Abstract
The MUC7 12-mer (RKSYKCLHKRCR) is a cationic antimicrobial peptide derived from the human salivary mucin MUC7. To study its effect/mechanism of action on fungi, we performed a fitness screen of a tagged, diploid, homozygous gene deletion mutant pool of the yeast Saccharomyces cerevisiae grown in the presence of the MUC7 peptide. Forty-five strains exhibiting reduced fitness and 13 strains exhibiting increased fitness (sensitivity or resistance, respectively) were identified by hybridization intensities to tag arrays. The strongest fitness defects were observed with deletions in genes encoding elements of the RIM101 signaling pathway (regulating response to alkaline and neutral pH and other environmental conditions) and of the endosomal sorting complex required for transport (ESCRT; functioning mainly in protein sorting for degradation, but also required for activation of the RIM101 pathway). Other deletions identified as conferring fitness defect or gain are in genes associated with a variety of functions, including transcription regulation, protein trafficking, transport, metabolism, and others. The results of the pool fitness screen were validated by a set of mutant strains tested individually in the presence of the MUC7 12-mer. All tested RIM101-related deletion strains showing fitness defects confirmed their sensitivities. Taken together, the results led us to conclude that deletions of genes associated with the RIM101 pathway confer sensitivity to the peptide by preventing activation of this pathway and that this stress response plays a major role in the protection of S. cerevisiae against damage inflicted by the MUC7 12-mer peptide.
Cationic antimicrobial peptides (CAMPs) are a large and diverse group of ribosomally synthesized molecules exhibiting killing and growth-inhibiting activities against a broad spectrum of microorganisms (3, 39). Cationic peptides of various lengths derived from the N-terminal nonglycosylated part of low-molecular-weight salivary mucin (MUC7) are active against several microorganisms, including the cariogenic bacterium Streptococcus mutans, the opportunistic fungal pathogen Candida albicans, and the nonpathogenic yeast Saccharomyces cerevisiae (2). The most potent among them is the 12-mer containing six cationic amino acids and forming amphipathic α-helix in a hydrophobic environment (28, 29). Despite intensive research, the mechanisms of action of CAMPs, including the MUC7 12-mer, are poorly understood.
An important distinction between antimicrobial peptides and “classical” antibiotics is that while the latter compounds usually have specific proteinaceous targets, CAMPs likely exert multiple effects on cells resulting in death or inhibition of growth. The apparent lack of a single protein target is underscored by the fact that for at least some CAMPs, including those derived from the MUC7 protein, the all-d amino acid forms are as effective as the native all-l forms (31), precluding involvement of stereospecific interactions with cellular macromolecules in their modes of operation.
It appears that the action of CAMPs against target cells is a multifaceted process involving attachment, delivery, inflicted damage, and potential defensive response of affected cells. Hence, interrogation of a large number of easily identifiable mutants for altered susceptibilities to peptides might at once provide information covering all aspects of peptide action. Such a tool is currently available, thanks to the development of genomewide collections of deletion mutants of the baker's yeast, Saccharomyces cerevisiae. Although this organism is not a pathogen, the extensive knowledge of its biology and high number of deletion mutants, not available for any other species of fungi, makes this approach a potential source of valuable data. It may form the basis for further studies of more clinically relevant fungi, such as Candida albicans.
A particularly valuable feature of the mutant collections is the inclusion of unique DNA sequence tags at the site of each deletion, which enables simultaneous monitoring of growth of all pooled mutant strains under the conditions of interest (5, 27). This methodology (chemical-genetic fitness profiling) has been successfully applied to identification of the cellular targets of known and prospective drugs and to study of their mechanisms of action and potential side effects (4, 6, 13, 18, 21, 35). Among available S. cerevisiae deletion collections are a homozygous collection of nonessential gene deletions and a heterozygous collection of deletions in both essential and nonessential genes. The latter, having only one copy of each gene deleted, is employed for studying the effects of reduced levels of encoded proteins (haploinsufficiency) on growth under the conditions of interest. It is particularly useful in direct identification of protein targets of antimicrobial compounds (6, 13). Homozygous deletion mutants, having both alleles of each gene deleted, are expected to have stronger phenotypes due to complete lack of activity encoded by the deleted gene. Hence, they appear to be better suited to investigate the effects exerted by antimicrobial peptides on target cells, although it needs to be noted that the genetic screen of the homozygous collection is limited to nonessential genes.
The presented project is aimed at gaining more insight into the mode of antifungal action of the antimicrobial peptide MUC7 12-mer by employing a genetic screen of S. cerevisiae genomewide diploid, homozygous deletion mutants treated with the peptide. The screen identified a variety of deletion mutants, a majority of which led to higher peptide sensitivity (fitness defect) and some to resistance (fitness gain).
MATERIALS AND METHODS
Strains and growth conditions.
The genomewide pool of diploid, homozygous deletion mutants of S. cerevisiae (4,653 strains; catalog no. 95401.H1Pool), the mis1Δ deletion mutant, and the parental wild-type strain BY4743 were purchased from Invitrogen (Carlsbad, CA). Other individual diploid deletion strains were obtained from Open Biosystems (Huntsville, AL). Haploid MATa deletion strains and parental strain S288C (Open Biosystems) were kindly provided by P. Cullen, University at Buffalo. Yeasts were grown at 30°C in full-strength Sabouraud dextrose broth (SDB), or in twofold-diluted SDB if used for the treatment with the MUC7 12-mer, since the peptide is ineffective against microorganisms in full-strength medium (32). The peptide (RKSYKCLHKRCR) was synthesized by Bio-Synthesis (Lewisville, TX).
Fitness profiling.
Two 200-μl vials containing 4 × 106 cells each (∼8.6 × 102 of each strain) were thawed and grown overnight in 12 ml of full-strength SDB medium. From the expanded culture, 1.5 × 108 cells were collected for DNA isolation (time = 0), and the remaining yeasts were used for fitness profiling and direct selection. For fitness profiling, 2.5 × 107 cells were inoculated into four tubes, each containing 25 ml of twofold-diluted SDB. The MUC7 12-mer was added to two of these tubes to a concentration of 5 μM; the cells in the other two tubes were left untreated. Following 24 h culture, 1.5 × 108 cells from each of the four cultures were collected for DNA isolation (24 h time point), and 2.5 × 107 cells from each culture were used to inoculate four new tubes containing 25 ml of twofold-diluted SDB for next round of 24 h competitive growth. Again, the peptide was added to two of the tubes to a concentration of 5 μM. After 24 h growth (48 h time point), cells were collected for DNA isolation.
DNA isolation.
The cells from each time point (0, 24, and 48 h) were harvested by centrifugation (5,000 rpm, 5 min at room temperature), and the DNA was isolated immediately. The supernatant was removed, and the cells were resuspended in 100 μl of breaking buffer (2% Triton X-100, 1% sodium dodecyl sulfate, 100 mM NaCl, 10 mM Tris-Cl [pH 8.0], 1 mM EDTA). The cells were then lysed as follows. One hundred microliters of phenol-chloroform-isoamyl alcohol mixture (25/24/1) and approximately 0.1 g of Biospec glass beads (0.5-mm diameter) were added to the cell suspension, and the suspension was vortexed for 4 minutes at high speed. Tris-EDTA (TE) buffer (100 μl) was then added to each sample; samples were mixed by vortexing and then centrifuged for 5 minutes at 13,000 rpm. The aqueous layers were transferred to a microcentrifuge tube, 250 μl of 100% ethanol was added, and samples were mixed by inverting the tubes to precipitate the DNA. The tubes were centrifuged again at 13,000 rpm at room temperature for 3 minutes. The supernatant was discarded, and the pellet was dried at 37°C for 30 min. DNA pellet was suspended in 100 μl of TE buffer and treated with 2 μl of 1 mg/ml RNase A at 37°C for 10 min. The DNA was then precipitated by addition of 2.5 μl of 4 M ammonium acetate and 250 μl ethanol. The content of tubes was mixed by inversion and the DNA recovered by centrifugation at 13,000 rpm for 3 min at room temperature. DNA was resuspended in 100 μl of TE buffer, and a portion was separated on 1% agarose gel to determine the amount and quantity of DNA isolated.
Asymmetric PCR.
PCR was performed with 200 ng of a genomic DNA template, using pairs of universal Uptag- and Downtag-specific primers (purchased from Bio-Synthesis), with one primer of each pair labeled with Cy3 or Cy5 fluorescent dye, at a final concentration of 0.5 and 5 μM for the unlabeled and labeled primers, respectively. Four PCRs were prepared to be combined for hybridization to one tag array: two Cy5 reactions (control samples, with Uptag- and Downtag-labeled primers) and two Cy3 reactions (treated samples, with Uptag and Downtag primers). The following primers were used: Reaction 1 (Uptag), 5′-(Cy5)GTCGACCTGCAGCGTACG-3′ and 5′-GATGTCCACGAGGTCTCT-3′; Reaction 2 (Uptag), 5′-(Cy3)GTCGACCTGCAGCGTACG-3′ and 5′-GATGTCCACGAGGTCTCT-3′; Reaction 3 (Downtag), 5′-(Cy5)CGAGCTCGAATTCATCGAT-3′ and 5′-CGGTGTCGGTCTCGTAG-3′; Reaction 4 (Downtag), 5′-(Cy3)CGAGCTCGAATTCATCGAT-3′ and 5′-CGGTGTCGGTCTCGTAG-3′.
The PCRs were also done with “a dye swap.”
Due to unequal concentrations of primers, the reaction produced predominantly single-stranded labeled DNA fragments complementary to probes on the tag arrays. Fifty cycles of amplification were performed using the following parameters: 94°C for 10 s, 50°C for 10 s, and 72°C for 20 s. Following amplification, blocking oligonucleotides complementary to the labeled DNA products at regions encompassing universal priming sites were added to each sample to a concentration of 100 μM. Samples were heated for 1 min at 100°C, cooled on ice, and ethanol precipitated. A small portion of each sample was run on 8% acrylamide gel (in Tris-acetate-EDTA buffer), and products were visualized by the Typhoon 9400 imager (GE Healthcare, Piscataway, NJ) for size, quality, and quantity.
Tag array preparation and hybridization.
The custom tag arrays were manufactured by NimbleGen Systems, Inc. (Madison, WI). Sequences of the probes were retrieved from the GEO database, accession number GPL1444 (37). Each Uptag and each Downtag is represented by six probes scattered randomly on the array. Hybridizations were performed in the MAUI hybridization station (BioMicro Systems, Salt Lake City, UT), following the manufacturer's instructions. The ethanol-precipitated Cy dye-labeled samples (as described above) were dissolved in approximately 28 μl of 1× SSTE buffer (1 M NaCl, 10 mM Tris-Cl [pH 7.5], 0.5% Triton X-100, and 1 mM dithiothreitol [DTT]) and injected into an X1 mixer chamber. Following hybridization (∼16 h at 42°C), slides were washed, first in 6× SSPE buffer (0.9 M NaCl, 60 mM NaH2PO4, 6 mM EDTA) supplemented with 0.05% Triton X-100 and 1 mM DTT, and then in 0.06× SSPE buffer with 1 mM DTT. The arrays were scanned using a GenePix professional 4200A microarray scanner (Molecular Devices, Sunnyvale, CA).
Tag array data analysis.
Individual scanned images were automatically aligned using NimbleScan software (NimbleGen). Raw intensities were further processed separately for Up- and Downtags (23, 24). For each hybridization, intensities in both channels were quantile normalized, and medians of untreated/treated ratios of six replicate probes per array were calculated. The fitness of deletion mutants at each time point is expressed as log2 of the median of six calculated intensity ratios (Up- and Downtags for three separate cultures). Statistical significance was determined by a two-tailed, paired t test. Positive log2 values of mean hybridization intensity ratios of untreated to treated samples correspond to fitness defect (sensitivity), while negative values correspond to fitness gain (resistance).
Direct selection.
Cells from the expanded 24 h culture of deletion pool were diluted to a density of 5 × 105/ml in 10 mM phosphate buffer (pH 7.4). Four 40-μl samples supplemented with the MUC7 12-mer at concentrations of 0, 5, 10, 30 μM were incubated at 30°C for 2 h, and then diluted to 1 ml in the phosphate buffer. Small sample aliquots were plated onto agar to estimate the fraction of killed cells, and the rest of the culture was resuspended in 9 ml of SDB medium and grown overnight. After six cycles of selection and growth, several clones plated onto agar were tested individually in a killing assay in 10 mM sodium phosphate buffer (pH 7.4) as described earlier (28).
Growth rate measurements.
Overnight cultures grown in SDB were diluted in twofold-diluted SDB to an optical density at 600 nm (OD600) of 0.02 or 0.01 (for diploid or haploid strains, respectively). The diluted suspensions (100 μl) were transferred into three 96-well microtiter plate wells containing 100 μl of twofold-diluted SDB medium supplemented with the MUC7 12-mer at a concentration of 20 μM or 40 μM (for diploid or haploid strains, respectively) and three control wells (without the peptide). Plates were incubated at 30°C, and ODs of cultures were measured at different time points in a microtiter reader. OD values of three wells were averaged. Plotted OD595 values for each time point are the means of the results for three separate experiments.
RESULTS
Fitness profiling.
To evaluate the effects of individual gene deletions on the susceptibility of S. cerevisiae to the antimicrobial peptide MUC7 12-mer, we screened the pool of homozygous diploid deletion mutants during growth in the presence of the peptide. A suitable concentration of the peptide was determined by testing the parental strain. At a concentration of 5 μM, growth was inhibited by approximately 50% compared to the untreated culture (not shown). Based on these results, the dose at a concentration of 5 μM was selected for the fitness profiling studies.
Yeasts were grown in the medium, with or without the peptide, for two consecutive 24 h cultures as described in Materials and Methods. At time zero and at the end of each culture, samples were collected for genomic DNA isolation, and the DNA was used as templates for asymmetric PCR amplification of “barcodes,” using Cy dye-labeled primers. The abundance of each strain within the pool was measured at each time point by hybridization of the PCR-amplified “barcodes” to DNA tag arrays. Each pair of untreated and treated cultures was replicated three times. The average ratio of hybridization intensities between the untreated and peptide-treated samples was the measure of fitness. Despite the growth of cells treated with the peptide being slower than that of the control, equal amounts of DNA were used for PCR. Therefore, strains whose growth was inhibited by the peptide to the same extent as the average of the pool had an intensity ratio of 1, defining lack of fitness defect or gain. Fitness defect (or increased sensitivity to the peptide) was defined as the intensity ratio of at least 4 (or log2 value of 2) at the 48 h time point, whereas fitness gain (or resistance to the peptide) was defined as a ratio of 1/2 or lower (expressed as log2 of −1 or less). The lower cutoff in the latter case was set due to a low number of deletion mutants exhibiting fitness gain. By these arbitrary criteria, 58 deletion mutant strains conferring differential fitness in the presence of the MUC7 12-mer have been identified. They are listed in Table 1.
TABLE 1.
Deletions of S. cerevisiae genes conferring reduced fitness (higher sensitivity) or increased fitness (resistance), to MUC7 12-mer peptide, identified in the fitness screen
| Open reading frame | Gene | Log2 intensity ratioa
|
P valueb | Function, component of a complex, or GO termc | |
|---|---|---|---|---|---|
| 24 h | 48 h | ||||
| Reduced fitness | |||||
| YPL065W | VPS28 | 3.92 | 4.87 | 0.003639 | ESCRT I |
| YPL002C | VPS22 | 2.98 | 4.79 | 0.000369 | ESCRT II |
| YJR102C | VPS25 | 3.72 | 4.59 | 0.007009 | ESCRT II |
| YCL008C | VPS23 | 3.92 | 4.57 | 0.008344 | ESCRT I |
| YGL045W | RIM8 | 3.68 | 4.52 | 0.009784 | RIM101 pathway |
| YMR077C | VPS20 | 3.61 | 4.49 | 0.010161 | ESCRT III |
| YLR025W | SNF7 | 3.26 | 4.47 | 0.010608 | ESCRT III |
| YOR030W | DFG16 | 3.75 | 4.33 | 0.006508 | RIM101 pathway |
| YNL294C | RIM21 | 3.52 | 4.33 | 0.010459 | RIM101 pathway |
| YLR417W | VPS36 | 2.99 | 4.33 | 0.008441 | ESCRT II |
| YOR275C | RIM20 | 3.83 | 4.30 | 0.003918 | RIM101 pathway |
| YGR122W | YGR122W | 3.12 | 4.03 | 0.003781 | RIM101 pathway |
| YHL027W | RIM101 | 3.28 | 3.99 | 0.006259 | RIM101 pathway |
| YCR081W | SRB8 | 2.97 | 3.99 | 0.028961 | Transcription regulation |
| YOR035C | SHE4 | 2.14 | 3.83 | 0.012647 | Actin cytoskeleton organization |
| YCR068W | ATG15 | 1.26 | 3.75 | 0.003086 | Autophagy |
| YBR291C | CTP1 | 1.27 | 3.69 | 0.015299 | Mitochondrial citrate transport |
| YNR052C | POP2 | 2.48 | 3.65 | 0.007818 | mRNA deadenylation |
| YNL147W | LSM7 | 1.76 | 3.52 | 0.000894 | mRNA catabolic process |
| YIL148W | RPL40A | 1.71 | 3.52 | 0.021529 | 60S ribosomal subunit-ubiquitin fusion |
| YHR021C | RPS27B | 2.33 | 3.49 | 0.010849 | 40S ribosomal subunit component |
| YGL250W | RMR1 | 2.34 | 3.43 | 0.004995 | Meiotic gene conversion and recombination |
| YCR045C | YCR045C | 1.49 | 3.40 | 0.029685 | Unknown |
| YCR087C-A | LUG1 | 1.28 | 3.39 | 0.006636 | Unknown |
| YCR050C | YCR050C | 2.05 | 3.37 | 0.038273 | Unknown |
| YNL025C | SSN8 | 3.26 | 3.35 | 0.025936 | Transcription regulation |
| YDR414C | ERD1 | 1.92 | 3.20 | 0.008131 | Protein retention in ER lumen |
| YCR036W | RBK1 | 1.76 | 3.18 | 0.040463 | Putative ribokinase |
| YMR154C | RIM13 | 2.43 | 3.09 | 0.078834 | RIM101 pathway |
| YHL009C | YAP3 | 0.64 | 3.00 | 0.037838 | Transcription regulation |
| YBR290W | BSD2 | 1.33 | 2.90 | 0.007485 | Metal ion transport, protein targeting to vacuole |
| YCL001W-A | YCL001W-A | 1.00 | 2.79 | 0.015056 | Unknown |
| YDR069C | DOA4 | 1.16 | 2.79 | 0.048266 | ESCRT III |
| YML097C | VPS9 | 2.28 | 2.74 | 0.027094 | Protein targeting to vacuole |
| YMR063W | RIM9 | 2.96 | 2.68 | 0.062849 | RIM101 pathway |
| YOL004W | SIN3 | 2.00 | 2.64 | 0.021773 | Histone deacylation |
| YDR323C | PEP7 | 0.81 | 2.64 | 0.022957 | Vacuolar protein sorting |
| YLR423C | ATG17 | 1.48 | 2.58 | 0.030774 | Autophagy |
| YDR443C | SSN2 | 1.32 | 2.49 | 0.000173 | Transcription regulation |
| YBR283C | SSH1 | 1.53 | 2.45 | 0.030825 | Cotranslational protein targeting to membrane |
| YBR298C | MAL31 | 2.54 | 2.41 | 0.027856 | Maltose transporter |
| YCR024C-A | PMP1 | 0.84 | 2.36 | 0.020819 | Cation transport |
| YLR330W | CHS5 | 1.28 | 2.23 | 0.022982 | Golgi to plasma membrane transport |
| YNL183C | NPR1 | 0.69 | 2.22 | 0.001061 | Regulation of nitrogen utilization |
| YNR051C | BRE5 | 0.54 | 2.20 | 0.017038 | Protein deubiquitination |
| YPL265W | DIP5 | 1.72 | 2.15 | 0.047433 | Amino acid transport |
| YPR101W | SNT309 | 1.03 | 2.13 | 0.044431 | Nuclear mRNA splicing via spliceosome |
| Increased fitness | |||||
| YBL101C | ECM21 | −0.12 | −2.68 | 0.021139 | Ubiquitin-dependent endocytosis |
| YJR056C | YJR056C | 0.13 | −2.02 | 0.004089 | Unknown |
| YDR276C | PMP3 | −0.67 | −1.80 | 0.020970 | Regulation of membrane potential |
| YDR516C | EMI2 | 0.01 | −1.73 | 0.040832 | Transcription regulation |
| YBL089W | AVT5 | 0.02 | −1.62 | 0.019372 | Amino acid vacuolar transport |
| YDR067C | OCA6 | −0.34 | −1.58 | 0.024253 | Unknown |
| YIL125W | KGD1 | −0.13 | −1.47 | 0.006007 | Tricarboxylic acid cycle |
| YDR346C | SVF1 | −0.09 | −1.43 | 0.029858 | Response to oxidative stress |
| YJR009C | TDH2 | 0.00 | −1.42 | 0.028543 | Glyceraldehyde-3-phosphate dehydrogenase |
| YDR310C | SUM1 | −0.38 | −1.41 | 0.031673 | Transcription regulation, DNA replication initiation |
| YJR014W | TMA22 | −0.06 | −1.23 | 0.030385 | Unknown, associates with ribosome |
| YJL110C | GZF3 | 0.04 | −1.20 | 0.008931 | Transcription regulation, nitrogen utilization |
| YMR109W | MYO5 | −0.32 | −1.11 | 0.008158 | Type I myosin |
Log2 hybridization intensity ratio between untreated and treated samples collected at the indicated time point.
For 48 h time point only.
Saccharomyces genome database (www.yeastgenome.org).
Deletion mutants displaying fitness defects.
Among 45 deletions conferring sensitivities to the peptide, two major groups of open reading frames could be distinguished. One represents genes in the RIM101 signaling pathway (22), the end product of which is activation of Rim101p (10), a transcription factor, responsible for regulation of a variety of cellular functions following changes in ambient pH and other environmental stimuli. The other group represents genes encoding elements of the endosomal sorting complex required for transport (ESCRT) (1, 33), a molecular machinery sorting ubiquitinated proteins to vacuole for degradation, with some of its subunits also required for activation of RIM101 pathway (22).
We have examined how the ESCRT-associated genes identified in our screen relate to the RIM101 pathway. We found that all eight ESCRT deletion strains exhibiting strong fitness defect in the presence of the MUC7 peptide (vps23Δ, vps28Δ, vps22Δ, vps25Δ, vps36Δ, vps20Δ, snf7Δ, and doa4Δ strains) (Table 1) are indeed in genes encoding proteins known to participate in the induction of the RIM101 pathway (7, 25, 36). In contrast, none of the deletions in all remaining known ESCRT-related genes not involved in the RIM101 induction were identified in our fitness screen as having an effect on the growth of yeasts in the presence of the peptide (25, 36). This finding strongly suggests that the observed hypersensitivity of mutant strains with deleted ESCRT genes is caused by lack of a defensive response induced by the RIM101 pathway. Other deletions conferring fitness defects are associated with a variety of functions, including transcription, transport, protein targeting, and others (as listed in Table 1).
Deletion mutants displaying fitness gains.
Thirteen deletions resulted in resistance to the MUC7 12-mer peptide, marked by at least a twofold increase of hybridization intensity between the treated and untreated samples (Table 1). Deletions in this group represent genes associated with diverse cellular functions, including ECM21, which is involved in regulation of endocytosis of membrane proteins; PMP3, which possibly regulates membrane potential; AVT5, a putative vacuolar transporter; and others.
Validation of fitness profiling results.
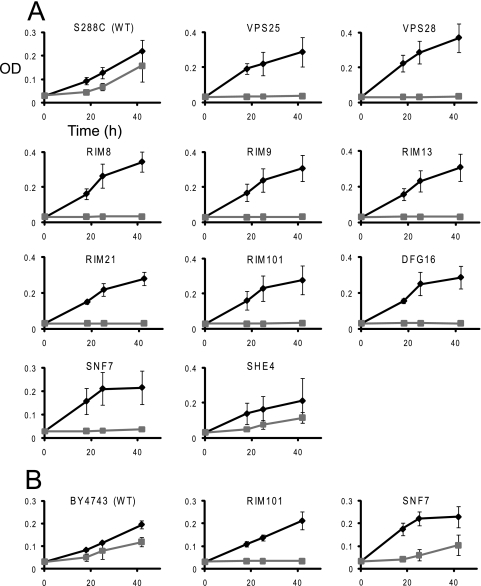
We have chosen a set of mutant strains, both diploid and haploid, to be tested individually. Diploid homozygous and haploid null mutants have been demonstrated to provide similar results in such tests (20). Selected strains were grown for 42 h in liquid cultures in the presence or absence of the MUC7 12-mer. All nine tested deletion strains in genes known to be indispensable for the Rim101p activation (7, 25, 36) and showing fitness defects in the MUC7 12-mer pool screen exhibited complete growth suppression by the peptide (Fig. 1), confirming their strong sensitivity.
FIG. 1.
Confirmation of sensitivity of individual deletion mutant strains to MUC7 12-mer. Growth of haploid (A) and diploid homozygous (B) mutants exhibiting fitness defects in the pool screen, in the presence (squares) or absence (diamonds) of the MUC7 12-mer peptide, monitored by OD595. Time points are means of three independent experiments. Error bars represent the standard deviation.
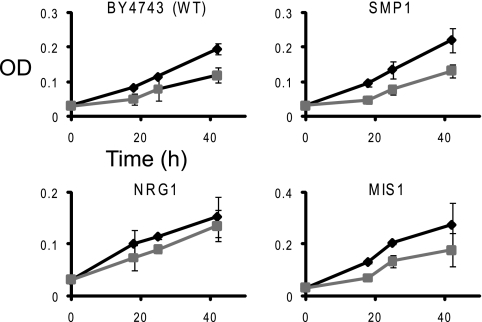
Activated transcription factor Rim101p controls signaling through repression of downstream regulatory proteins Nrg1p and Smp1p that, being repressors themselves, control effector genes (8). Thus, we hypothesized that if some of these downstream genes play significant roles in defense against the MUC7 peptide, deletions of NRG1 or SMP1 might lead to a fitness gain missed by the large-scale profiling. The individual growth tests with nrg1Δ and smp1Δ mutants in the presence of the peptide, shown in Fig. 2, suggest that Nrg1p, but not Smp1p, might indeed control some of the genes needed for response to the peptide.
FIG. 2.
Growth of individual strains not identified in the pool screen but related to the RIM101 pathway or directly selected (mis1Δ strain), in the presence (squares) or absence (diamonds) of the MUC7 12-mer peptide. All strains shown are diploid homozygous mutants. Other details are as described in the legend for Fig. 1.
We also tested one other strain, the she4Δ mutant with deletion in a gene not related to the RIM101 pathway or ESCRT, but exhibiting comparable fitness defect in the genetic screen. In contrast, when grown individually, this strain did not show visibly stronger sensitivity to the peptide than that shown by the wild-type strain (Fig. 1).
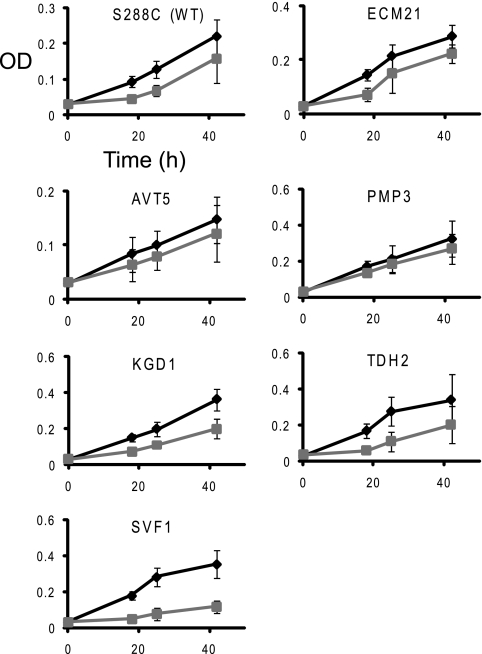
Six deletion strains identified as having increased fitness (thus resistance) were also tested individually for growth in the presence of the peptide. Two (pmp3Δ and avt5Δ mutants) confirmed their resistance (Fig. 3), while the growth of three other strains (ecm21Δ, kgd1Δ, and tdh2Δ mutants) was difficult to distinguish from that of the wild type. In one case, the svf1Δ strain, the peptide had a somewhat opposite effect than that during the pool screen, inhibiting its growth to a larger extent than the growth of the wild type (Fig. 3). Similar apparent discrepancies between the large-scale screen and testing of individual strains have been noted previously (34).
FIG. 3.
Confirmation of resistance of individual deletion mutant strains to MUC7 12-mer. Growth of mutants exhibiting fitness gain in the pool screen, in the presence (squares) or absence (diamonds) of the MUC7 12-mer peptide. All strains shown are haploid mutants. Other details are as described in the legend for Fig. 1.
Direct selection for resistant strains.
In addition to fitness profiling, we performed direct selection for resistant strains among the pool of deletion mutants. In contrast to the fitness profiling, where mutant strains competed during continuous growth in the presence of the MUC7 peptide, the direct selection was carried out by repeatedly challenging members of the pool by 2 hours of exposure to the peptide in a buffer. During this time, a fraction of cells, proportional to the peptide concentration, is killed. Following each treatment, the surviving cells were cultured overnight in the absence of the peptide. One clone, which exhibited elevated survival rates, was identified by sequencing of its barcodes as carrying deletion in the MIS1 gene encoding mitochondrial C1-tetrahydrofolate synthase. Interestingly, this strain did not exhibit fitness gain during the large-scale screen, nor did it show consistent resistance in continuous growth culture (Fig. 2). In contrast, both, the originally isolated strain and the separately purchased mis1Δ mutant consistently exhibited much higher resistance to the peptide than did the parental strain in killing assay. For example, at the 10 μM peptide concentration, the parental strain showed ∼10% survival, compared to ∼44% and ∼53% for the purchased and selected mis1Δ clones, respectively (data not shown).
DISCUSSION
The strains identified in the pool screen are mutants with genes associated with a variety of cellular processes, including transcription regulation, small molecule and protein transport, and others. Particularly striking was the identification of two groups of strains characterized by strong fitness defects, namely deletions in genes encoding proteins associated the RIM101 signaling pathway and with the ESCRT complexes (Table 1).
RIM101 pathway regulates, in response to alkaline pH, a variety of physiological functions, from meiosis and sporulation to invasive growth. It also controls overall adaptation to high pH, as well as to other environmental conditions including Na+ and Li+ ions and low temperatures (8, 9, 22). The signal, originating in the integral plasma membrane proteins Rim21p and Dfg16p, is conveyed to the vacuolar sorting complex ESCRT III, where transcription regulator Rim101p is activated by cleavage of its C-terminal portion. Proteolytically activated Rim101p acts as a repressor, by blocking transcription of downstream genes whose protein products are themselves repressors inhibiting expression of effector genes further downstream. Among genes directly affected by Rim101p are NRG1, which controls alkaline growth and sodium and lithium tolerance, and SMP1, which is responsible for the control of meiosis, sporulation, and invasive growth (8). Deletion in the former appears to partially reduce sensitivity to the peptide compared to that of the parental strain (Fig. 2). This may suggest that at least some of the genes controlled by the repressor Nrg1p play roles in defense against the 12-mer.
ESCRT complexes, localized to endosomes, function in sorting of monoubiquitinated proteins to vacuoles for degradation (1, 33). The ESCRT machinery plays important roles in a variety of cellular functions by regulating surface receptors and other membrane proteins. As noted above, it also participates in the RIM101 signaling. In fact, our screen identified only those genes encoding ESCRT subunits that have been demonstrated to be indispensable for activation of the RIM101 pathway (7, 25, 36). This suggests that the process of protein trafficking for degradation per se is not needed for the defense against MUC7 peptide. Rather, the ESCRT machinery appears to play a role in response to the peptide only as far as it is required for functioning of the RIM101 pathway.
Among the deletions conferring fitness gain, three—pmp3Δ, ecm21Δ and avt5Δ—are particularly interesting. PMP3 encodes a small, highly hydrophobic plasma membrane proteolipid of unknown function. Its deletion results in the hyperpolarization of membrane potential and, consequently, in hypersensitivity to sodium ions and toxic cations, hygromycin B, and tetramethylammonium (19). Plant homologues of Pmp3p play a role in tolerance to salt, cold, and dehydration (15, 16). Despite cationic character of the MUC7 12-mer, the S. cerevisiae pmp3Δ strain is resistant to the peptide, as revealed both by fitness profiling and by evaluation of individual mutants (Table 1 and Fig. 3). A possible explanation may be that the presence or absence of this polypeptide may affect physicochemical properties of plasma membrane. It would be interesting, for example, to determine how the deletion of PMP3 affects binding to cell surface and/or internalization of the peptide.
ECM21, also known as ART2, encodes one of several cargo-specific ubiquitin ligase adaptors, regulating turnover of membrane proteins. Ecm21p mediates downregulation of the lysine transporter Lyp1p in response to stress (11). Mutation in this gene has also been identified in a transposon-mediated genetic screen as hypersensitive to calcofluor white, an agent interfering with cell wall organization (14). Resistance of the ecm21Δ mutant to the MUC7 peptide may be related to retention of the lysine transporter in the cell membrane. Alternatively, Ecm21p may also regulate another and still unknown membrane protein whose presence on the cell surface may affect MUC7 12-mer functions.
AVT5 belongs to a family of seven S. cerevisiae genes encoding membrane proteins related to GABA-glycine vacuolar transporters (26). Some Avt proteins are involved in vacuolar transport of amino acids, but neither substrate nor cellular localization of Avt5p has been determined. It would be interesting to look into a possible vacuolar localization of the MUC7 peptide and to determine how it may be affected by AVT5 deletion. Nonlethal vacuolar localization of another CAMP, histatin 5, has been recently observed in C. albicans at low peptide concentrations, whereas cytoplasmic localization at higher concentrations led to death of cells (17).
Another approach employed in this study was direct selection for mutant strains resistant to temporary exposure to the peptide in a buffer. We have previously developed and partially characterized a C. albicans mutant highly resistant to killing in the buffer, but not in medium during continuous growth (12). We hypothesize that the resistance was caused by changes in properties of the plasma membrane, resulting in reduced efficiency of internalization of the peptide by the cells, an event concurrent with their death. Here, we attempted to select an S. cerevisiae mutant(s) characterized by a similar type of resistance. The selected mutant has deletion in the MIS1 gene encoding mitochondrial C-1-tetrahydrofolate synthase. The mis1Δ mutant has been previously reported to exhibit reduced growth on nonfermentable carbon sources (30), a property reminiscent of the C. albicans mutant mentioned above, which was characterized by complete inability to grow on nonfermentable carbon sources and by significant changes in metabolism (12).
We are aware of only one other report describing yeast fitness profiling in response to antimicrobial peptides dermaseptin and magainin (18). No deletion strains suggesting involvement of the RIM101 pathway in the defense against these peptides have been identified in that screen, suggesting that their modes of action may differ from that of the MUC7 12-mer.
In another study (38), the effects of deletion mutations of S. cerevisiae genes on sensitivity to chitosan were investigated. This chitin-derived polymer shares some crucial properties with cationic antimicrobial peptides, such as net positive charge and perturbations inflicted on cellular membranes of bacteria and fungi (38). One group of reported deletions leading to chitosan sensitivity was in genes encoding proteins involved in endosomal processes. Although the authors did not specifically point to the RIM101 signaling, analysis of the provided supplemental material revealed that all deletions associated with this pathway including elements of ESCRT, which are shown in Table 1 (except doa4Δ), are also sensitive to chitosan (with log2 intensity ratios ranging from 1.1 to 1.9, at 9 h of treatment) (38). This suggests that some of the effects of both compounds (chitosan and the MUC7 12-mer) may be similar. It needs to be pointed out, however, that deletions in genes associated with the high-osmolarity pathway conferred much stronger fitness defects (up to log2 intensity ratio of 4.46) (38) in the presence of chitosan. In contrast, results of our screen did not suggest involvement of the high-osmolarity pathway in response to the MUC7 peptide.
In conclusion, the importance of the RIM101 stress response and other identified deletions conferring both hypersensitivity and resistance to the MUC7 peptide points to the direction of further studies. They are needed to understand the nature of the stress imposed by this and possibly other antimicrobial peptides, especially on clinically important fungal pathogens, such as C. albicans.
Acknowledgments
This work was supported by NIH/NIDCR grant DE009820 (L.A.B.) and DE009820 supplement (J.R.F.).
We thank Paul Cullen, Department of Biological Sciences, University at Buffalo, for providing the S. cerevisiae haploid deletion mutants.
Footnotes
Published ahead of print on 13 July 2009.
REFERENCES
- 1.Babst, M. 2005. A protein's final ESCRT. Traffic 6:2-9. [DOI] [PubMed] [Google Scholar]
- 2.Bobek, L. A., and H. Situ. 2003. MUC7 20-mer: investigation of antimicrobial activity, secondary structure, and possible mechanism of antifungal action. Antimicrob. Agents Chemother. 47:643-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brogden, K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238-250. [DOI] [PubMed] [Google Scholar]
- 4.Ericson, E., M. Gebbia, L. E. Heisler, J. Wildenhain, M. Tyers, G. Giaever, and C. Nislow. 2008. Off-target effects of psychoactive drugs revealed by genome-wide assays in yeast. PLoS Genet. 4:e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 6.Giaever, G., D. D. Shoemaker, T. W. Jones, H. Liang, E. A. Winzeler, A. Astromoff, and R. W. Davis. 1999. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet. 21:278-283. [DOI] [PubMed] [Google Scholar]
- 7.Ikeda, M., A. Kihara, A. Denpoh, and Y. Igarashi. 2008. The rim101 pathway is involved in rsb1 expression induced by altered lipid asymmetry. Mol. Biol. Cell 19:1922-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb, T. M., and A. P. Mitchell. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamb, T. M., W. Xu, A. Diamond, and A. P. Mitchell. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276:1850-1856. [DOI] [PubMed] [Google Scholar]
- 10.Li, W., and A. P. Mitchell. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin, C. H., J. A. MacGurn, T. Chu, C. J. Stefan, and S. D. Emr. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135:714-725. [DOI] [PubMed] [Google Scholar]
- 12.Lis, M., and L. A. Bobek. 2008. Proteomic and metabolic characterization of a Candida albicans mutant resistant to the antimicrobial peptide MUC7 12-mer. FEMS Immunol. Med. Microbiol. 54:80-91. [DOI] [PubMed] [Google Scholar]
- 13.Lum, P. Y., C. D. Armour, S. B. Stepaniants, G. Cavet, M. K. Wolf, J. S. Butler, J. C. Hinshaw, P. Garnier, G. D. Prestwich, A. Leonardson, P. Garrett-Engele, C. M. Rush, M. Bard, G. Schimmack, J. W. Phillips, C. J. Roberts, and D. D. Shoemaker. 2004. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116:121-137. [DOI] [PubMed] [Google Scholar]
- 14.Lussier, M., A. M. White, J. Sheraton, T. di Paolo, J. Treadwell, S. B. Southard, C. I. Horenstein, J. Chen-Weiner, A. F. Ram, J. C. Kapteyn, T. W. Roemer, D. H. Vo, D. C. Bondoc, J. Hall, W. W. Zhong, A. M. Sdicu, J. Davies, F. M. Klis, P. W. Robbins, and H. Bussey. 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147:435-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medina, J., M. L. Ballesteros, and J. Salinas. 2007. Phylogenetic and functional analysis of Arabidopsis RCI2 genes. J. Exp. Bot. 58:4333-4346. [DOI] [PubMed] [Google Scholar]
- 16.Mitsuya, S., M. Taniguchi, H. Miyake, and T. Takabe. 2005. Disruption of RCI2A leads to over-accumulation of Na+ and increased salt sensitivity in Arabidopsis thaliana plants. Planta 222:1001-1009. [DOI] [PubMed] [Google Scholar]
- 17.Mochon, A. B., and H. Liu. 2008. The antimicrobial peptide histatin-5 causes a spatially restricted disruption on the Candida albicans surface, allowing rapid entry of the peptide into the cytoplasm. PLoS Pathog. 4:e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morton, C. O., A. Hayes, M. Wilson, B. M. Rash, S. G. Oliver, and P. Coote. 2007. Global phenotype screening and transcript analysis outlines the inhibitory mode(s) of action of two amphibian-derived, alpha-helical, cationic peptides on Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 51:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navarre, C., and A. Goffeau. 2000. Membrane hyperpolarization and salt sensitivity induced by deletion of PMP3, a highly conserved small protein of yeast plasma membrane. EMBO J. 19:2515-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pagé, N., M. Gerard-Vincent, P. Menard, M. Beaulieu, M. Azuma, G. J. Dijkgraaf, H. Li, J. Marcoux, T. Nguyen, T. Dowse, A. M. Sdicu, and H. Bussey. 2003. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163:875-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsons, A. B., A. Lopez, I. E. Givoni, D. E. Williams, C. A. Gray, J. Porter, G. Chua, R. Sopko, R. L. Brost, C. H. Ho, J. Wang, T. Ketela, C. Brenner, J. A. Brill, G. E. Fernandez, T. C. Lorenz, G. S. Payne, S. Ishihara, Y. Ohya, B. Andrews, T. R. Hughes, B. J. Frey, T. R. Graham, R. J. Andersen, and C. Boone. 2006. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126:611-625. [DOI] [PubMed] [Google Scholar]
- 22.Peñalva, M. A., J. Tilburn, E. Bignell, and H. N. Arst, Jr. 2008. Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 16:291-300. [DOI] [PubMed] [Google Scholar]
- 23.Peyser, B. D., R. Irizarry, and F. A. Spencer. 2008. Statistical analysis of fitness data determined by TAG hybridization on microarrays. Methods Mol. Biol. 416:369-381. [DOI] [PubMed] [Google Scholar]
- 24.Pierce, S. E., R. W. Davis, C. Nislow, and G. Giaever. 2007. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat. Protoc. 2:2958-2974. [DOI] [PubMed] [Google Scholar]
- 25.Rothfels, K., J. C. Tanny, E. Molnar, H. Friesen, C. Commisso, and J. Segall. 2005. Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 25:6772-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russnak, R., D. Konczal, and S. L. McIntire. 2001. A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J. Biol. Chem. 276:23849-23857. [DOI] [PubMed] [Google Scholar]
- 27.Shoemaker, D. D., D. A. Lashkari, D. Morris, M. Mittmann, and R. W. Davis. 1996. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat. Genet. 14:450-456. [DOI] [PubMed] [Google Scholar]
- 28.Situ, H., G. Wei, C. J. Smith, S. Mashhoon, and L. A. Bobek. 2003. Human salivary MUC7 mucin peptides: effect of size, charge and cysteine residues on antifungal activity. Biochem. J. 375:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, C. J., and L. A. Bobek. 2001. Bactericidal and fungicidal activity of salivary mucin (MUC7) peptide fragments. J. Dent. Res. 80:601. [Google Scholar]
- 30.Steinmetz, L. M., C. Scharfe, A. M. Deutschbauer, D. Mokranjac, Z. S. Herman, T. Jones, A. M. Chu, G. Giaever, H. Prokisch, P. J. Oefner, and R. W. Davis. 2002. Systematic screen for human disease genes in yeast. Nat. Genet. 31:400-404. [DOI] [PubMed] [Google Scholar]
- 31.Wei, G. X., and L. A. Bobek. 2005. Human salivary mucin MUC7 12-mer-l and 12-mer-d peptides: antifungal activity in saliva, enhancement of activity with protease inhibitor cocktail or EDTA, and cytotoxicity to human cells. Antimicrob. Agents Chemother. 49:2336-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei, G. X., A. N. Campagna, and L. A. Bobek. 2007. Factors affecting antimicrobial activity of MUC7 12-mer, a human salivary mucin-derived peptide. Ann. Clin. Microbiol. Antimicrob. 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams, R. L., and S. Urbe. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8:355-368. [DOI] [PubMed] [Google Scholar]
- 34.Wright, R., M. L. Parrish, E. Cadera, L. Larson, C. K. Matson, P. Garrett-Engele, C. Armour, P. Y. Lum, and D. D. Shoemaker. 2003. Parallel analysis of tagged deletion mutants efficiently identifies genes involved in endoplasmic reticulum biogenesis. Yeast 20:881-892. [DOI] [PubMed] [Google Scholar]
- 35.Xia, L., L. Jaafar, A. Cashikar, and H. Flores-Rozas. 2007. Identification of genes required for protection from doxorubicin by a genome-wide screen in Saccharomyces cerevisiae. Cancer Res. 67:11411-11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, W., F. J. Smith, Jr., R. Subaran, and A. P. Mitchell. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 15:5528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan, D. S., X. Pan, S. L. Ooi, B. D. Peyser, F. A. Spencer, R. A. Irizarry, and J. D. Boeke. 2005. Improved microarray methods for profiling the yeast knockout strain collection. Nucleic Acids Res. 33:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zakrzewska, A., A. Boorsma, D. Delneri, S. Brul, S. G. Oliver, and F. M. Klis. 2007. Cellular processes and pathways that protect Saccharomyces cerevisiae cells against the plasma membrane-perturbing compound chitosan. Eukaryot. Cell 6:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]