.125 μg/ml) but with susceptibility (MIC < 32 μg/ml) or only low-level resistance (MIC = 32 μg/ml) to nalidixic acid (12). Since 2003, 36 Salmonella isolates demonstrating this novel, nonclassical resistance phenotype have been found in our laboratory, and all were isolated from travelers returning from Thailand or Malaysia (18).
Resistance to fluoroquinolones is typically mediated by alterations in the target enzymes DNA gyrase (GyrA and GyrB) and topoisomerase IV (ParE and ParC) or by changes in drug entry and efflux (14). Recently, plasmid-mediated quinolone resistance (PMQR) has emerged in Salmonella and in other Enterobacteriaceae, and the prevalence of PMQR is increasing worldwide (27). The first PMQR determinant identified was the qnrA gene (20). Subsequently, further qnr alleles, e.g., qnrB, qnrS, qnrC, and qnrD, have been described (3, 19). In addition, two other PMQR genes have been described: aac(6′)-Ib-cr, an aminoglycoside acetyltransferase variant gene which confers resistance to aminoglycosides in addition to reduced susceptibility to ciprofloxacin (28), and qepA (quinolone efflux pump) which extrudes hydrophilic fluoroquinolones including ciprofloxacin from the bacterial cell (35). Although all of these PMQR determinants confer only low-level resistance to fluoroquinolones, it is possible that PMQR is responsible for the rapid increase of fluoroquinolone resistance among Enterobacteriaceae (27, 31). It is also of concern that there seems to be a correlation between qnr-positive and extended-spectrum β-lactamase-positive isolates (2, 22, 25).
Since 2003, S. enterica isolates with a novel quinolone resistance phenotype, isolated from Finnish patients, have been detected. These isolates do not carry mutations within gyrA, and the molecular mechanism responsible for this nonclassical phenotype has remained unclear (12). The aim of the present study was to characterize the molecular mechanisms of quinolone resistance present in various serovars of S. enterica exhibiting the nonclassical quinolone resistance phenotype isolated from Finnish patients returning from Thailand and Malaysia.
(This work was presented in part at the 18th European Congress of Clinical Microbiology and Infectious Diseases [ECCMID], Barcelona, Spain, 2008 [P1536].)
MATERIALS AND METHODS
Salmonella strains.
Ten S. enterica strains belonging to seven serovars and showing the nonclassical quinolone resistance phenotype were chosen for detailed investigation. The strains were a sample from a total of 36 S. enterica strains, which showed reduced susceptibility (MIC ≥ 0.125 μg/ml) to ciprofloxacin and were either susceptible or had only low-level resistance (MIC ≤ 32 μg/ml) to nalidixic acid, collected from Finnish travelers returning from Thailand or Malaysia between 2003 and 2007. The strains were chosen to include different serovars, years of collection, and origins present in the collection. An isolate was designated to be of Malaysian or Thai origin if the patient had reported travel to those destinations within 1 month before the specimen date. The strains were serotyped in the Gastrointestinal Infection Unit of the National Institute for Health and Welfare, and they belonged to seven serovars: Braenderup (n = 1), Corvallis (n = 2), Mbandaka (n = 1), Montevideo (n = 2), Stanley (n = 1), Typhimurium (n = 2), and Virginia (n = 1). As controls, we used six clinical, quinolone-susceptible strains of the same serovars (Corvallis [from Brazil], Stanley [Thailand], Mbandaka [Finland], Braenderup [India], Montevideo [Finland], and Typhimurium [Vietnam]) and one laboratory strain (S. Typhimurium SL1344).
Antimicrobial susceptibility testing.
The MICs of the antibiotics were determined by the standard plate agar dilution method according to Clinical and Laboratory Standards Institute (CLSI) guidelines (5). Susceptibilities to nalidixic acid (Sigma, Steinheim, Germany), ciprofloxacin (Sigma), and five additional fluoroquinolones (enrofloxacin [Bayer, Elberfeld, Germany], norfloxacin and ofloxacin [Sigma], levofloxacin [Hoechst Marion Roussel, Romainville Cedex, France], and moxifloxacin [Bayer, Wuppertal, Germany]) were determined. In addition, six β-lactams (ampicillin, amoxicillin [amoxicilline]-clavulanic acid, cephalothin [cefalotin], cefuroxime, cefotaxime, and ceftazidime [all from Sigma]) were tested.
Mueller-Hinton II agar (BBL; Becton Dickinson and Company, Cockeysville, MD) was used as the culture medium. On the basis of results from earlier publications (1, 9, 10), the MIC breakpoint value for reduced ciprofloxacin susceptibility of ≥0.125 μg/ml was chosen. The breakpoint concentrations for the other tested antimicrobials were chosen according to CLSI guidelines (6).
Staphylococcus aureus ATCC 29213, Escherichia coli ATCC 25922, E. coli ATCC 35218, Pseudomonas aeruginosa ATCC 27853, and one local highly ciprofloxacin-resistant S. enterica strain were used as controls for susceptibility testing.
Detection of mutations in the topoisomerase genes gyrA, gyrB, parC, and parE.
PCR was used to amplify the quinolone resistance-determining regions (QRDRs) of target genes to identify mutations in each isolate. DNA was prepared and the QRDRs of gyrA, gyrB, parC, and parE were amplified with previously described primers and protocols (7, 8). Mutations in the target genes were identified with a denaturing high-pressure liquid chromatography method as previously described (7).
Detection of PMQR.
Screening for the qnrA, qnrB, and qnrS genes was carried out by multiplex PCR using a previously described method and specific primers (29). The positive controls were E. coli J53 carrying pMG298 (positive for qnrB; strain kindly provided by George Jacoby) and pMG252 (positive for qnrA; kindly provided by Neil Woodford). The qnrD allele was amplified by PCR using a previously described method and primers (3), and as a positive control, we used Klebsiella pneumoniae (U58), an isolate in our collection previously confirmed as carrying qnrD. Any qnr-positive results were confirmed by the direct sequencing of both strands of amplicons using specific PCR primers. The sequencing was performed as previously described (23), and Vector NTI software (Informax, Inc., Bethesda, MD) was used for the DNA sequence analysis and translation into amino acids. Closest matches to these amino acid sequences were then obtained using the BLAST search engine (http://www.ebi.ac.uk/Tools/blast/).
Isolates were also screened for the other PMQR determinants. The aac(6′)-Ib-cr and qepA genes were amplified as previously described in references 24 and 34, respectively, using previously described primers. As positive controls, we used E. coli J53(pEK499) [positive for aac(6′)-Ib-cr; kindly provided by Neil Woodford] and E. coli KAM32(pSTVqepA) (positive for qepA; kindly provided by Kunikazu Yamane).
Accumulation of ciprofloxacin and Hoechst 33342.
The efflux activity of the nonclassical quinolone-resistant isolates was assessed by determining the accumulation of ciprofloxacin and the fluorescent substrate Hoechst 33342 (bis-benzimide) (Sigma). The uptake of Hoechst 33342 was determined as described previously (32). The fluorescence of Hoechst 33342 was measured using excitation and emission wavelengths of 355 and 365 nm, respectively, using a FLUOstar Optima fluorescence microplate reader (BMG Labtech, Aylesbury, United Kingdom). Ciprofloxacin accumulation was measured as previously described (21) and ciprofloxacin concentration measured using an LS45 fluorospectrophotometer (Perkin Elmer, Cambridge, United Kingdom) at excitation and emission wavelengths of 311 and 447 nm, respectively. For both substrates, accumulation was measured in the presence and absence of 100 μM CCCP (carbonyl cyanide m-chlorophenylhydrazone; added to cell suspensions prior to the addition of Hoechst 33342 or ciprofloxacin). The results for the serovar control strains were found to be very similar (data not shown), and as a result, all data from the nonclassical quinolone-resistant strains were compared with S. Typhimurium SL1344, as this strain has been used extensively in accumulation assays in our laboratory. The Student t test was used to analyze statistically significant differences in accumulation between strains.
Plasmid DNA analysis.
Plasmid analysis was performed for all 10 qnr-positive isolates. Plasmid DNA was extracted using the QIAprep spin miniprep kit (Qiagen Finland, Helsinki, Finland). The presence of qnr genes was ensured using Southern hybridization as previously described (26).
Qnr plasmids were further analyzed by restriction mapping. The plasmid DNA was restricted with HindIII enzyme (New England Biolabs; Finnzymes, Espoo, Finland) to observe the restriction pattern, and the resulting fragment sizes were determined by gel electrophoresis.
Transformation experiment.
Electroporation was used to transfer the plasmid DNA suspected to carry qnr alleles into E. coli DH5α competent cells. Transformants were selected on LB agar plates containing 0.03 μg/ml ciprofloxacin. E. coli transformants were assessed using both phenotypic and genotypic methods. Ciprofloxacin and nalidixic acid susceptibilities were determined using the agar dilution method, and multiplex PCR was used to detect qnr determinants. The results were compared with those of the corresponding donor strain.
Detection of β-lactamase genes.
Multiplex PCR was used to detect β-lactamase genes (TEM, SHV, and CTX-M genes) from all 10 Salmonella strains. The amplification of the TEM, CTX-M, and SHV genes was performed using previously described primers and protocols (23), and the exact sequences of the PCR-positive strains were analyzed by direct sequencing (23).
RESULTS
Antimicrobial susceptibility testing.
All 10 Salmonella strains showed reduced fluoroquinolone susceptibility with ciprofloxacin MICs between 0.25 and 4 μg/ml but were either susceptible or only resistant to a low level of nalidixic acid with a MIC range between 16 and 32 μg/ml. Reduced susceptibility to additional fluoroquinolones (norfloxacin, ofloxacin, enrofloxacin, levofloxacin, and moxifloxacin) was also detected compared to ciprofloxacin-susceptible strains (Table 1). Three of the strains (Mbandaka, Montevideo, and Typhimurium) were also resistant to ampicillin and amoxicillin-clavulanic acid. However, cephalosporin resistance was not detected among these novel quinolone phenotype strains.
TABLE 1.
QRDR mutations, PMQR genes, and MICs of antimicrobial agents for ten non-classical phenotype S. enterica strains, their corresponding transformants and seven control strains
| Isolate no.a | Serovar | Year of isolation | parC QRDR mutationb | PMQR genec
|
Plasmid size (kb) (est)d | MICs (μg/ml) of antimicrobial agents for:e
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| qnrA | qnrB | qnrS | qnrD | qepA | aac(6′)- Ib-cr | Isolates
|
Transformants
|
||||||||||||
| NAL | NOR | OFX | ENR | CIP | LVX | MFX | NAL | CIP | |||||||||||
| s2017 | S. Corvallis | 2003 | Thre57→Ser | − | − | + | − | − | − | 13 | 16 | 2 | 2 | 2 | 0.25 | 1 | 2 | 8 | 0.25 |
| s2018 | S. Stanley | 2003 | Thre57→Ser | − | − | + | − | − | − | ND | 16 | 2 | 2 | 1 | 0.5 | 1 | 2 | ND | ND |
| s2052 | S. Corvallis | 2003 | Thre57→Ser | − | − | + | − | − | − | 13 | 16 | 1 | 1 | 1 | 0.5 | 0.5 | 2 | 8 | 0.25 |
| s2093 | S. Mbandaka | 2003 | Thre57→Ser | + | − | − | − | − | − | 14.5 | 16 | 16 | 1 | 0.5 | 4 | 1 | 0.5 | ND | ND |
| s2209 | S. Montevideo | 2004 | Thre57→Ser | − | − | + | − | − | − | 30 | 32 | 2 | 2 | 2 | 0.5 | 1 | 2 | 8 | 0.25 |
| s2219 | S. Virginia | 2004 | Thre57→Ser | − | − | + | − | − | − | 13 | 32 | 2 | 2 | 2 | 0.5 | 1 | 2 | 8 | 0.5 |
| s2425 | S. Typhimurium | 2005 | WT | − | − | + | − | − | − | 13 | 16 | 2 | 2 | 2 | 0.5 | 1 | 2 | 8 | 0.5 |
| s2705 | S. Typhimurium | 2006 | WT | − | − | + | − | − | − | 13 | 16 | 2 | 2 | 2 | 0.5 | 1 | 2 | 8 | 0.5 |
| s2856 | S. Braenderup | 2007 | Thre57→Ser | − | − | + | − | − | − | 15 | 16 | 2 | 2 | 2 | 0.5 | 1 | 2 | ND | ND |
| s2944 | S. Montevideo | 2007 | Thre57→Ser | − | − | + | − | − | − | ND | 16 | 2 | 2 | 2 | 0.5 | 1 | 2 | 8 | 0.25 |
| s2043 | S. Stanley | 2003 | Thre57→Ser | − | − | − | − | − | − | ND | 4 | 0.125 | 0.125 | 0.06 | 0.015 | 0.06 | 0.06 | ND | ND |
| s2064 | S. Corvallis | 2003 | Thre57→Ser | − | − | − | − | − | − | ND | 4 | 0.06 | 0.03 | 0.03 | 0.015 | 0.06 | 0.06 | ND | ND |
| s2159 | S. Mbandaka | 2003 | Thre57→Ser | − | − | − | − | − | − | ND | 4 | 0.125 | 0.125 | 0.06 | 0.008 | 0.06 | 0.125 | ND | ND |
| s2317 | S. Montevideo | 2004 | Thre57→Ser | − | − | − | − | − | − | ND | 4 | 0.125 | 0.125 | 0.06 | 0.015 | 0.06 | 0.06 | ND | ND |
| s2878 | S. Typhimurium | 2007 | WT | − | − | − | − | − | − | ND | 16 | ND | ND | ND | 0.03 | ND | ND | ND | ND |
| s2906 | S. Braenderup | 2007 | Thre57→Ser | − | − | − | − | − | − | ND | 4 | ND | ND | ND | 0.015 | ND | ND | ND | ND |
| SL1344 | S. Typhimurium | WT | − | − | − | − | − | − | ND | 4 | ND | ND | ND | 0.06 | ND | ND | ND | ND | |
Test strains are isolate numbers s2017 to s2944, and the control strains are s2043 to SL1344.
WT, wild-type allele (no mutation).
+, present; −, absent.
est, estimated; ND, not determined.
NAL, nalidixic acid; NOR, norfloxacin; OFX, ofloxacin; ENR, enrofloxacin; CIP, ciprofloxacin; LVX, levofloxacin; MFX, moxifloxacin. MICs tested with standard agar plate dilution method.
Mutations in topoisomerase genes.
None of the common mutations associated with fluoroquinolone resistance in the QRDRs of the gyrA, gyrB, or parE genes were detected by denaturing high-pressure liquid chromatography analysis. All strains with the nonclassical resistance phenotype except S. Typhimurium strains possessed a Thr57-Ser substitution in ParC. However, it is unlikely that this mutation contributes to quinolone resistance, as it was also present in all control strains with wild-type quinolone susceptibility except S. Typhimurium strains (Table 1). This indicates that this substitution is likely to be a polymorphism common in serovars other than Typhimurium.
PMQR.
Multiplex PCR results showed that the S. Mbandaka strain contained the qnrA1 gene (GenBank accession number AY070235); the remainder of the strains all carried the qnrS1 gene (GenBank accession number AB187515). The qnrB, qnrD, aac(6′)-Ib-cr, and qepA genes were not detected among any of the tested Salmonella strains (Table 1). All qnr-positive PCR results were confirmed by sequencing.
Plasmid analysis.
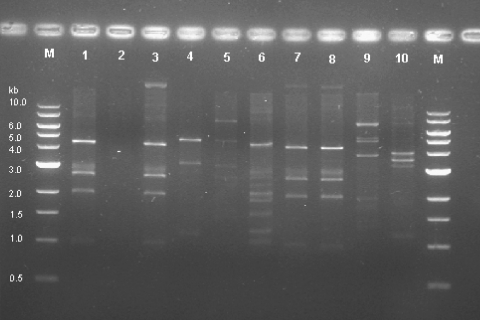
Plasmid analysis of the qnr-positive strains showed that the qnrA1 and qnrS1 genes were located in plasmids ranging in size from 13 to 30 kb. Plasmid fingerprinting performed with HindIII revealed that four of the strains had a similar restriction pattern (the S. Typhimurium and S. Corvallis strains), whereas five had unique restriction patterns and no restriction pattern could be obtained from the S. Stanley strain (Fig. 1). Southern blotting confirmed that the qnr alleles were present on the isolated plasmids (data not shown).
FIG. 1.
HindIII plasmid restriction pattern of 10 nonclassical quinolone resistance phenotype strains. Lane M, molecular weight marker (1-kb DNA ladder); lane 1, S. Corvallis s2017; lane 2, S. Stanley s2018; lane 3, S. Corvallis s2052; lane 4, S. Mbandaka s2093; lane 5, S. Montevideo s2209; lane 6, S. Virginia s2219; lane 7, S. Typhimurium s2425; lane 8, S. Typhimurium s2705; lane 9, S. Braenderup s2856; lane 10, S. Montevideo s2944.
Plasmids carrying qnr genes were transferable in vitro. Plasmids from 7 of 10 qnr-positive strains could be transferred by electroporation to E. coli DH5α, resulting in seven qnrS-positive transformants. Amplification using PCR confirmed that the transformants harbored the same qnrS1 gene as their donor strains. Susceptibility testing showed that the MICs of ciprofloxacin and nalidixic acid for all transformants were similar to the corresponding host strains (Table 1).
Efflux activity.
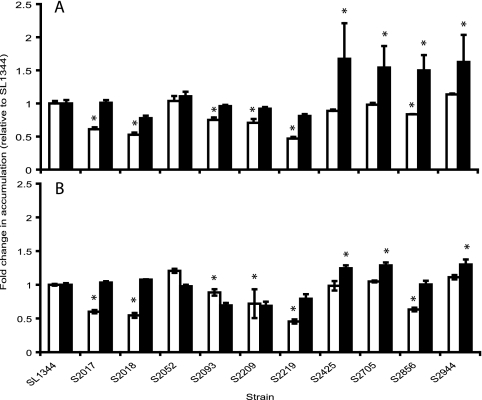
Six of the 10 isolates accumulated significantly less of both ciprofloxacin and Hoechst 33342 than the wild-type control SL1344 (Fig. 2), indicating that increased active efflux activity was present in these strains which may contribute to the resistance phenotype observed. Of these six strains, two (s2093 and s2209) did not increase the level of ciprofloxacin accumulated upon the addition of CCCP, indicating that the low accumulation in these strains is not dependent on the proton motive force.
FIG. 2.
Accumulation of Hoechst 33342 by all 10 strains relative to that by S. Typhimurium SL1344 (control strains) after 10 min (A) and accumulation of ciprofloxacin after 10 min (B). Unfilled bars indicate accumulation without efflux pump inhibitor, and filled bars indicate accumulation in the presence of CCCP. Asterisks indicate values statistically significantly different from the corresponding results for SL1344 (P < 0.05 by t test).
β-Lactamase genes.
By PCR, two of the strains (highly resistant to ampicillin) were TEM positive but CTX-M and SHV negative. The other eight strains were PCR negative for all β-lactamase genes tested. The two TEM genes were confirmed as TEM-1 genes (non-extended-spectrum β-lactamase genes) by DNA sequencing.
DISCUSSION
We have previously described a novel population of S. enterica isolates from Finnish patients returning from southeast Asia (12, 18). In the present study, 10 S. enterica strains showing reduced ciprofloxacin susceptibility and either susceptibility or only low-level resistance to nalidixic acid were analyzed for the presence of all known mechanisms causing quinolone resistance. The qnr genes were the only PMQR determinants found among these nonclassical resistance phenotype isolates. The qnr plasmids were successfully transferred to a susceptible recipient, and the same resistance phenotype was seen in both the host and recipient strains. The plasmid profile patterns indicated that the horizontal transfer of PMQR does occur, since the same plasmid profile was seen in different serovars. Collectively, these results indicated that the plasmids containing qnr genes are spreading among isolates from different locations, times, and serovars. In addition, more than one plasmid was detected, indicating that qnr genes are not limited to one single successful plasmid.
It has been suggested that the Qnr proteins protect DNA gyrase and topoisomerase IV from quinolone inhibition and that isolates with a qnr gene may be less likely to develop topoisomerase mutations than other strains (4). Our findings are in line with these observations. The QRDRs of the gyrase and topoisomerase IV genes were screened for the most commonly occurring mutations, but no resistance-associated substitutions were detected. Although a Thr57-Ser substitution in ParC was detected, the same substitution was present in quinolone-susceptible control strains, indicating that this mutation is likely to have no role in nonclassical quinolone resistance (33). We have also previously shown by pyrosequencing that none of the isolates with the nonclassical resistance phenotype tested had a mutation at codon Ser83 or Asp87 of the QRDR of gyrA (12).
The aac(6′)-Ib-cr variant has been shown to be more prevalent among qnr-positive than qnr-negative strains in E. coli and Klebsiella (15). However, our data for Salmonella do not support this finding. We did not find any other PMQR genes among isolates with the nonclassical resistance phenotype. Although all tested strains were negative for the qepA gene, it is possible that enhanced efflux may play a role in this nonclassical resistance phenotype since six strains accumulated less ciprofloxacin and Hoechst 33342 than the control isolates.
Isolates such as we describe here may be hard to identify in clinical laboratories since this phenotype is difficult to recognize using conventional methods (13, 22). In many laboratories, the nalidixic acid disc diffusion test is used to screen for reduced fluoroquinolone susceptibility in Salmonella, since it has been shown that high resistance to nalidixic acid usually correlates with reduced fluoroquinolone susceptibility (10). However, the nalidixic acid screening may miss isolates showing reduced fluoroquinolone susceptibility but being susceptible or only demonstrating a low-level resistance to nalidixic acid. Therefore, it is important to revise fluoroquinolone susceptibility testing practice. Since the results from a nalidixic acid Etest or disc diffusion test may be misleading, a ciprofloxacin Etest or disc diffusion test should always be examined in parallel.
In conclusion, this study demonstrates the emergence of a highly mobile qnr genotype that confers a nonclassical quinolone resistance phenotype in S. enterica isolates. It is notable that isolates with the nonclassical resistance phenotype persist in the absence of topoisomerase mutations. Although efflux may have some role in the nonclassical phenotype, the qnr genes alone are adequate to produce isolates with reduced fluoroquinolone susceptibility. This is of concern since nonclassical quinolone resistance is plasmid mediated and therefore mobilizable.
Acknowledgments
We are grateful to George Jacoby, Neil Woodford, and Kunikazu Yamane for kindly providing the qnrB-, qnrA-, aac(6′)-Ib-cr-, and qepA-positive control strains.
This work has been financially supported by the Turku University Central Hospital Research Fund, and M.A.W. is supported by a BBSRC David Phillips fellowship.
Footnotes
Published ahead of print on 13 July 2009.
REFERENCES
- 1.Aarestrup, F. M., C. Wiuff, K. Molbak, and E. J. Threlfall. 2003. Is it time to change fluoroquinolone breakpoints for Salmonella spp.? Antimicrob. Agents Chemother. 47:827-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cattoir, V., L. Poirel, V. Rotimi, C. J. Soussy, and P. Nordmann. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60:394-397. [DOI] [PubMed] [Google Scholar]
- 3.Cavaco, L. M., H. Hasman, S. Xia, and F. M. Aarestrup. 2009. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 53:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesaro, A., R. R. Bettoni, C. Lascols, A. Merens, C. J. Soussy, and E. Cambau. 2008. Low selection of topoisomerase mutants from strains of Escherichia coli harbouring plasmid-borne qnr genes. J. Antimicrob. Chemother. 61:1007-1015. [DOI] [PubMed] [Google Scholar]
- 5.CLSI. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M07-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.CLSI. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Eaves, D. J., E. Liebana, M. J. Woodward, and L. J. Piddock. 2002. Detection of gyrA mutations in quinolone-resistant Salmonella enterica by denaturing high-performance liquid chromatography. J. Clin. Microbiol. 40:4121-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaves, D. J., L. Randall, D. T. Gray, A. Buckley, M. J. Woodward, A. P. White, and L. J. Piddock. 2004. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob. Agents Chemother. 48:4012-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hakanen, A., P. Kotilainen, P. Huovinen, H. Helenius, and A. Siitonen. 2001. Reduced fluoroquinolone susceptibility in Salmonella enterica serotypes in travelers returning from Southeast Asia. Emerg. Infect. Dis. 7:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakanen, A., P. Kotilainen, J. Jalava, A. Siitonen, and P. Huovinen. 1999. Detection of decreased fluoroquinolone susceptibility in salmonellas and validation of nalidixic acid screening test. J. Clin. Microbiol. 37:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakanen, A. J., P. Kotilainen, S. Pitkanen, S. Huikko, A. Siitonen, and P. Huovinen. 2006. Reduction in fluoroquinolone susceptibility among non-typhoidal strains of Salmonella enterica isolated from Finnish patients. J. Antimicrob. Chemother. 57:569-572. [DOI] [PubMed] [Google Scholar]
- 12.Hakanen, A. J., M. Lindgren, P. Huovinen, J. Jalava, A. Siitonen, and P. Kotilainen. 2005. New quinolone resistance phenomenon in Salmonella enterica: nalidixic acid-susceptible isolates with reduced fluoroquinolone susceptibility. J. Clin. Microbiol. 43:5775-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopkins, K. L., M. Day, and E. J. Threlfall. 2008. Plasmid-mediated quinolone resistance in Salmonella enterica, United Kingdom. Emerg. Infect. Dis. 14:340-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacoby, G. A. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41(Suppl. 2):S120-S126. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, Y., Z. Zhou, Y. Qian, Z. Wei, Y. Yu, S. Hu, and L. Li. 2008. Plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-cr in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 61:1003-1006. [DOI] [PubMed] [Google Scholar]
- 16.Kehrenberg, C., S. Friederichs, A. de Jong, G. B. Michael, and S. Schwarz. 2006. Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J. Antimicrob. Chemother. 58:18-22. [DOI] [PubMed] [Google Scholar]
- 17.Kotilainen, P., S. Pitkanen, A. Siitonen, P. Huovinen, and A. J. Hakanen. 2005. In vitro activities of 11 fluoroquinolones against 816 non-typhoidal strains of Salmonella enterica isolated from Finnish patients with special reference to reduced ciprofloxacin susceptibility. Ann. Clin. Microbiol. Antimicrob. 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindgren, M. M., P. Kotilainen, P. Huovinen, S. Hurme, S. Lukinmaa, M. A. Webber, L. J. Piddock, A. Siitonen, and A. J. Hakanen. 2009. Reduced fluoroquinolone susceptibility in Salmonella enterica isolates from travelers, Finland. Emerg. Infect. Dis. 15:809-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Martinez, L., M. Eliecer Cano, J. Manuel Rodriguez-Martinez, J. Calvo, and A. Pascual. 2008. Plasmid-mediated quinolone resistance. Expert Rev. Anti-Infect. Ther. 6:685-711. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 21.Mortimer, P. G., and L. J. Piddock. 1991. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 28:639-653. [DOI] [PubMed] [Google Scholar]
- 22.Murray, A., H. Mather, J. E. Coia, and D. J. Brown. 2008. Plasmid-mediated quinolone resistance in nalidixic-acid-susceptible strains of Salmonella enterica isolated in Scotland. J. Antimicrob. Chemother. 62:1153-1155. [DOI] [PubMed] [Google Scholar]
- 23.Nyberg, S. D., M. Osterblad, A. J. Hakanen, P. Huovinen, J. Jalava, and T. F. Resistance. 2007. Detection and molecular genetics of extended-spectrum beta-lactamases among cefuroxime-resistant Escherichia coli and Klebsiella spp. isolates from Finland, 2002-2004. Scand. J. Infect. Dis. 39:417-424. [DOI] [PubMed] [Google Scholar]
- 24.Park, C. H., A. Robicsek, G. A. Jacoby, D. Sahm, and D. C. Hooper. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel, L., J. D. Pitout, L. Calvo, J. M. Rodriguez-Martinez, D. Church, and P. Nordmann. 2006. In vivo selection of fluoroquinolone-resistant Escherichia coli isolates expressing plasmid-mediated quinolone resistance and expanded-spectrum β-lactamase. Antimicrob. Agents Chemother. 50:1525-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randall, L. P., S. W. Cooles, M. K. Osborn, L. J. Piddock, and M. J. Woodward. 2004. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 53:208-216. [DOI] [PubMed] [Google Scholar]
- 27.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6:629-640. [DOI] [PubMed] [Google Scholar]
- 28.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 29.Robicsek, A., J. Strahilevitz, D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2006. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50:2872-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevenson, J. E., K. Gay, T. J. Barrett, F. Medalla, T. M. Chiller, and F. J. Angulo. 2007. Increase in nalidixic acid resistance among non-Typhi Salmonella enterica isolates in the United States from 1996 to 2003. Antimicrob. Agents Chemother. 51:195-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strahilevitz, J., D. Engelstein, A. Adler, V. Temper, A. E. Moses, C. Block, and A. Robicsek. 2007. Changes in qnr prevalence and fluoroquinolone resistance in clinical isolates of Klebsiella pneumoniae and Enterobacter spp. collected from 1990 to 2005. Antimicrob. Agents Chemother. 51:3001-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webber, M. A., L. P. Randall, S. Cooles, M. J. Woodward, and L. J. Piddock. 2008. Triclosan resistance in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 62:83-91. [DOI] [PubMed] [Google Scholar]
- 33.Weill, F. X., S. Bertrand, F. Guesnier, S. Baucheron, A. Cloeckaert, and P. A. Grimont. 2006. Ciprofloxacin-resistant Salmonella Kentucky in travelers. Emerg. Infect. Dis. 12:1611-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamane, K., J. Wachino, S. Suzuki, and Y. Arakawa. 2008. Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan. Antimicrob. Agents Chemother. 52:1564-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamane, K., J. Wachino, S. Suzuki, K. Kimura, N. Shibata, H. Kato, K. Shibayama, T. Konda, and Y. Arakawa. 2007. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51:3354-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]