Abstract
We compared the propensities of Enterococcus faecalis JH2-2 and of the recombination-deficient JH2-2 recA strain to develop mutational resistance to linezolid. In both organisms, a mutation in a single rrl copy conferred resistance to linezolid. Delay in acquisition of the mutation by other rrl copies in JH2-2 recA showed that gene conversion contributed to the acquisition of resistance.
The oxazolidinone antimicrobial agent linezolid interferes with protein synthesis and is active against multidrug-resistant gram-positive organisms (11).
Linezolid resistance in Enterococcus faecalis has been associated with a variety of mutations in domain V of the 23S rRNA genes, in particular G2447T, T2500A, G2505A, G2512U, G2513U, G2576U, and C2610G (4, 5, 7, 10) (Escherichia coli numbering). Reports of in vitro investigations and clinical studies assigned a major role to the G2576U mutation in linezolid resistance in E. faecalis, Enterococcus faecium, and Staphylococcus aureus (4, 6, 8, 10). Further, studies showed a direct correlation between the level of resistance and the proportion of mutated rrl (23S rRNA) genes (4, 6). Mutation in one, several, or all of the rrl gene copies may be associated with gradually increasing levels of resistance. The gene copy number ranges from four in Streptococcus pneumoniae and E. faecalis to six in E. faecium and five or six in S. aureus. However, the frequency with which clinical isolates having the mutation in multiple gene copies are found suggests that homologous recombination between mutated and wild-type 23S rRNA genes may promote the development of high-level resistance under antimicrobial selective pressure.
Gene conversion has already been implicated in increasing the levels of resistance of Mycobacterium smegmatis to aminoglycosides (9) and of S. pneumoniae to evernimicin (1). To test the role of homologous recombination in the acquisition of resistance, we compared the abilities of E. faecalis JH2-2 and its recombination-deficient relative E. faecalis JH2-2 recA to acquire in vitro stepwise resistance to linezolid.
To inactivate the recA gene in E. faecalis JH2-2, we used the thermosensitive shuttle vector pMAD, which confers resistance to erythromycin (3). We constructed a derivative of the pMAD vector carrying a 404-bp DNA fragment from the 5′ end of the recA gene, an antibiotic (spectinomycin) resistance cassette, and a 398-bp DNA fragment from the 3′ end of the gene. The recombinant pMAD/recA vector was extracted from E. coli, and a two-step procedure was then used for allele replacement in E. faecalis. Briefly, the double-recombination event was obtained by subjecting the strain to several subcultures at 44.5°C under the selective pressure of spectinomycin, as described previously (3). The recombinant E. faecalis recA strain was finally retrieved from among spectinomycin-resistant, erythromycin-susceptible colonies (recognized by counterselection), indicating successful replacement recombination. Efficient inactivation was verified by PCR.
The strain JH2-2 recA was tested for the ability to repair DNA damaged by UV-induced degradation. For the UV sensitivity assay, serial dilutions of E. faecalis JH2-2 and JH2-2 recA were spotted onto brain heart infusion (BHI) plates and exposed to UV as described previously (12). After 10 s of exposure, survivor counts for E. faecalis JH2-2 recA were 200-fold lower than those for the parental strain. This result confirmed the deficiency in recombinational repair related to RecA protein inactivation. No evidence for the impairment of the biological fitness was found, since no significant difference between the growth curves for E. faecalis JH2-2 and E. faecalis JH2-2 recA in BHI medium was observed.
Linezolid-resistant mutants of E. faecalis JH2-2 and JH2-2 recA were obtained by serial passages in BHI broth (Becton-Dickinson, Le-Pont-de-Claix, France) containing increasing concentrations (0.25 to 64 μg/ml) of linezolid. A minimum of 11 passages was performed.
At each selection step, 50 μl of bacterial culture growing at the highest antibiotic concentration was streaked onto antibiotic-free BHI agar. Five to 10 colonies were then suspended in saline for PCR and sequencing and for the determination of MICs of linezolid by the agar dilution method (http://www.sfm.asso.fr).
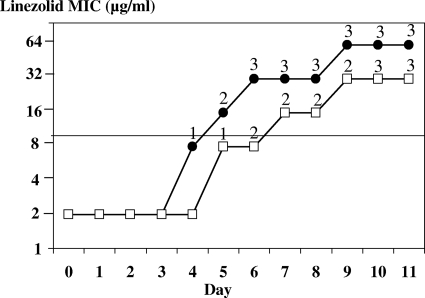
The initial linezolid MIC for both JH2-2 and JH2-2 recA was 2 μg/ml. Serial passages on doubling concentrations of linezolid resulted in progressive MIC increases for both strains (Table 1). A first level of resistance, with the MIC equal to 8 μg/ml, was reached after 4 days for JH2-2 and 5 days for JH2-2 recA. After this point, the MIC for JH2-2 increased daily to reach 32 μg/ml, remained stable for 3 days, and then increased to 64 μg/ml (Fig. 1). For JH2-2 recA, the MIC of linezolid also increased in a stepwise pattern but only to a maximum of 32 μg/ml and it remained stable after 17 passages (data not shown). In addition, for each increase in the MIC for JH2-2 recA, a minimum of 2 days was required.
TABLE 1.
Correlation of the presence of point mutation G2576T in the 23S rRNA genes with the level of linezolid resistance in E. faecalis
| E. faecalis strain | No. of passagesa | Linezolid MIC (μg/ml) | Nucleotide at position 2576b in:
|
|||
|---|---|---|---|---|---|---|
| rrlA | rrlB | rrlC | rrlD | |||
| JH2-2 | 0 | 2 | G | G | G | G |
| 4 | 8 | G | G | G | T | |
| 5 | 16 | T | G | G | T | |
| 6, 8 | 32 | T | G | G/T | T | |
| 9, 10, 11 | 64 | T | G | T | T | |
| JH2-2 recA | 0 | 2 | G | G | G | G |
| 5 | 8 | G | G | G | T | |
| 6 | 8 | G | G | G/T | T | |
| 7, 8, 9 | 16 | G | G | T | T | |
| 10, 11 | 32 | T | G | T | T | |
Number of passages under linezolid selective pressure.
E. coli numbering. Mutations are in boldface.
FIG. 1.
Selection of mutants of E. faecalis JH2-2 (closed circles) and E. faecalis JH2-2 recA (open squares) resistant to linezolid by serial passages in BHI broth containing linezolid. The numbers above the symbols indicate the numbers of mutated rrl gene copies.
To determine the number of rrl copies that carried mutations, a DNA fragment encompassing the region associated with linezolid resistance (nucleotides 2254 to 2683; E. coli numbering) in each rrl gene copy from the wild type and mutants was amplified. One common primer and one operon-specific primer for every copy were designed on the basis of the published E. faecalis V583 genome (GenBank accession number AE016830). The primers used are those described previously (4), except for the specific primer for rrlD, which was changed to 5′-CCTTTTTAAAACTTAGTCAATC-3′. Amplified DNA fragments were then sequenced in both directions.
DNA sequences of rrl copies showed that JH2-2 and JH2-2 recA mutants contained the major G2576T mutation (Table 1). No other mutation in positions critical for linezolid resistance (2505, 2512, 2513, and 2610) could be detected, in contrast to the data in previous reports (4, 5, 7, 10). The first G2576T mutation occurred on the rrlD copy in both JH2-2 and JH2-2 recA and was sufficient for the acquisition of linezolid resistance (MIC = 8 μg/ml). This result confirms previous data obtained from an animal model of enterococcal infection by Bourgeois-Nicolaos et al. (4). In contrast, Lobritz et al. (5) were unable to identify a resistant JH2-2 mutant selected in vitro with the G2576T mutation at only one locus. In our study, additional mutations occurred in rrlA and rrlC copies. The relationship between the number of mutated copies and the expression of resistance was unclear, since the level of linezolid resistance did not strictly correlate with the number of 23S rRNA gene copies carrying the nucleotide exchange.
At certain intermediate mutation steps, the selected bacterial population contained mixtures of wild-type and mutated rrlC copies (Table 1): the plating of culture samples and the sequencing of rrlC copies in 10 individual colonies showed that only a small proportion (2 of 10) of the colonies were mutated. This result may reflect partial replacement of the parental population by secondary-step mutants under linezolid selective pressure.
The delays in the selection of the secondary mutations in the recombination-deficient strain were only one passage for the acquisition of the first and second mutations but six passages for the acquisition of the mutation on the third copy. This finding suggested a role for gene conversion in the acquisition of resistance to linezolid. However, the observed delays were relatively minor, suggesting that recombination systems other than recA were active in this strain and could compensate partially for recA inactivation.
A previous study by Lobritz et al. (5) was designed to explore the role of recombination in linezolid resistance in E. faecalis and led to similar conclusions. However, that study was carried out with the E. faecalis UV202 strain, which is a recombination-deficient mutant of JH2-2 (12). The UV202 mutant was obtained from the JH2-2 strain by chemical mutagenesis with N-methyl-N′-nitro-N-nitrosoguanidine on the basis of sensitivity to UV irradiation. Although the derivative was found to contain a single amino acid mutation in the RecA protein (2), the presence of other mutations, including some in genes other than recA involved in recombinational repair, could not be ruled out. In addition, revertants are easily obtained at a frequency of 10−8 (12). Therefore, the use of a strain in which the recA gene has been specifically and definitely inactivated seemed preferable. Moreover, Lobritz et al. (5) selected a G2576U mutation in JH2-2, as in our study, but only a G2505A mutation in E. faecalis UV202, which renders comparisons between the two strains difficult.
The G2576T mutations were still present in an E. faecalis JH2-2 mutant and a JH2-2 recA mutant after 15 serial passages in antibiotic-free medium.
In conclusion, the data presented here confirm that the successive accumulation of a single point mutation, G2576T, in the rrl gene copies of E. faecalis is associated with a stepwise increase in the level of linezolid resistance and show that this accumulation is favored by homologous recombination.
Acknowledgments
We thank M. Debarbouillé for the generous gift of plasmid pMAD.
Footnotes
Published ahead of print on 22 June 2009.
REFERENCES
- 1.Adrian, P. V., C. Mendrick, D. Loebenberg, P. McNicholas, K. J. Shaw, K. P. Klugman, R. S. Hare, and T. A. Black. 2000. Evernimicin (SCH27899) inhibits a novel ribosome target site: analysis of 23S ribosomal DNA mutants. Antimicrob. Agents Chemother. 44:3101-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, H., L. Ke, S. M. Poppe, T. J. Poel, E. A. Weaver, R. C. Gadwood, R. C. Thomas, D. L. Shinabarger, and M. C. Ganoza. 2002. Oxazolidinone antibiotics target the P site on Escherichia coli ribosomes. Antimicrob. Agents Chemother. 46:1080-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgeois-Nicolaos, N., L. Massias, B. Couson, M. J. Butel, A. Andremont, and F. Doucet-Populaire. 2007. Dose dependence of emergence of resistance to linezolid in Enterococcus faecalis in vivo. J. Infect. Dis. 195:1480-1488. [DOI] [PubMed] [Google Scholar]
- 5.Lobritz, M., R. Hutton-Thomas, S. Marshall, and L. B. Rice. 2003. Recombination proficiency influences frequency and locus of mutational resistance to linezolid in Enterococcus faecalis. Antimicrob. Agents Chemother. 47:3318-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall, S. H., C. J. Donskey, R. Hutton-Thomas, R. A. Salata, and L. B. Rice. 2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 46:3334-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meka, V. G., and H. S. Gold. 2004. Antimicrobial resistance to linezolid. Clin. Infect. Dis. 39:1010-1015. [DOI] [PubMed] [Google Scholar]
- 8.Pillai, S. K., G. Sakoulas, C. Wennersten, G. M. Eliopoulos, R. C. Moellering, Jr., M. J. Ferraro, and H. S. Gold. 2002. Linezolid resistance in Staphylococcus aureus: characterization and stability of resistant phenotype. J. Infect. Dis. 186:1603-1607. [DOI] [PubMed] [Google Scholar]
- 9.Prammananan, T., P. Sander, B. Springer, and E. C. Bottger. 1999. RecA-mediated gene conversion and aminoglycoside resistance in strains heterozygous for rRNA. Antimicrob. Agents Chemother. 43:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prystowsky, J., F. Siddiqui, J. Chosay, D. L. Shinabarger, J. Millichap, L. R. Peterson, and G. A. Noskin. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45:2154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinabarger, D. L., K. R. Marotti, R. W. Murray, A. H. Lin, E. P. Melchior, S. M. Swaney, D. S. Dunyak, W. F. Demyan, and J. M. Buysse. 1997. Mechanism of action of oxazolidinones: effects of linezolid and eperezolid on translation reactions. Antimicrob. Agents Chemother. 41:2132-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yagi, Y., and D. B. Clewell. 1980. Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143:966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]