Abstract
One essential prerequisite for genotypic drug susceptibility testing of human cytomegalovirus (HCMV) is the phenotypic characterization of mutations identified in the viral protein kinase gene UL97 and the viral DNA polymerase gene UL54 regarding their quantitative impact on drug susceptibility. We developed a new method for phenotypic characterization of UL54 mutations with regard to polymerase activity, viral replication, and drug susceptibility. To determine the most suitable viral indicator gene, enhanced green fluorescence protein was C-terminally fused to the HCMV early-late protein UL83 (pp65) or the late proteins UL32 (pp150) and UL99 (pp28), resulting in reporter viruses vTB65g, vTB150g, and vTB28g. vTB65g proved to be superior to the other constructs due to its favorable signal-to-noise ratio and was therefore used to establish the optimum conditions for our assay. The UL54 E756K and D413E mutations were introduced into vTB65g by markerless bacterial artificial chromosome mutagenesis, resulting in virus strains vE756Kg and vD413Eg. The drug susceptibility phenotypes of vE756Kg and vD413Eg were comparable to those previously reported. Furthermore, we found a reduced replicative fitness of vE756Kg by measuring fluorescence intensity as well as by conventional virus growth kinetics. Decreased fluorescence signals of vE756Kg- and vD413Eg-infected cells at late times of infection suggested a reduced polymerase activity, which was confirmed by real-time PCR quantification of the newly synthesized viral DNAs. This new fluorescence-based assay is a highly reproducible method for the phenotypic characterization of mutations potentially influencing drug susceptibility, viral replicative fitness, and polymerase activity of HCMV after marker transfer.
Human cytomegalovirus (HCMV) is a major opportunistic pathogen in individuals with an immature or compromised immune system (17). Three drugs are commonly used for treatment of HCMV infections, including the nucleoside analogue ganciclovir (GCV), the nucleotide analogue cidofovir (CDV), and the pyrophosphate analogue foscarnet (FOS), all targeting the viral polymerase (4). However, application of these drugs for a prolonged period, especially if plasma drug levels are below optimum therapeutic concentrations, leads to the selection of drug-resistant virus mutants, which may in turn result in failure of therapy (1, 2, 7, 31).
The viral protein kinase, pUL97, and the viral DNA polymerase, pUL54, are involved in the selective action of the mentioned drugs and the selection of drug-resistant HCMV mutants (19). pUL97 mediates the initial monophosphorylation of GCV, which is necessary for the antiviral activity of GCV (6, 24). Since CDV and FOS act independently from phosphorylation by pUL97, mutations in UL97 lead to resistance exclusively against the first-line drug GCV. Most mutations occurring in UL97 have already been characterized phenotypically regarding their quantitative influence on GCV drug susceptibility (reviewed in reference 6).
In contrast, the impact of many mutations found in different functional domains of the UL54 polymerase gene, potentially capable of conferring resistance against single (or even all three) antiviral drugs (9), has still to be elucidated. The fact that UL54 is quite polymorphic (8), at least outside its seven conserved catalytic domains (9), makes it even more important to determine which of the many mutations in UL54 confer drug resistance. More than 120 UL54 mutations that are potentially associated with drug resistance have been published so far, but only half of these have been characterized phenotypically. The fact that some of these mutations have been found not to contribute to reduced drug susceptibility strengthens the importance of phenotypic characterization of all individual mutations after marker transfer into genetically defined viral backgrounds.
Another possible implication of amino acid exchanges in the polymerase, apart from changes in drug susceptibility, is modulation of the polymerase activity (36), resulting in reduced viral replicative fitness, which might even be clinically relevant. An assay for phenotypic characterization of UL54 mutations should also allow determination of the polymerase activity in addition to the investigation of drug susceptibility.
A variety of assays have been used for the characterization of quantitative drug susceptibility, which is generally expressed as the drug concentration that inhibits viral replication by 50% (IC50). Classical phenotypic drug sensitivity assays analyze virus isolates obtained from patients and grown in cell culture. Other assays use recombinant viruses, where the mutation to be characterized has been introduced into a sensitive wild-type parental strain (marker transfer). Today, the gold standard for phenotypic characterization of drug susceptibility of HCMV is still the plaque reduction assay (PRA). However, interassay and especially interlaboratory standardization of the PRA has been shown to be very difficult (22). Many efforts have been made to develop new assays which are easier to perform and allow for better standardization. Some groups have generated reporter cell lines that allow determination of IC50s by measuring HCMV spread in cell culture, reflected by luciferase activity (18) or green fluorescent protein (GFP) intensity (35). Other assays are based on recombinant viruses expressing reporter proteins, such as GFP (23), enhanced yellow fluorescent protein (12; Sarah Straschewski and Michael Winkler, personal communication), or secreted alkaline phosphatase (11). Most of these reporter viruses indicate the spread of virus in cell culture. By measuring the diminishing HCMV major-immediate-early promoter activity in newly infected cells in the presence of different drug concentrations, IC50s can be determined (11, 12, 23). However, polymerase activity itself cannot be assayed by these approaches, except in a nonradioactive in vitro polymerase assay (14, 15). Very recently, a new method based on quantitative real-time PCR was published, allowing both the investigation of IC50s and the investigation of changes in polymerase activity in cell culture by comparing the numbers of genome copies in cells infected with HCMV strains (30).
We have chosen a different approach based on the fact that inhibition of the polymerase leads to reduced synthesis of viral late proteins. Labeling of such late proteins in a reporter virus would enable not only the determination of drug susceptibility but also the indirect evaluation of polymerase activity. We tested this hypothesis by generating three recombinant viruses, using markerless bacterial artificial chromosome (BAC) mutagenesis, in which enhanced GFP (EGFP) was C-terminally fused to the early-late protein UL83 (pp65) and the late proteins UL32 (pp150) and UL99 (pp28). The UL83-labeled strain vTB65g was characterized in detail and thereafter was used in marker transfer experiments in which 50% effective concentrations (EC50s) were determined for GCV, CDV, and FOS by measuring the decrease in fluorescence intensity at different drug concentrations. Results for phenotypic drug susceptibility obtained by our fluorescence assay were comparable to those obtained by PRA. In addition, viral growth and polymerase activity were determined by measuring fluorescence intensities in infected cells and were similar to results from quantitative real-time PCR or titration. Our data demonstrate that the new assay provides a useful tool for connecting so far uncharacterized HCMV polymerase genotypes to phenotypes.
MATERIALS AND METHODS
Viruses, cells, and transfection.
Three recombinant HCMV strains—vTB65g, vTB28g, and vTB150g—were generated by C-terminal fusion of EGFP to UL83 (pp65), UL99 (pp28), and UL32 (pp150), respectively, using a markerless two-step RED-GAM recombination protocol (34). Briefly, a kanamycin resistance gene with an adjoining I-SceI cleavage site flanked with homologous regions was used to introduce the desired target modification into the BAC in the first recombination step. The kanamycin resistance gene was then removed from the BAC in the second step by a combination of I-SceI cleavage and intramolecular Red recombination, using the previously introduced sequence duplication and leaving only the desired new sequence. The primers used to generate the PCR product for the first recombination were ep_pp65-xFP_for (5′-AAGCCCTGCCCGGGCCATGCATCGCCTCGACGCCCAAAAAGCATCGAGGTGTGAGCAAGGGCGAGGAG-3′) and ep_pp65_gfp-in_rev (5′-ACGTGGGTTTTTATAGAGTCGTCTTAAGCGCGTGCGCGGCGGGTGGCTCACTTGTACAGCTCGTCCATGCCG-3′) for vTB65g, ep_UL99CxFP_for (5′-AACGTCCACCCACCCCCGGGACAAAAAAGCCCGCCGCCCCCTTGTCCTTTGTGAGCAAGGGCGAGGAG-3′) and ep_UL99CxFP_rev (5′-CCCATTCCCGACTCGCGAATCGTACGCGAGACCTGAAAGTTTATGAGTTACTTGTACAGCTCGTCCATGCCG-3′) for vTB28g, and ep_UL32CxFP_for (5′-CCGTGCAGAACATCCTCCAAAAGATCGAGAAGATTAAGAAAACGGAGGAAGTGAGCAAGGGCGAGGAG-3′) and ep_UL32CxFP_rev (5′-TATCCGATGATTTCATTAAAAAGTACGTCTGCGTGTGTGTTTCTTAACTACTTGTACAGCTCGTCCATGCCG-3′) for vTB150g. The primers contain homologous sequences of HCMV both upstream and downstream of the site of insertion and are homologous to the pEP-EGFP_in template plasmid (underlined) (34). RED-GAM recombination was performed with Escherichia coli GS1783 (kindly provided by Gregory Smith, Northwestern University Medical School, Chicago, IL) harboring an infectious BAC clone of the endotheliotropic HCMV strain TB40-BAC4 (33). Key genetic attributes of E. coli GS1783 are a heat shock-inducible promoter driving the expression of Red recombination proteins (necessary for both recombination steps) and an arabinose-inducible promoter for I-SceI expression (needed for the cleavage prior to the second recombination step). The insertion of egfp was confirmed by PCR and by sequencing of the mutated regions. Restriction analyses of the recombinant bacmids ensured that no spurious recombinations had occurred. To generate a UL54 mutant virus bearing the point mutation E756K, markerless BAC mutagenesis was performed using recombinant bacmid clone TB65g. The primers used for the generation of vE756Kg were ep_54E756K_for (5′-ACCCCGTGGACCCCCGCCGATGTATACAGCGTCACGCTAAAGAACGGCGTGACTCACCGAGGATGACGACGATAAGTAGGG-3′) and ep_54E756K_rev (5′-CACCGAAGCACGCACAAAGCGGTGAGTCACGCCGTTCTTTAGCGTGACGCTGTATACACAACCAATTAACCAATTCTGATTAG-3′), and those used for the generation of vD413Eg were ep_D413E_for (5′-CCTTATACAGGTACTCGAGGCGCGTGAGGATGTACTTCAACTCAAAAGAGTTGATGTTGTAAGGATGACGACGATAAGTAGGG-3′) and ep_D413E_rev (5′-CGCCGGCCTTTGTGACCGGTTACAACATCAACTCTTTTGAGTTGAAGTACATCCTCACGCGCAACCAATTAACCAATTCTGATTAG-3′). The primers are homologous to the UL54 gene both upstream and downstream of the sequence to be mutated and are homologous to the pEPkan-S template plasmid (underlined) (34). The resulting recombinant E756K and D413E clones were confirmed by PCR and sequencing of the whole polymerase gene. Again, restriction analyses of the recombinant bacmids ensured that no unwanted recombinations had occurred during the mutagenesis process.
MRC-5 cells were used for virus reconstitution from bacmid DNA, as previously described (5). Human foreskin fibroblasts (HFF), maintained as previously described (5), were used for all following infection experiments. Cell-free virus stocks were prepared after propagation of the viruses on HFF and were titrated for infectivity by enumeration of cells staining positive at 48 h postinfection for HCMV IE1/2 and UL44 antigens (monoclonal mouse anti-CMV clones CCH2 and DDG9; Dako). The UL54 gene of every stock virus was sequenced prior to the experiments.
Phenotypic assays of recombinant strains.
All phenotypic assays were performed in 96-well plates. HFF (1.3 × 104 per well) were seeded 2 days prior to the experiments and were used at 100% confluence (optical control). For determination of EC50s, either by fluorescence intensity reduction (FIR) assay or by PRA, HFF grown in 96-well plates were infected at a multiplicity of infection (MOI) of 1 for the FIR assay and 0.001 for PRA. At the same time, drugs were added to the wells in twofold serial dilutions, starting from 800 μM FOS (Foscavir; Astra Zeneca), from 10 μM GCV (Cymeven; Roche), and from 4 μM CDV (Vistide; Pfizer). Cells were then centrifuged at 1,800 rpm for 30 min and incubated at 37°C. After 3 days, drug-containing medium was renewed, and the assays were stopped at day 5 postinfection. For the FIR assay, cells were fixed with 4% paraformaldehyde, washed, and stored in 0.01 M phosphate-buffered saline (PBS) before measurement of the relative fluorescence intensity by a plate spectrofluorometer (Flx-Xenius; Safas, Monaco). The relative fluorescence intensity of mock-infected HFF was used as the baseline. For PRA, cells were fixed with methanol and stained for pp65 (mouse anti-pp65 monoclonal antibody [MAb] 28-77; kindly provided by William Britt, University of Alabama, Birmingham), and the plaques, defined as at least four neighboring pp65-positive cells, were counted. The drug concentration (EC50) required to reduce the relative fluorescence intensity or the number of plaques by 50% was calculated by exponential regression of the values. The use of the term EC50 instead of IC50 was recommended by the FDA in 2007 (http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm071173.pdf) for cell-based drug susceptibility testing. EC50s were determined for at least four independent experiments for each virus and drug.
Viral growth curves were performed by inoculating the cells with the respective viruses at the indicated MOIs. Cells were then centrifuged at 1,800 rpm for 30 min and incubated at 37°C. After 2 h, the virus-containing medium was removed, the cells were washed twice with 0.01 M PBS, and fresh medium was added to the cells. The culture supernatants were collected at the indicated time points and stored at −80°C, and the removed medium was replaced with fresh medium. The stored aliquots were subsequently titrated on HFF. For growth curves obtained by measuring fluorescence intensities, HFF were cultivated in minimum essential medium without phenol red (Gibco/Invitrogen, Karlsruhe, Germany), and the relative fluorescence intensity (in arbitrary units [AU]) of the infected cells was measured in a plate spectrofluorometer (Flx-Xenius) at the indicated time points. The relative fluorescence intensity of mock-infected HFF in minimum essential medium without phenol red was used as the baseline.
Fluorescence microscopy of HFF infected with vTB65g, vTB150g, and vTB28g was performed in 96-well plates, using a Zeiss Axiovert 200 microscope.
Western blot analyses.
For Western blot analyses, HFF were infected using an MOI of 3; the amount of virus used in the experiment was controlled by titration of the inocula. Cells were centrifuged at 1,800 rpm for 30 min, incubated at 37°C for 2 h, and washed twice with 0.01 M PBS, and fresh medium was added to the cells. Samples of infected cells were taken at the indicated time points, and cell lysates were prepared and analyzed by Western blotting as previously described (5). HCMV-encoded proteins were detected with MAbs. MAbs used in this study included those directed against HCMV pp65 (UL83; MAb 65-33), HCMV major capsid protein (MCP) (UL86; MAb 28-4) (both kindly provided by William Britt, University of Alabama, Birmingham), HCMV IE1/2 (Argene, North Massapequa, NY), and β-actin (Sigma, St. Louis, MO).
Real-time PCR analyses.
For the determination of HCMV genome copy numbers by real-time PCR, HFF were infected in a 96-well plate with vTB65g and vE756Kg, using an MOI of 1. The amount of infectious virus in the inoculum was controlled by titration. After 2 h of incubation, the inoculum was removed and the cells washed three times with 0.01 M PBS. Cells were harvested in 200 μl of 0.01 M PBS at day 1 to 6 postinfection and frozen at −80°C until further preparation. Viral DNA for determination of growth kinetics was prepared from the cells by use of a High Pure nucleic acid kit (Roche) according to the manufacturer's instructions. Viral DNA for the determination of polymerase activity was prepared from the cells as previously described (30). Real-time PCR for relative determination of HCMV genome copies was performed using an Mx3000P qPCR machine (Stratagene), brilliant Sybr green quantitative PCR mix (Stratagene), and HCMV-specific primers as previously described (28), with modifications. Standard curves were generated by using tenfold dilution series of the samples. Identical efficiencies were found, which allowed comparison of the values for relative quantification of genome copies. For the dilution series of vTB65g-infected cells from day 1 postinfection, the cycle threshold value for the highest dilution was set to one HCMV genome copy. All other values for relative genome copies were derived from this standard curve. As a control for cellular DNA, values for glyceraldehyde-3-phosphate dehydrogenase, determined as previously described (21), with modifications, were used to normalize the HCMV-specific values.
RESULTS
Protein alignment of pUL54 proteins from HCMV laboratory strains.
Although the HCMV polymerase, pUL54, is an essential protein, it has been shown to be unexpectedly polymorphic, at least outside its seven conserved catalytic domains (reviewed in reference 8). As a consequence, the polymerases of HCMV strains commonly used in laboratories show several differences in protein sequence (Table 1). Here we compared the pUL54 amino acid sequence of strain TB40-BAC4, used as a reference and drug-sensitive strain in this study, to the sequences of strains Towne, AD169, Toledo, FIX-BAC, and Merlin. All amino acid differences in pUL54 proteins of these strains are located outside the catalytic domains. Some amino acid positions in pUL54 are more variable than others, e.g., positions 685 and 1122 (Table 1). Changes at these two positions are also frequently found in sensitive clinical isolates (8). Interestingly, the polymerase amino acid sequence of strain TB40-BAC4 shows the highest homology to the sequence of strain Towne, which has been used before as a sensitive virus for antiviral drug susceptibility testing (10). pUL54 of TB40-BAC4 differs from the Towne protein only at amino acid positions 655 and 1164. The amino acid exchange S655L has been found in 20 of 40 tested sensitive clinical isolates (8) and can therefore be classified as not relevant to antiviral drug resistance. The amino acid exchange A1146V detected in our TB40-BAC4 strain has not yet been reported but is located outside the region involved in antiviral drug resistance (9) and is therefore probably not relevant. In addition, EC50s of all three drugs for the TB40-BAC4 strain (see Table 3) were comparable to those for other sensitive strains, such as AD169 or Towne. We therefore concluded that HCMV strain TB40-BAC4 is a drug-sensitive wild-type strain which can be used as a reference for the investigation of polymerase point mutations in marker transfer experiments.
TABLE 1.
Amino acid differences in UL54 proteins of commonly used HCMV strains
| HCMV strain or sequence | Amino acid at HCMV UL54 position
|
GenBank accession number | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 655 | 685 | 874 | 885 | 890 | 897 | 898 | 1012 | 1108 | 1122 | 1147 | 1154 | 1164 | ||
| Consensus | S | S/N | G | T | L | S | D | A | T | T/A | N | A | A | |
| TB40-BAC4 | L | S | T | V | EF999921 | |||||||||
| Towne | S | T | P | AY315197 | ||||||||||
| AD169 | N | A | N | A | X17403 | |||||||||
| Toledo | N | L | A | AC146905 | ||||||||||
| FIX-BAC | L | S | R | F | V | A | S | AC146907 | ||||||
| Merlin | N | L | N | A | T | AY446894 | ||||||||
TABLE 3.
Genotypes and phenotypes of HCMV TB40-BAC4 and derived strainsb
| HCMV strain | Genotype
|
EC50 (μM) by FIR assaya
|
EC50 (μM) by PRAa
|
|||||
|---|---|---|---|---|---|---|---|---|
| UL97 | UL54 | GCV | CDV | FOS | GCV | CDV | FOS | |
| vTB40-BAC4 | wt | wt | ND | ND | ND | 3 ± 1.4 | 0.15 ± 0.05 | 43 ± 11 |
| vTB65g | wt | wt | 0.9 ± 0.1 | 0.17 ± 0.06 | 38 ± 5 | 2.7 ± 0.8 (0.9) | 0.19 ± 0.05 (1.3) | 41 ± 8 (0.90) |
| vE756Kg | wt | E756K | 2.0 ± 0.3 (2.3) | 0.26 ± 0.07 (1.5) | 137 ± 22 (3.6) | 5.6 ± 2.9 (2.1) | 0.31 ± 0.07 (1.6) | 144 ± 21 (3.5) |
| vD413Eg | wt | D413E | 4.3 ± 1.0 (5.0) | 2.54 ± 0.84 (15) | 35 ± 7 (0.90) | ND | ND | ND |
EC50s are means ± standard deviations for at least four assays performed in triplicate. Ratios of EC50s to those of the sensitive parental strains are given in parentheses.
wt, wild type; ND, not determined.
HCMV recombinant strains vTB65g, vTB150g, and vTB28g.
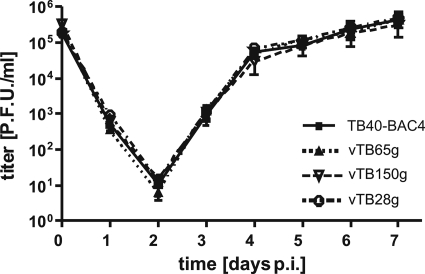
In order to establish a new fluorescence-based phenotypic assay for HCMV drug susceptibility testing, we generated three recombinant HCMV mutants based on the endotheliotropic strain TB40-BAC4 (33) by markerless BAC mutagenesis (Table 2). In our new assay, the dependence of viral protein expression on viral polymerase activity is measured by fluorescence. In the recombinant viruses vTB150g, vTB28g, and vTB65g, the true late proteins pp150 and pp28 and the early-late protein pp65, respectively, are C-terminally fused to EGFP. From a theoretical point of view, we expected that viruses expressing labeled true late proteins should be most suitable for our assay. In all three recombinant viruses, the fusion proteins were expressed correctly (data not shown). The insertion of EGFP had no impact on viral replication, as determined by high-MOI (Fig. 1) and low-MOI (data not shown) growth kinetics.
TABLE 2.
TB40-BAC4-derived recombinant HCMV strains
| Recombinant virus | Parental virus | Selection marker |
|---|---|---|
| vTB65g | TB40-BAC4 | egfp |
| vTB150g | TB40-BAC4 | egfp |
| vTB28g | TB40-BAC4 | egfp |
| vE756Kg | vTB65g | egfp; FOSr GCVr |
| vD413Eg | vTB65g | egfp; GCVr CDVr |
FIG. 1.
Growth kinetics of HCMV strains TB40-BAC4, vTB65g, vTB150g, and vTB28g. HFF were infected at an MOI of 5, culture supernatants were sampled at the indicated time points, and infectivity was determined subsequently on HFF. Day zero values represent the input infectivity. Each point and error bar represent the mean ± standard deviation for four independent experiments. p.i., postinfection.
Signals of the EGFP-labeled proteins were detected by fluorescence microscopy in the same temporal and spatial patterns as those previously described for pp65, pp150, and pp28 (3, 20, 27, 29; data not shown). Input pp65EGFP was observed in the nucleus shortly after infection, and newly synthesized protein was detectable in the nucleus at 3 days postinfection and in both the nucleus and cytoplasm at 4 days postinfection. pp150EGFP and pp28EGFP fluorescence accumulated in the HCMV assembly compartment starting from day 4 postinfection.
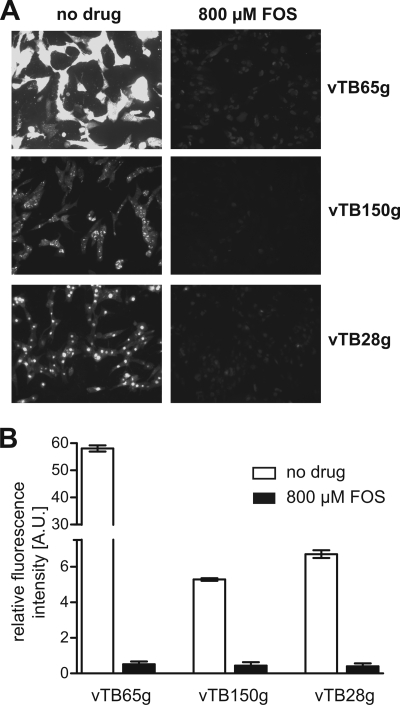
A suitable reporter virus should provide high fluorescence signal intensities and a clear and complete inhibition of the signals by effective antiviral drugs. We therefore measured fluorescence signals at 5 days postinfection in HFF infected with vTB65g, vTB150g, and vTB28g in the presence and absence of antiviral drugs. HFF infected with vTB65g displayed by far the broadest range between maximum and minimum fluorescence signals (150-fold) compared to cells infected with vTB150g (12-fold) or vTB28g (16-fold) (Fig. 2). Blocking the progression from early to late stages of infection by application of 800 μM FOS, which in turn results in a decreased expression of late genes, led to a strong inhibition of HCMV polymerase activity. In fact, signal intensities of infected cells treated with 800 μM FOS were only slightly higher than those of uninfected cells. Although pp65EGFP expression follows early-late kinetics (13), the minimum signals from drug-treated cells infected with vTB65g were not significantly higher than those from cells infected with strain vTB150g or vTB28g (Fig. 2B).
FIG. 2.
(A) Fluorescence microscopy (magnification, ×100) of HFF infected with EGFP-labeled HCMV strains vTB65g, vTB150g, and vTB28g. Shown are representative pictures of cells 5 days after infection at an MOI of 1 with subsequent incubation without or with 800 μM FOS. All pictures were taken with the same exposure time in order to allow comparison of the fluorescence signals. (B) Signal-to-noise comparison for strains vTB65g, vTB150g, and vTB28g. Fluorescence signals of infected cells were measured using Flx-Xenius (Safas), and each bar and error bar represent the mean ± standard deviation for values from six wells.
In a comparative analysis, we tested whether EC50s could be obtained reproducibly for all three reporter viruses. By applying increasing concentrations of antiviral drugs, EC50s can be determined by measuring the exponentially decreasing fluorescence signals. We found that only the strong basal fluorescence signal of vTB65g-infected cells was suitable to provide reproducible results for the EC50s, whereas vTB150g or vTB28g gave less reliable results in most of these experiments (data not shown).
Because of the best signal-to-noise ratio, an efficient inhibition of fluorescence signals after the addition of antiviral drugs, and the good reproducibility of EC50s, vTB65g suited our requirements best and was chosen for all following FIR assays.
Characterization of the FIR assay.
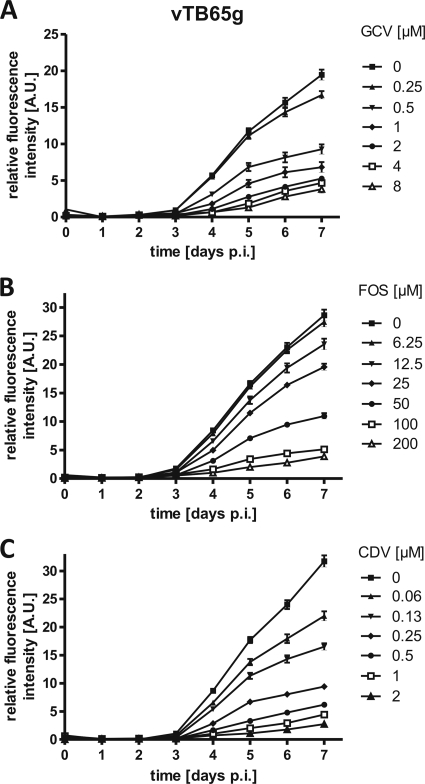
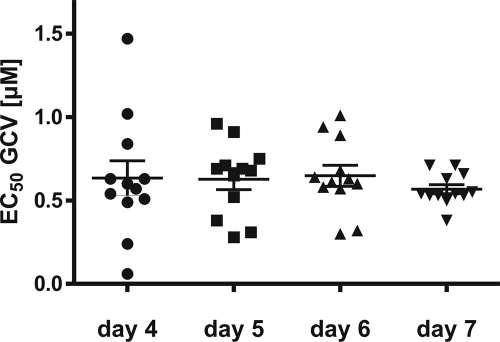
In order to determine the optimum time for measurement of fluorescence, we determined the growth kinetics with strain vTB65g, where the fluorescence intensity of pp65EGFP was measured at the indicated time points (Fig. 3). Differences in fluorescence intensities between cells infected with vTB65g virus at various concentrations of GCV, FOS, or CDV were observed starting at day 4 postinfection. With increasing doses of the drugs, fluorescence intensities decreased, and EC50s were calculated on days 4, 5, 6, and 7 postinfection (shown for GCV in Fig. 4). It appeared that EC50s remained relatively stable throughout these 4 days, but the number of outliers was higher at day 4 postinfection, probably due to the weaker fluorescence signal. Day 5 postinfection was shown to be the earliest time to obtain reproducible EC50s. On day 5 postinfection, the range of EC50s seemed to be higher than on days 6 and 7, probably due to the special conditions of this evaluation experiment, such as removing the cell culture plates from the incubator every day for measurement and measuring live cells in medium instead of fixed cells in PBS. This problem was solved (Table 3) by applying tightly standardized conditions, as defined by the final standard operation procedure of our FIR assay. Therefore, all further measurements were performed at 5 days postinfection.
FIG. 3.
Fluorescence growth curves for HCMV strain vTB65g grown with GCV (A), FOS (B), and CDV (C). HFF were infected at an MOI of 1 and grown at various drug concentrations. Day zero values represent the relative fluorescence intensity after a 2-h inoculation period and two subsequent washes of the cells with PBS. The relative fluorescence intensity (AU) of the infected cells was measured at the indicated time points. Each point and error bar represent the mean ± standard deviation for values from six wells. p.i., postinfection.
FIG. 4.
Comparison of GCV EC50s of HCMV strain vTB65g on different days postinfection. HFF were infected at an MOI of 1 and grown at various GCV concentrations. Each point represents the value for one well. Shown are means and standard errors for 12 values obtained from two independent experiments.
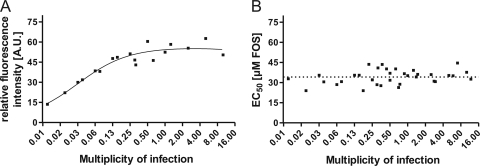
We then investigated whether determination of the EC50 in the FIR assay was sensitive to the MOI used for infection. HFF were infected with vTB65g virus at MOIs ranging from 0.009 to 12. We found that EC50s remained nicely stable (shown for FOS in Fig. 5B), although the intensity of maximum fluorescence signals was dependent on the MOI until an MOI of about 0.25 (Fig. 5A). For following FIR assays, an MOI of 1 was used.
FIG. 5.
MOI dependence of EC50s determined by FIR assay. HFF were infected with vTB65g virus at different FOS concentrations and various MOIs ranging from 0.01 to 12. Relative fluorescence intensities (A) and EC50s (B) were determined on day 5 postinfection.
Introduction of UL54 E756K mutation into strain vTB65g.
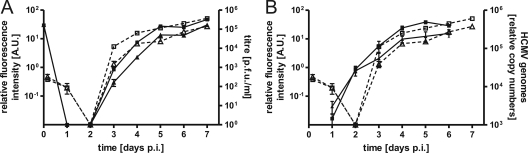
For a more thorough evaluation of our FIR assay regarding the determination of EC50s, viral growth, and polymerase activity, the known polymerase mutation E756K (25) was introduced into vTB65g by markerless BAC mutagenesis (34), resulting in recombinant strain vE756Kg. It was shown previously that the E756K mutation confers reduced drug susceptibility to FOS, GCV, and CDV and leads to a slight defect in replication (9). We determined the growth kinetics of vE756Kg as well as of its parental strain, vTB65g, at high MOIs by titrating the virus (PFU/ml) from supernatants and by measuring, in parallel, the relative fluorescence intensity (AU) of the infected cells at the indicated time points (Fig. 6A). Fluorescence signals of pp65EGFP were detected as early as 4 h postinfection, probably resulting from the incoming virus particle during infection. These signals decreased until day 2 postinfection. Starting from day 3 postinfection, the fluorescence signals increased exponentially, and they reached their maximum on day 5 postinfection. Growth curves obtained by titration of infectious virus from the supernatants were similar to those obtained by measuring fluorescence, with few exceptions. Virus particles in the supernatant started to accumulate and reached the maximum slightly later than in the fluorescence curves for pp65EGFP (Fig. 6A). This can be explained by the fact that the early-late protein pp65 is expressed earlier, before progeny virions are released into the supernatant. Viral replication kinetics obtained by determination of the amount of viral DNA by quantitative real-time PCR (Fig. 6B) again correlated with fluorescence growth curves, except for day 2 postinfection, on which the HCMV genome copy number started to increase, corresponding to the onset of viral DNA replication.
FIG. 6.
Growth curves for HCMV strains vTB65g and vE756Kg. HFF were infected with the indicated viruses at an MOI of 1. The relative fluorescence intensity (AU) of the cells was measured at the indicated time points. p.i., postinfection. (A) In parallel, the culture supernatants were sampled, and triplicate samples were pooled and titrated on HFF. Day zero values represent the fluorescence intensity or infectivity after a 2-h inoculation and two subsequent washes of the cells with PBS. Each point and error bar represent the mean ± standard deviation for three independent experiments. □, vTB65g fluorescence; ▵, vE756Kg fluorescence; ▪, vTB65g titer; ▴, vE756Kg titer (fluorescence is plotted on the left y axis, and titers are plotted on the right y axis). (B) In parallel, cells were harvested at the indicated time points, and quantitative real-time PCR was performed in order to determine relative HCMV genome copy numbers. Each point and error bar represent the mean ± standard deviation for three independent experiments. □, vTB65g fluorescence; ▵, vE756Kg fluorescence; ▪, vTB65g genome copies; ▴, vE756Kg genome copies (fluorescence is plotted on the left y axis, and titers are plotted on the right y axis).
Using all three different methods, we showed that the introduction of the polymerase mutation E756K caused a slight growth defect, as described previously (9). This growth defect resulted in up to fivefold reduced virus titers compared to those of vTB65g virus (Fig. 6A). In conclusion, the investigation of viral growth by measuring fluorescence intensity leads to results which are very comparable to those obtained by determination of virus titers or relative genome copy numbers.
Drug susceptibility phenotypes of vTB65g, vE756Kg, and vD413Eg viruses.
Next, drug susceptibility phenotypes of vTB65g and vE756Kg viruses were analyzed by PRA and compared with the results of our FIR assay. To ensure that the generation of vTB65g did not alter its drug susceptibility, EC50s were determined for all three drugs, GCV, FOS, and CDV, by PRA. As shown in Table 3, EC50s for vTB65g virus were similar to those for its parental strain, TB40-BAC4. In contrast, EC50s for the mutant virus vE756Kg were elevated. For CDV, GCV, and FOS, 1.5-fold, 2.3-fold, and 3.6-fold increases of the EC50 were determined for vE756Kg virus by the FIR assay (Table 3). EC50s for the UL54 E756K mutant in the background of strain Towne, determined by PRA, have already been published to be 2.2-fold elevated for CDV, 3.5-fold elevated for GCV, and >8-fold elevated for FOS (9). Our EC50 results are comparable, but the ratios between mutant and parental viruses are different. These differences are probably due to the different viral backgrounds (TB40-BAC4 in our study) and not to our FIR assay, because similar ratios of the EC50s for our viruses were obtained irrespective of the method used for determination (FIR assay or PRA) (Table 3). EC50s of vTB65g and vE756Kg viruses determined by FIR assay were slightly lower than those obtained by PRA. Also, standard deviations of the EC50s assayed by FIR assay were lower than those for values obtained by PRA (Table 3), and severe outliers such as those obtained by PRA were not observed in the FIR assay (data not shown).
As an additional control, we introduced the polymerase mutation D413E (16) into vTB65g, resulting in strain vD413Eg. It was previously shown by marker transfer experiments using strain Towne and PRA phenotyping that this mutation confers reduced susceptibility to both nucleoside analogues, GCV and CDV, and leads to a modest attenuation of viral growth (9). The increase in EC50 for vD413Eg compared to vTB65g was 5.0-fold for GCV and 15-fold for CDV (Table 3), thereby confirming the tendency of the published increases of 4.8-fold for GCV and 4.3-fold for CDV (9). These data demonstrate the feasibility of our FIR assay for determination of drug susceptibility phenotypes.
Indirect determination of polymerase activity in HCMV strains vTB65g, vE756Kg, and vD413Eg.
We have shown so far that our approach to use a recombinant virus expressing the early-late pp65 gene fused with EGFP was successful for the analysis of viral growth kinetics and drug susceptibility. Another consequence of changes in the polymerase protein sequence can be an alteration of polymerase activity, resulting in changes in viral replicative fitness. In theory, such an alteration could lead to a change in the expression of genes depending on polymerase activity, such as the pp65 gene, and should therefore be measurable by changes in fluorescence intensity in our system.
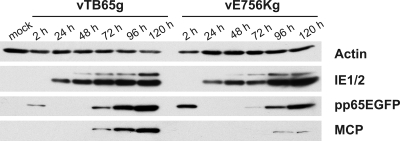
The growth kinetics of the vTB65g and vE756Kg viruses revealed a growth defect of vE756Kg in HFF (Fig. 6A and B). It also has been described that the D413E mutation results in a slight growth defect (9). We hypothesized that these growth defects are caused by decreased activity of the mutated polymerases. In order to detect possible changes in the amounts of proteins whose expression is dependent on polymerase activity, we performed Western blot analyses of cells infected with the vTB65g and vE756Kg viruses over a period of 5 days (Fig. 7). As expected, expression profiles for HCMV protein IE1/2, which is independent of polymerase activity, did not differ for the two viruses. In contrast, expression of the true late protein MCP was delayed, and the amounts of MCP at 96 and 120 h postinfection were drastically reduced in cells infected with vE756Kg virus. Expression of the early-late protein pp65EGFP started simultaneously at 48 h postinfection for the two viruses (only visible with longer exposure times; data not shown), but the increase in expression of pp65EGFP was lower in vE756Kg-infected cells at later time points (Fig. 7). These results again indicated that the polymerase activity in the vE756Kg mutant might be reduced.
FIG. 7.
Western blot analyses of cell lysates from HFF infected with HCMV strains vTB65g and vE756Kg at an MOI of 3. Cells were harvested at the indicated time points.
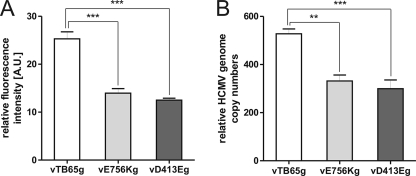
We then wanted to test whether we could indirectly measure a decrease in polymerase activity of vE756Kg and vD413Eg by FIR assay. Therefore, HFF were infected with the vTB65g, vE756Kg, and vD413Eg viruses, without adding antiviral drugs, and fluorescence intensities were measured at day 5 postinfection (Fig. 8A). Fluorescence intensities of vE756Kg- and vD413Eg-infected cells were 1.8-fold and 2.0-fold lower, respectively, than those of cells infected with vTB65g virus. Confirming these results, the E756K mutation led to a 1.6-fold reduced genome copy number in vE756Kg-infected cells and the D413E mutation led to a 1.8-fold reduced copy number in vD413Eg-infected cells compared to vTB65g-infected cells, as determined by quantitative real-time PCR (Fig. 8B). These results clearly show that our new fluorescence-based assay is a highly reproducible method, not only for the analysis of viral drug susceptibility but also for analysis of viral growth and the indirect determination of polymerase activity in marker transfer experiments.
FIG. 8.
(A) Relative fluorescence intensities of vTB65g, vE756Kg, and vD413Eg viruses at day 5 postinfection. HFF were infected with the indicated viruses at an MOI of 1. Each column and error bar represent the mean ± standard deviation for at least four independent experiments. (B) Numbers of genome copies in vTB65g-, vE756Kg-, and vD413Eg-infected cells at day 5 postinfection. HFF were infected at an MOI of 1 and collected at the indicated time points. Equal inputs of virus were controlled by titration of the inocula and determination of genome copy numbers from infected cells at 48 h postinfection. The number of HCMV genome copies was determined by real-time PCR. Each column and error bar represent the mean ± standard deviation for four independent experiments.
DISCUSSION
All three drugs currently available for treatment of HCMV target the viral polymerase, thereby reducing the synthesis of viral DNA. We developed a new assay for phenotypic characterization of HCMV drug susceptibility. This assay is based on the fact that expression of multiple HCMV genes is dependent on polymerase activity, which is reduced in the presence of antiviral drugs and results in measurably lower expression of such genes.
The fusion of the EGFP reporter protein with HCMV proteins depending on viral polymerase activity enabled the establishment of an assay providing highly reproducible results for the determination of EC50s. Among the three drug-sensitive reporter viruses that were generated, the pp65-labeled reporter virus, vTB65g, showed the strongest fluorescence signals. High levels of fluorescence, present only in vTB65g-infected cells, were obviously necessary to reliably and reproducibly determine EC50s. We then measured the fluorescence of infected cells after blocking of viral DNA synthesis by the addition of 800 μM FOS. The baseline signals after early gene expression in cells infected with vTB65g were not significantly higher than those of the two reporter viruses expressing true late proteins, pp150EGFP and pp28EGFP. These experiments proved that early expression of pp65 was not relevant for our test. In consequence, we chose vTB65g as the reporter virus for our FIR assay.
An accurate standardization of assays for EC50 determination and a low susceptibility of results to slight variations within the test are very important. For our FIR assay, we chose day 5 postinfection as the optimum day for fluorescence measurement and determination of EC50s, as it was the earliest point combining reproducible results and few outliers. However, since EC50s remained stable until day 7 postinfection, EC50 determination could alternatively be conducted at later times. This could become important if an inserted polymerase mutation caused a growth defect and high fluorescence signals may be detectable only on day 5 postinfection.
In daily virological routine, one of the very difficult tasks is the tight control of the MOI used in an assay. Therefore, it was of utmost importance to determine the impact of the MOI on the EC50s obtained. It has been shown previously that in the PRA, EC50s are dependent on the MOI (22, 26). In contrast, EC50s determined by FIR assay remained remarkably stable within a broad range of MOIs. This allows standardization of the assay, since differences in inoculums do not affect the EC50s. A similar stability in response to variations in the MOI was observed in a recent study determining EC50s by quantitative analysis of newly synthesized viral DNA by real-time PCR (30). Unlike other assays, including PRA, which are based on measuring the spread of progeny virus in cell culture (11, 18, 22, 23, 35), our FIR assay allows the determination of EC50s from single-cycle virus replication. This accelerates testing and allows indirect evaluation of polymerase activity at higher MOIs. In addition, using single-cycle infection in our assay allows simultaneous determination of the polymerase activity.
By introduction of the well-characterized polymerase mutations E756K (9, 25) and D413E (9, 16), the feasibility of our FIR assay was tested. We confirmed the decrease of susceptibility to FOS due to the E756K mutation, and we also found a slightly decreased susceptibility to GCV and CDV, as previously published (9). However, the previously published higher EC50 ratios between the E756K mutant and its parental virus, especially for FOS, could not be found. In contrast, we could confirm the decrease of susceptibility to GCV and CDV due to the D413E mutation, and EC50 ratios were even higher than those previously published (9). The latter results indicate that the lower EC50 ratios for the E756K mutation did not reveal a general problem of our assay but may have been due to a combination of this specific mutation and our clonal viral background if they were not a consequence of the low interlaboratory standardization level of the PRA. The EC50s determined by FIR assay were highly reproducible, and standard deviations were lower than those with conventional PRA. Interestingly, overall EC50s were consistently lower when measured by FIR assay than those measured by PRA. A similar effect was found using a real-time PCR-based assay (30). EC50s determined by PRA might be higher because PRA measures the reduction in the number of plaques only, irrespective of the effect of the drug on plaque size. Since our FIR assay and the real-time PCR-based assay take into account small changes in DNA synthesis due to antiviral drugs, they are more sensitive than PRA, although using a higher MOI, explaining the consistently lower EC50s.
We then analyzed vTB65g and vE756Kg viruses regarding viral replicative fitness. The E756K mutation is situated in the palm domain of the polymerase, the main region for polymerase activity (32). It is believed that the variation of glutamic acid to lysine interferes with the stability of the palm domain (32). Therefore, it is not surprising that the E756K mutation results in a growth defect (9; this report). The D413E mutation is located in the center of the exonuclease active site and has already been associated with a modest attenuation of viral growth (9). We were able to show for the first time that these growth defects are probably due to a reduction in polymerase activity. This reduction was measured by reduced fluorescence intensity in our system and was confirmed by determination of newly synthesized viral DNA. These results show that our approach of labeling HCMV late proteins is also suitable for indirectly measuring polymerase activity. Although pathogenetic variants of HCMV have not been defined clearly for patients' isolates, determination of viral replicative fitness may provide important information in view of potential differences in pathogenicity and clinical outcome.
The FIR assay as it is used now, with a relatively high MOI and single-cycle replication, is not suitable for the characterization of UL97 mutations or the determination of EC50s for drugs that do not inhibit the polymerase. However, a new standardization of this assay in order to measure EC50s from the second cycle of replication is surely possible and thus will overcome this limitation.
The general advantages of our newly established FIR assay are its reproducibility, rapidity (compared to other marker transfer experiments), and suitability for different applications. Samples in our FIR assay can be measured nondestructively and with ongoing virus replication, allowing measurements at multiple days postinfection. Yet cells can also be fixed and stored for a longer period until being measured. Also, the condition of the cell monolayer (important for a stable fluorescence measurement) and the EGFP intensity in the cells can easily be controlled by microscopy. In addition, unlike the case for all other published assays, the samples do not need further processing before measurement, thereby minimizing explicitly hands-on time, costs, and errors due to additional handling.
The combination of the FIR assay with powerful markerless BAC mutagenesis (34) allows a rapid analysis of polymerase mutations to be characterized by marker transfer analyses. Such marker transfer analyses allow the characterization of single mutations or a defined combination of several mutations in a known viral background, in contrast to the characterization of clinical isolates, where unknown mutations may influence the drug susceptibility phenotype and the viral replicative fitness. The generation of recombinant BACs bearing polymerase mutations takes about 2 weeks, and reconstitution of the mutant viruses can be completed in another 3 to 4 weeks. However, the reconstitution may take longer depending on the viral fitness of the generated mutant. Finally, the determination of EC50s by FIR assay takes 1 week per experiment.
The slow growth of HCMV in cell culture makes phenotypic resistance testing difficult for clinical application, because results are often needed within days. Genotypic resistance testing is very fast and is therefore the preferable approach for clinical application (31). However, it definitely requires a preceding phenotypic characterization of the mutation, ideally by marker transfer analyses. It would also be interesting for diagnostic purposes to directly assay clinical isolates for antiviral drug susceptibility. Therefore, we are currently developing methods to quickly transfer HCMV polymerases from patients into the defined viral backbone of our FIR assay, thus allowing the analysis of entire polymerase sequences regarding their drug susceptibility and viral replicative fitness phenotypes.
Acknowledgments
This work was supported by the University Hospital Ulm and by the Robert-Koch-Institut, Berlin, Germany. Meike Chevillotte is a participant in the International Graduate School in Molecular Medicine Ulm, funded by the excellence initiative of the German federal and state governments.
We thank Gregory Smith and William Britt for providing us with reagents. The technical assistance of Ina Ersing is gratefully acknowledged.
Footnotes
Published ahead of print on 22 June 2009.
REFERENCES
- 1.Avery, R. K. 2007. Management of late, recurrent, and resistant cytomegalovirus in transplant patients. Transpl. Rev. 21:65-76. [Google Scholar]
- 2.Baldanti, F., N. Lurain, and G. Gerna. 2004. Clinical and biologic aspects of human cytomegalovirus resistance to antiviral drugs. Hum. Immunol. 65:403-409. [DOI] [PubMed] [Google Scholar]
- 3.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biron, K. K. 2006. Antiviral drugs for cytomegalovirus diseases. Antivir. Res. 71:154-163. [DOI] [PubMed] [Google Scholar]
- 5.Chevillotte, M., S. Landwehr, L. Linta, G. Frascaroli, A. Luske, C. Buser, T. Mertens, and J. von Einem. 2009. Major tegument protein pp65 of human cytomegalovirus is required for the incorporation of pUL69 and pUL97 into the virus particle and for viral growth in macrophages. J. Virol. 83:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou, S. 2008. Cytomegalovirus UL97 mutations in the era of ganciclovir and maribavir. Rev. Med. Virol. 18:233-246. [DOI] [PubMed] [Google Scholar]
- 7.Chou, S. W. 2001. Cytomegalovirus drug resistance and clinical implications. Transpl. Infect. Dis. 3:20-24. [DOI] [PubMed] [Google Scholar]
- 8.Chou, S., N. S. Lurain, A. Weinberg, G. Y. Cai, P. L. Sharma, C. S. Crumpacker, and the Adult Aids Clinical Trials Group. 1999. Interstrain variation in the human cytomegalovirus DNA polymerase sequence and its effect on genotypic diagnosis of antiviral drug resistance. Antimicrob. Agents Chemother. 43:1500-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou, S., N. Lurain, K. Thompson, R. Miner, and W. Drew. 2003. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32-39. [DOI] [PubMed] [Google Scholar]
- 10.Chou, S., R. Miner, and W. Drew. 2000. A deletion mutation in region V of the cytomegalovirus DNA polymerase sequence confers multidrug resistance. J. Infect. Dis. 182:1765-1768. [DOI] [PubMed] [Google Scholar]
- 11.Chou, S., L. C. Van Wechel, H. M. Lichy, and G. I. Marousek. 2005. Phenotyping of cytomegalovirus drug resistance mutations by using recombinant viruses incorporating a reporter gene. Antimicrob. Agents Chemother. 49:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dal Pozzo, F., G. Andrei, D. Daelemans, M. Winkler, J. Piette, E. De Clercq, and R. Snoeck. 2008. Fluorescence-based antiviral assay for the evaluation of compounds against vaccinia virus, varicella zoster virus and human cytomegalovirus. J. Virol. Methods 151:66-73. [DOI] [PubMed] [Google Scholar]
- 13.Depto, A. S., and R. M. Stenberg. 1989. Regulated expression of the human cytomegalovirus pp65 gene: octamer sequence in the promoter is required for activation by viral gene products. J. Virol. 63:1232-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducancelle, A., S. Alain, F. Petit, M.-J. Sanson Le Pors, and M. C. Mazeron. 2007. Development and validation of a non-radioactive DNA polymerase assay for studying cytomegalovirus resistance to foscarnet. J. Virol. Methods 141:212-215. [DOI] [PubMed] [Google Scholar]
- 15.Ducancelle, A., J. Gravisse, S. Alain, A. M. Fillet, F. Petit, M.-J. S. L. Pors, and M. C. Mazeron. 2005. Phenotypic characterisation of cytomegalovirus DNA polymerase: a method to study cytomegalovirus isolates resistant to foscarnet. J. Virol. Methods 125:145-151. [DOI] [PubMed] [Google Scholar]
- 16.Erice, A., C. Gil-Roda, J.-L. Pérez, H. H. Balfour, K. J. Sannerud, M. N. Hanson, G. Boivin, and S. W. Chou. 1997. Antiviral susceptibilities and analysis of UL97 and DNA polymerase sequences of clinical cytomegalovirus isolates from immunocompromised patients. J. Infect. Dis. 175:1087-1092. [DOI] [PubMed] [Google Scholar]
- 17.Gerna, G., F. Baldanti, and M. G. Revello. 2004. Pathogenesis of human cytomegalovirus infection and cellular targets. Hum. Immunol. 65:381-386. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, C., and G. Boivin. 2005. New reporter cell line to evaluate the sequential emergence of multiple human cytomegalovirus mutations during in vitro drug exposure. Antimicrob. Agents Chemother. 49:4860-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert, C., and G. Boivin. 2005. Human cytomegalovirus resistance to antiviral drugs. Antimicrob. Agents Chemother. 49:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hensel, G., H. Meyer, S. Gartner, G. Brand, and H. F. Kern. 1995. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol. 76:1591-1601. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, K. L., K. A. Dukes, J. Vidaver, E. S. LeShane, I. Ramirez, W. D. Weber, F. Z. Bischoff, S. Hahn, A. Sharma, D. X. Dang, L. M. Hire, D. W. Bianchi, J. L. Simpson, W. Holzgreve, S. Elias, and K. W. Klinger. 2004. Interlaboratory comparison of fetal male DNA detection from common maternal plasma samples by real-time PCR. Clin. Chem. 50:516-521. [DOI] [PubMed] [Google Scholar]
- 22.Landry, M. L., S. Stanat, K. Biron, D. Brambilla, W. Britt, J. Jokela, S. Chou, W. L. Drew, A. Erice, B. Gilliam, N. Lurain, J. Manischewitz, R. Miner, M. Nokta, P. Reichelderfer, S. Spector, A. Weinberg, B. Yen-Lieberman, C. Crumpacker, and the AIDS Clinical Trials Group CMV Resistance Working Group. 2000. A standardized plaque reduction assay for determination of drug susceptibilities of cytomegalovirus clinical isolates. Antimicrob. Agents Chemother. 44:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marschall, M., M. Freitag, S. Weiler, G. Sorg, and T. Stamminger. 2000. Recombinant green fluorescent protein-expressing human cytomegalovirus as a tool for screening antiviral agents. Antimicrob. Agents Chemother. 44:1588-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michel, D., and T. Mertens. 2004. The UL97 protein kinase of human cytomegalovirus and homologues in other herpesviruses: impact on virus and host. Biochim. Biophys. Acta 1697:169-180. [DOI] [PubMed] [Google Scholar]
- 25.Mousavi-Jazi, M., L. Schloss, W. L. Drew, A. Linde, R. C. Miner, J. Harmenberg, B. Wahren, and M. Brytting. 2001. Variations in the cytomegalovirus DNA polymerase and phosphotransferase genes in relation to foscarnet and ganciclovir sensitivity. J. Clin. Virol. 23:1-15. [DOI] [PubMed] [Google Scholar]
- 26.Prix, L., J. Maierl, G. Jahn, and K. Hamprecht. 1998. A simplified assay for screening of drug resistance of cell-associated cytomegalovirus strains. J. Clin. Virol. 11:29-37. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sassenscheidt, J., J. Rohayem, T. Illmer, and D. Bandt. 2006. Detection of [beta]-herpesviruses in allogenic stem cell recipients by quantitative real-time PCR. J. Virol. Methods 138:40-48. [DOI] [PubMed] [Google Scholar]
- 29.Schmolke, S., H. F. Kern, P. Drescher, G. Jahn, and B. Plachter. 1995. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 69:5959-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnepf, N., N. Boiteau, F. Petit, S. Alain, M.-J. Sanson-Le Pors, and M. C. Mazeron. 2009. Rapid determination of antiviral drug susceptibility of human cytomegalovirus by real-time PCR. Antivir. Res. 81:64-67. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber, A., G. Haerter, A. Schubert, D. Bunjes, T. Mertens, and D. Michel. 2009. Antiviral treatment of cytomegalovirus infection and resistant strains. Expert Opin. Pharmacother. 10:191-209. [DOI] [PubMed] [Google Scholar]
- 32.Shi, R., A. Azzi, C. Gilbert, G. Boivin, and S. X. Lin. 2006. Three-dimensional modeling of cytomegalovirus DNA polymerase and preliminary analysis of drug resistance. Proteins 64:301-307. [DOI] [PubMed] [Google Scholar]
- 33.Sinzger, C., G. Hahn, M. Digel, R. Katona, K. L. Sampaio, M. Messerle, H. Hengel, U. Koszinowski, W. Brune, and B. Adler. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89:359-368. [DOI] [PubMed] [Google Scholar]
- 34.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40:191-197. [DOI] [PubMed] [Google Scholar]
- 35.Ueno, T., Y. Eizuru, H. Katano, T. Kurata, T. Sata, S. Irie, and K. Ogawa-Goto. 2006. Novel real-time monitoring system for human cytomegalovirus-infected cells in vitro that uses a green fluorescent protein-PML-expressing cell line. Antimicrob. Agents Chemother. 50:2806-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye, L. B., and E. S. Huang. 1993. In vitro expression of the human cytomegalovirus DNA polymerase gene: effects of sequence alterations on enzyme activity. J. Virol. 67:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]