Abstract
Analysis of 15 European clinical Enterobacteriaceae isolates showed that differences in the genetic context of blaCMY-2-like genes reflected the replicon type, usually IncA/C or IncI1. These blaCMY-2 loci may originate from the same ISEcp1-mediated mobilization from the Citrobacter freundii chromosome as structures described in earlier studies.
Among plasmid-mediated AmpC enzymes, CMY-2 is the most prevalent both in France and worldwide, especially in Salmonella species (1, 17). Previous studies in the United States revealed that large plasmids carrying blaCMY-2 have been transmitted between different genera of bacteria, and notably from enteric Salmonella strains to pathogenic Escherichia coli strains (22). American studies of plasmids encoding CMY-2 using mixed-plasmid microarrays showed that the plasmids fell into two clusters (3); likewise, replicon-typing experiments revealed two replicon types, I1 and A/C (11). Similar studies in Taiwan confirmed that plasmids from diverse isolates carrying blaCMY-2 genes have common restriction patterns, evidence of interspecies spread (23). Further investigations of Taiwanese isolates allowed the identification of a specific transposon-like element responsible for the spread of blaCMY-2 among members of the family Enterobacteriaceae (6, 19). In the American and Taiwanese studies, ISEcp1 has been presumed to be involved both in the mobilization of blaCMY-2 from the Citrobacter freundii chromosome and in the expression of ampC lacking chromosomal ampR (6, 10, 12, 14, 15, 19). Poirel et al. have shown the involvement of ISEcp1B, an ISEcp1-like element, in the expression and mobilization of the β-lactamase gene blaCTX-M-19 (18).
We characterized 15 CMY-type-producing isolates, including eight E. coli isolates, four Klebsiella pneumoniae isolates, two Proteus mirabilis isolates, and one Salmonella enterica isolate. We describe the genetic organization of blaCMY-2-like genes in these European isolates and compare our findings with those from the United States and Taiwan.
Table 1 lists the 15 clinical isolates and their sources. Six isolates were previously reported (4, 7, 9, 13, 21). Total DNA was extracted by using a QIAamp DNA Mini kit (Qiagen, Courtaboeuf, France). Using repetitive-element PCR, we showed that three of the eight E. coli clinical isolates were clonally related (profile A: TN2106, TN38148, TN386) according to the interpretation criteria of van Belkum et al. (Table 1) (16, 20). Enterobacterial repetitive intergenic consensus PCR of the four K. pneumoniae isolates gave different amplification patterns (Table 1) (16).
TABLE 1.
Clinical isolates, origins, epidemiological features, and exploration by PCR mapping
| Strain | Reference | Collection yr | Country of origin (city, hospital)a | PCR profile | CMY plasmid transferb | Replicon typing | CMY type | PCRe
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | ||||||||
| S. enterica serovar Senftenberg SENF | 13 | 1994 | Algeria | ND | Tc E. coli C1a Nalr | A/C | 2 | + | + | + | − | − | + | + | + | + | − | − | − |
| K. pneumoniae 169 | 7 | 1999 | France (Paris, H2) | Wc | Tc E. coli J53 | A/C | 2 | + | + | + | − | − | + | + | + | + | − | − | − |
| K. pneumoniae BM2974 | 4 | 1998 | Sweden | Xc | Tc E. coli J53 | A/C | 4 | +del | +del | +del | − | − | + | + | + | + | − | − | − |
| K. pneumoniae LMCMY | This study | France (Colombes, H3) | Yc | Tc E. coli J53 | A/C | 2 | + | + | + | − | − | + | + | + | + | − | − | − | |
| K. pneumoniae 9701 | 7 | 1997 | France (Paris, H1) | Zc | Tc E. coli J53 | A/C | 4 | +ins | +ins | +ins | − | − | + | + | + | + | − | − | − |
| P. mirabilis H223b | 21 | 1996 | Tunisia | ND | Tc E. coli J53 | A/C | 4 | + | + | + | − | − | + | + | + | + | − | − | − |
| P. mirabilis RPCMY | This study | 2002 | France (Garches, H6) | ND | Tc E. coli J53 | Negative | 2 | − | − | + | − | − | + | + | + | + | + | + | − |
| E. coli TN2106 | This study | 2003 | France (Paris, H4) | Ad | Tc E. coli J53 | A/C | 2 | + | + | + | − | − | + | + | + | − | − | − | − |
| E. coli TN38148 | This study | 2004 | France (Paris, H4) | Ad | Tc E. coli J53 | I1 | 2 | − | − | + | + | + | + | − | − | − | − | − | + |
| E. coli TN10 | This study | 2002 | France (Paris, H4) | Bd | Tc E. coli J53 | I1 | 2 | − | − | + | + | + | + | − | − | − | − | − | + |
| E. coli TN13 | 9 | 2002 | France (Paris, H4) | Cd | Ep E. coli DH10B | I1 | 2 | − | − | + | + | + | + | − | − | − | − | − | + |
| E. coli TN386 | This study | 2002 | France (Paris, H4) | Ad | Tc E. coli J53 | I1 | 2 | − | − | + | + | + | + | − | − | − | − | − | + |
| E. coli IGR4801 | This study | 2005 | France (Villejuif, H5) | Dd | Tc E. coli C600 | Negative | 2 | − | − | + | + | + | + | − | − | − | − | − | + |
| E. coli IGR4872 | This study | 2005 | France (Villejuif, H5) | Ed | Tc E. coli C600 | Negative | 2 | − | − | + | + | + | +ins | − | − | − | − | − | +ins |
| E. coli TN44889 | This study | 2004 | France (Paris, H4) | Fd | NT | ND | 2 | − | + | + | − | − | − | − | − | − | − | − | − |
H1, Hôpital Cochin; H2, Hôpital Saint-Antoine; H3, Hôpital Louis Mourier; H4, Hôpital Tenon; H5, Institut Gustave Roussy; H6, Hôpital Raymond Poincaré.
Abbreviations: Tc, transconjugant; Ep, transformant; NT, no transfer; ND, not determined.
Enterobacterial repetitive intergenic consensus PCR profile.
Repetitive-element PCR profile.
−, negative; +, positive; +ins, positive with an insertion; +del, positive with a deletion.
Except for one strain, SENF, which was previously transferred in E. coli C1a Nalr, E. coli K-12 strain J53-2 (Rifr) or C600 (Nalr) transconjugants were obtained by mating and selection on cefoxitin (10 μg/ml) and either rifampin (rifampicin) (250 μg/ml) or nalidixic acid (50 μg/ml). For isolates with which mating was unsuccessful, E. coli strain DH10B (Invitrogen SARL, Cergy-Pontoise, France) was transformed with plasmid DNA by electroporation (Bio-Rad, Marnes la Coquette, France). Transformants were selected with cefoxitin (10 μg/ml). All of the transformants or transconjugants were successfully typed by PCR-based replicon typing, with the exception of three isolates (5). Most of the isolates (11/15) carried an A/C or an I1 replicon plasmid (Table 1).
Except for the six isolates previously published, the blaCMY gene was amplified by PCR experiments with primers ampC1 and ampC2, as previously described (7), and sequenced. Three isolates (BM2974, 9701, and H223b) carried blaCMY-4, a CMY-2 variant; all of the others carried blaCMY-2 (Table 1).
The genetic context of blaCMY was explored both by PCR mapping and cloning experiments. A 13-kb sequence surrounding blaCMY-2 from S. enterica serovar Newport was available in GenBank (accession number DQ164214); we used several PCRs (B, C, F, G, H, I, J, and K) to characterize the genetic context of blaCMY in our collection of 15 isolates (see the supplemental material). Table 1 shows several PCR profiles. PCR products B and F did not have the expected length for three isolates. Four isolates (169, TN44889, TN38148, and RPCMY) were chosen as representative of most of the PCR profiles, and cloning experiments were performed to study the genetic organization of blaCMY more extensively.
Plasmid DNA was partially digested with Sau3AI, and the fragments were ligated into the BamHI site of pACYC184. E. coli DH10B was transformed with the resulting plasmids, and transformants were selected with cefoxitin (10 μg/ml) and chloramphenicol (50 μg/ml). The largest plasmids with a blaCMY insert were selected, and both strands were sequenced. PCR primers were designed (PCRs A, D, E, and L), and PCR experiments were performed with all of the isolates (Table 1; see the supplemental material). The sequenced regions and DNA sequence analyses are shown in Fig. 1.
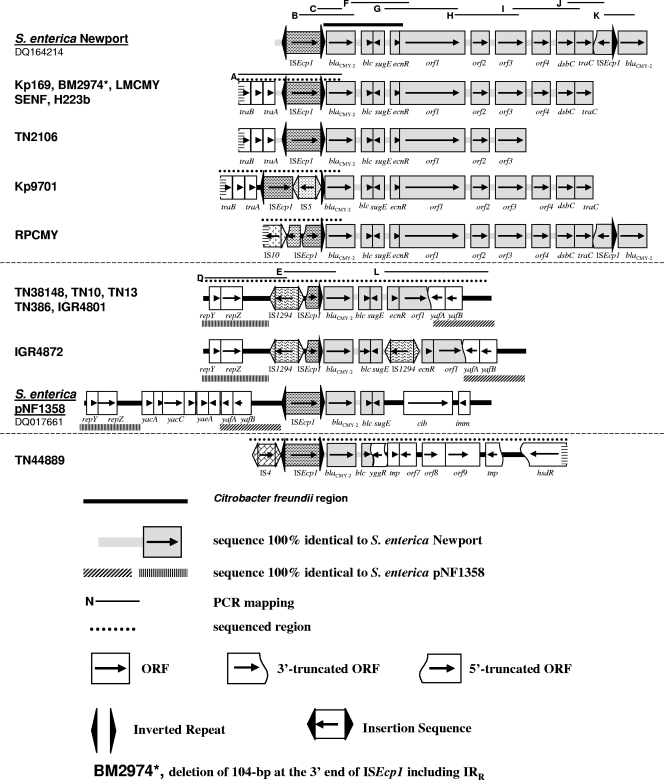
FIG. 1.
Comparison of regions surrounding blaCMY-2. IRR, imperfect right inverted repeat.
Previous American and Taiwanese studies suggested that ISEcp1 is involved in mobilization of the blaCMY-2-like gene from the C. freundii chromosome, with ISEcp1-IRR consistently 117 bp upstream from blaCMY-2 (6, 10, 12). By comparing the sequence of S. enterica serovar Newport DQ164214 with that of C. freundii chromosome U21727, a 2,823-bp region including a blaCMY-2-like gene, blc, sugE, and ΔecnR could be delimited. Interestingly, the last 14 bp of this region, CCACACAATTCAGG, could have been recognized as an imperfect right inverted repeat-like sequence by the transposase of ISEcp1 during the mobilization process. In the 15 isolates in this study, at least a part of ISEcp1 was present upstream from the blaCMY-2-like gene. Nevertheless, the ISEcp1-blaCMY-2-like gene region was in the same configuration in six isolates only: 169, LMCMY, SENF, H223b, TN2106, and TN44889. For the other nine isolates, ISEcp1 was present with various deletions/insertions which do not separate the blaCMY-2-like gene from its putative promoter in ISEcp1 (Fig. 1). These modifications are likely to have occurred after ISEcp1 had mobilized the blaCMY-2-like gene from the C. freundii chromosome to a plasmid.
The plasmids from most of the isolates shared various structural similarities with the 13-kb type I structure described in S. enterica serovar Newport (accession number DQ164214), where the blaCMY-2 gene is duplicated (12); the second copy follows a partial copy of ISEcp1 in the opposite orientation (Fig. 1).
Downstream from the blaCMY-2-like gene in this 13-kb type I structure, several open reading frames (ORFs) were described between the two copies of blaCMY-2, from blc to traC (Fig. 1). In 8 of our 15 isolates, RPCMY, 169, BM2974, LMCMY, SENF, H223b, 9701, and TN2106, the genetic organization downstream from the blaCMY-2-like gene was very similar to this 13-kb type I structure. Only RPCMY had two copies of the blaCMY-2-like gene. For the other isolates, the sequence remains unknown either downstream from traC or downstream from orf3 (Fig. 1).
Upstream from the blaCMY-2-like gene, ISEcp1 was present with deletions/insertions in its sequence for three isolates. (i) In RPCMY, there is a deletion of the first 1,140 bp due to the insertion of a separate copy of ISEcp1 in the reverse orientation; this second copy has a 1,296-bp deletion due to the insertion of IS10. Interestingly, IS10 has previously been identified as being responsible for the disruption of ISEcp1 upstream from blaCTX-M-14 in E. coli TN13, but in a different position (8). (ii) In BM2974, there is a deletion of a 104 bp-segment at the 3′ end of ISEcp1. (iii) In Kp9701, there is an insertion of IS5 at position 1509 in ISEcp1 (Fig. 1).
In 169, BM274, LMCMY, SENF, H223b, TN2106, and 9701, an additional 1,560-bp segment was characterized upstream from ISEcp1. It is 100% identical to the region comprising the traB, traV, and traA genes in S. enterica serovar Newport SL254 (accession no. CP000604).
Of the 15 isolates, 8 had a genetic organization around the blaCMY-2-like gene highly similar to that in the 13-kb type I structure and, except for RPCMY, carried an A/C replicon plasmid.
A second group included six isolates less similar to the 13-kb type I structure (TN38148, TN10, TN13, TN386, IGR4801, and IGR4872): the blaCMY-2-like gene was followed by blc, sugE, ecnR, and orf1 only. Indeed, orf1 had 1,078 bp deleted at the 3′ end and was followed by yafA, which had a 3′ deletion and was in the reverse orientation. The truncated yafA gene was followed by yafB, and both genes belonged to a 1,983-bp segment which is 98% identical to a region of S. enterica plasmid pNF1358 (accession number no. DQ017661) (Fig. 1). This segment in pNF1358 is located directly upstream from ISEcp1-blaCMY-2.
In all six isolates of this second group, a deletion of the first 1,281 bp in the 5′ part of ISEcp1 was due to the insertion of IS1294 in the reverse orientation (Fig. 1). IS1294 belongs to the IS91 family and is a putative transposable element capable of mediating one-ended transposition (www-is.biotoul.fr). Downstream from IS1294, a 2,265-bp segment comprising the repY and repZ genes involved in replication initiation is 95% identical to a region of S. enterica pNF1358 (Fig. 1). In IGR4872, an additional IS1294 was inserted between sugE and ecnR, leading to the duplication of the GTTC target site.
Four isolates of this second group carried an IncI1 plasmid; two isolates from the same hospital could not be replicon typed.
In the 15th isolate, TN44889, the genetic organization of the ISEcp1-blaCMY-2-like gene region was unique with the presence of IS4 upstream from a complete copy of ISEcp1 (Fig. 1). Downstream from the blaCMY-2-like gene, 467 bp of blc was deleted by the insertion of a truncated ORF, yggR. That is followed by a 4,591-bp region including five ORFs, which is 95% identical to the Shigella flexneri 2a SRL pathogenicity island. This system is linked to a cluster of multiple antibiotic resistance determinants (accession number no. AF326777).
Our findings are consistent with the study of Hopkins et al. (11); there is a predominance of A/C and I1 replicons among plasmids carrying blaCMY-2-like genes in 15 nonrepetitive Enterobacteriaceae isolates. Each replicon type reflected the genetic organization surrounding the blaCMY-2-like gene, with several minor rearrangements, including small insertions or deletions. Using phylogenetic methods, Barlow and Hall demonstrated that all CMY-2-type β-lactamase variants had a common ancestor which was plasmid borne (2). This suggests that a region from the C. freundii chromosome (ampC, blc, sugE, ecnR) has been mobilized only once. This study is consistent with results of Barlow and Hall, as ISEcp1 seems to have been involved in the mobilization of the blaCMY-2-like gene, irrespective of the replicon type and the plasmid backbone.
Nucleotide sequence accession numbers.
The nucleotide sequences in strains 169, 9701, RPCMY, TN38148, and TN44889 have been submitted to GenBank and have been assigned accession numbers FM246880, FM246881, FM246882, FM246883, and FM246884, respectively.
Supplementary Material
Acknowledgments
This work was supported by grants from the Faculté de Médecine Saint-Antoine, Université Pierre et Marie Curie Paris VI, and from the European Community (6th PCRD contract LSHM-CT 2003-503335).
Footnotes
Published ahead of print on 13 July 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Arlet, G., T. J. Barrett, P. Butaye, A. Cloeckaert, M. R. Mulvey, and D. G. White. 2006. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8:1945-1954. [DOI] [PubMed] [Google Scholar]
- 2.Barlow, M., and B. G. Hall. 2002. Origin and evolution of the AmpC β-lactamases of Citrobacter freundii. Antimicrob. Agents Chemother. 46:1190-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Call, D. R., M. S. Kang, J. Daniels, and T. E. Besser. 2006. Assessing genetic diversity in plasmids from Escherichia coli and Salmonella enterica using a mixed-plasmid microarray. J. Appl. Microbiol. 100:15-28. [DOI] [PubMed] [Google Scholar]
- 4.Cao, V. T., G. Arlet, B. M. Ericsson, A. Tammelin, P. Courvalin, and T. Lambert. 2000. Emergence of imipenem resistance in Klebsiella pneumoniae owing to combination of plasmid-mediated CMY-4 and permeability alteration. J. Antimicrob. Chemother. 46:895-900. [DOI] [PubMed] [Google Scholar]
- 5.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219-228. [DOI] [PubMed] [Google Scholar]
- 6.Chiu, C. H., L. H. Su, C. Chu, J. H. Chia, T. L. Wu, T. Y. Lin, Y. S. Lee, and J. T. Ou. 2004. Isolation of Salmonella enterica serotype choleraesuis resistant to ceftriaxone and ciprofloxacin. Lancet 363:1285-1286. [DOI] [PubMed] [Google Scholar]
- 7.Decré, D., C. Verdet, L. Raskine, H. Blanchard, B. Burghoffer, A. Philippon, M. J. Sanson-Le-Pors, J. C. Petit, and G. Arlet. 2002. Characterization of CMY-type β-lactamases in clinical strains of Proteus mirabilis and Klebsiella pneumoniae isolated in four hospitals in the Paris area. J. Antimicrob. Chemother. 50:681-688. [DOI] [PubMed] [Google Scholar]
- 8.Eckert, C., V. Gautier, and G. Arlet. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14-23. [DOI] [PubMed] [Google Scholar]
- 9.Eckert, C., V. Gautier, M. Saladin-Allard, N. Hidri, C. Verdet, Z. Ould-Hocine, G. Barnaud, F. Delisle, A. Rossier, T. Lambert, A. Philippon, and G. Arlet. 2004. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 48:1249-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giles, W. P., A. K. Benson, M. E. Olson, R. W. Hutkins, J. M. Whichard, P. L. Winokur, and P. D. Fey. 2004. DNA sequence analysis of regions surrounding blaCMY-2 from multiple Salmonella plasmid backbones. Antimicrob. Agents Chemother. 48:2845-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopkins, K. L., E. Liebana, L. Villa, M. Batchelor, E. J. Threlfall, and A. Carattoli. 2006. Replicon typing of plasmids carrying CTX-M or CMY β-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob. Agents Chemother. 50:3203-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang, M. S., T. E. Besser, and D. R. Call. 2006. Variability in the region downstream of the blaCMY-2 β-lactamase gene in Escherichia coli and Salmonella enterica plasmids. Antimicrob. Agents Chemother. 50:1590-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koeck, J. L., G. Arlet, A. Philippon, S. Basmaciogullari, H. V. Thien, Y. Buisson, and J. D. Cavallo. 1997. A plasmid-mediated CMY-2 β-lactamase from an Algerian clinical isolate of Salmonella Senftenberg. FEMS Microbiol. Lett. 152:255-260. [DOI] [PubMed] [Google Scholar]
- 14.Li, W. C., F. Y. Huang, C. P. Liu, L. C. Weng, N. Y. Wang, N. C. Chiu, and C. S. Chiang. 2005. Ceftriaxone resistance of nontyphoidal Salmonella enterica isolates in Northern Taiwan attributable to production of CTX-M-14 and CMY-2 β-lactamases. J. Clin. Microbiol. 43:3237-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindberg, F., L. Westman, and S. Normark. 1985. Regulatory components in Citrobacter freundii ampC β-lactamase induction. Proc. Natl. Acad. Sci. USA 82:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel, L., J.-W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su, L.-H., H.-L. Chen, J.-H. Chia, S.-Y. Liu, C. Chu, T.-L. Wu, and C.-H. Chiu. 2006. Distribution of a transposon-like element carrying blaCMY-2 among Salmonella and other Enterobacteriaceae. J. Antimicrob. Chemother. 57:424-429. [DOI] [PubMed] [Google Scholar]
- 20.van Belkum, A., P. T. Tassios, L. Dijkshoorn, S. Haeggman, B. Cookson, N. K. Fry, V. Fussing, J. Green, E. Feil, P. Gerner-Smidt, S. Brisse, and M. Struelens for the European Society of Clinical Microbiology and Infectious Diseases Study Group on Epidemiological Markers. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. Dis. 13(Suppl. 3):1-46. [DOI] [PubMed] [Google Scholar]
- 21.Verdet, C., G. Arlet, S. Ben Redjeb, A. Ben Hassen, P. H. Lagrange, and A. Philippon. 1998. Characterisation of CMY-4, an AmpC-type plasmid-mediated β-lactamase in a Tunisian clinical isolate of Proteus mirabilis. FEMS Microbiol. Lett. 169:235-240. [DOI] [PubMed] [Google Scholar]
- 22.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan, J. J., W. C. Ko, C. H. Chiu, S. H. Tsai, H. M. Wu, and J. J. Wu. 2003. Emergence of ceftriaxone-resistant Salmonella isolates and rapid spread of plasmid-encoded CMY-2-like cephalosporinase, Taiwan. Emerg. Infect. Dis. 9:323-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.