Abstract
The prevalence of heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA) among 1,012 vancomycin-susceptible methicillin (meticillin)-resistant S. aureus isolates collected from 14 cities in China from 2005 to 2007 was 13 to 16%, as determined by a combination of (i) measurement by the modified population analysis profile-area under the curve method (PAP-AUC) and (ii) estimation from the measured sensitivity and specificity of a screening method. Two hundred isolates from blood were chosen as a subset for measurement of the sensitivities and the specificities of several previously described screening methods by using the results of PAP-AUC as the reference. During this testing, one isolate was found to be a vancomycin-intermediate S. aureus (VISA) strain so was not used in the evaluation of the screening tests. Of the other 199 isolates, 26 (13.1%) were hVISA, as assessed by PAP-AUC. A screening cascade of culturing the isolates on brain heart infusion agar containing teicoplanin (5 mg/liter) and then subjecting the positive isolates to a macro-Etest method was applied to the 812 non-blood isolates, yielding 149 positive results. From these results and by adjusting for sensitivity (0.423) and specificity (0.861), the prevalence was estimated to be 15.7%. The precision of that estimate was assessed by reapplying the screening cascade to 120 randomly selected isolates from the 812 non-blood isolates and simultaneously determining their heterogeneous vancomycin-intermediate susceptibility status by PAP-AUC. Because PAP-AUC is impractical for use with large numbers of isolates, the screening-based estimation method is useful as a first approximation of the prevalence of hVISA. Of the 27 VISA or hVISA isolates from blood, 22.2% and 74.1% were staphylococcal chromosome cassette mec types II and III, respectively, while 77.8% and 22.2% were agr type 1 and agr type 2, respectively; the MIC ranges were 0.5 to 4 mg/liter for vancomycin and 0.25 to 1 mg/liter for daptomycin.
Since their first report in Japan in 1997 (11), heterogeneous vancomycin-intermediate (heteroresistant) Staphylococcus aureus (hVISA) strains have been a new concern with respect to methicillin (meticillin)-resistant S. aureus (MRSA) strains that display reduced susceptibility to glycopeptides (12, 13), because such strains are potentially associated with the clinical failure of vancomycin treatment (4). Vancomycin heteroresistance among S. aureus isolates was one of the reasons that Tenover and Moellering cited for the Clinical and Laboratory Standards Institute decision in 2006 to lower the vancomycin MIC breakpoints for S. aureus from ≤4 mg/liter to ≤2 mg/liter for susceptible, from 8 to 16 mg/liter to 4 to 8 mg/liter for intermediate, and from ≥32 mg/liter to ≥16 mg/liter for resistant (24).
These new MIC breakpoints were not sensitive enough to distinguish hVISA strains from vancomycin-susceptible S. aureus strains (31). Currently, a particular isolate of S. aureus is defined as hVISA if the large majority of cells of a population cannot grow in the presence of vancomycin concentrations in excess of 2 mg/liter but if some proportions of cells can form colonies, e.g., at frequencies as low as 10−6, in the presence of higher concentrations (13, 30). Many traditional methods have proved unreliable at detecting hVISA (30). Agar screening methods with different media, inocula, and glycopeptide concentrations have been investigated; but their sensitivities and specificities have varied considerably, making them also unreliable for hVISA detection (8, 30). The macro-Etest method (MET) has shown high degrees of sensitivity and specificity (27), but its use for routine clinical work has been hampered by its high cost. In 2001, a modified population analysis profile (PAP)-area under the curve (AUC) method (PAP-AUC) was proposed as a reliable, albeit labor-intensive, test for the identification of hVISA (29).
With the development of the various detection methods, the prevalence of hVISA has been reported worldwide, e.g., in France (0.7%); Australia (9.4%); the United States (0.3 to 2.3%); and several Asian countries, including Japan (1.3% to 20%), India (6.3%), South Korea (6.1%), and Singapore (2.3%) (3, 4, 12, 21a, 23). However, little information from China has been available. In a study conducted in 2004, a total of 84 MRSA isolates obtained from 1997 to 2000 from China were investigated, but none of them was identified as hVISA (23). Given the high prevalence of MRSA strains (32) and the increasing use of glycopeptides in China, the emergence of hVISA, vancomycin-intermediate S. aureus (VISA), and vancomycin-resistant S. aureus strains in this region might be anticipated. Therefore, we have conducted a survey using the PAP-AUC method with more MRSA isolates and isolates from more study centers to investigate the epidemiology of hVISA in mainland China. As part of the study, we evaluated several agar screening methods and MET for their potential wider use in more clinical microbiology laboratories in China.
The 26 hVISA isolates and the 1 VISA isolate identified were further characterized by antibiotic susceptibility testing and molecular typing. These data provide baseline information on the epidemiology of hVISA isolated from patients in mainland China.
MATERIALS AND METHODS
Bacterial isolates and reagents.
A total of 1,012 isolates of MRSA were collected from six groups in China from 2002 to 2007 (Table 1): (i) MRSA isolates from Gram Positive Cocci Resistance Surveillance (GPRS) and Surveillance by Etest and Agar Dilution of Nationwide Isolate Resistance (SEANIR) programs in 2005 (n = 272), (ii) MRSA isolates from the GPRS program in 2006 (n = 123), (iii) MRSA isolates from the SEANIR program in 2006 (n = 189), (iv) MRSA isolates from blood specimens collected at the Peking Union Medical College Hospitals (PUMCH) from 2002 to 2007 (n = 76), (v) MRSA isolates from the Chinese Antimicrobial Resistance Surveillance (CARES) program in 2007 (n = 168), and (vi) MRSA isolates from the GPRS program in 2007 (n = 184). The GPRS, SEANIR, and CARES programs are different national surveillance networks organized by PUMCH, which includes different hospitals in China. Each year the participant hospitals sent the defined number of clinical isolates to PUMCH according to the respective protocols. All MRSA isolates from blood infections (n = 200) from the six groups described above were defined as a subgroup (Table 1). Brain heart infusion (BHI) medium was from Oxoid, Ltd., Basingstoke, United Kingdom; teicoplanin was from Sanofi-Aventis, Romainville, France; vancomycin was from Eli Lilly & Company Limited, Indianapolis, IN.
TABLE 1.
Origins of MRSA isolates used in the present study
| Group | Yr of isolation | Program | No. of hospitals | No. of MRSA isolates from:
|
Region in China (no. of isolates) | |
|---|---|---|---|---|---|---|
| All specimens | Blood | |||||
| 1 | 2005 | GPRS-SEANIR | 16 | 272 | 26 | Northeast (54), southeast (35), south (35), north (50), northwest (40), southwest (30), central (28) |
| 2 | 2006 | GPRS | 7 | 123 | 19 | Northeast (16), southeast (32), south (26), north (17), northwest (21), southwest (0), central (11) |
| 3 | 2006 | SEANIR | 14 | 189 | 21 | Northeast (41), southeast (33), south (33), north (23), northwest (19), southwest (6), central (34) |
| 4 | 2002-2007 | Blood specimens in PUMCH | 1 | 76 | 76 | North (76) |
| 5 | 2007 | CARES | 14 | 168 | 41 | Northeast (15), southeast (20), south (36), north (47), northwest (16), southwest (6), central (28) |
| 6 | 2007 | GPRS | 10 | 184 | 17 | Northeast (23), southeast (36), south (39), north (38), northwest (25), southwest (12), central (11) |
| Total | 1,012 | 200 | ||||
Study design.
In order to estimate the sensitivities and the specificities of the several screening methods, described in more detail below, we used them to screen the subgroup of 200 blood isolates of MRSA for which we had determined a definitive result by the reference PAP-AUC method (29). While we were conducting the study, repeat susceptibility testing showed that the MIC of vancomycin for one of these isolates was 4 mg/liter, thus classifying it as a VISA strain. Data for this strain were excluded from the calculations of the sensitivities and the specificities of the screening methods because those screening methods would normally be applied to isolates that display vancomycin MICs of ≤2 mg/liter. Hence, the denominator for the calibration of the screening methods against the reference PAP-AUC method was 199. There were two agar screening methods: one with BHI medium containing teicoplanin at 5 mg/liter (BHIT5) and one with BHI medium containing vancomycin at 6 mg/liter (BHIV6). A third screening method was MET (8). Note also that the result of MET allowed different definitions of positivity for hVISA, depending on the values of the Etest readings defined as thresholds; these are also explained in detail below. The larger subgroup of MRSA isolates, i.e., those from non-blood specimens (n = 812), was then screened by a cascade of BHIT5 followed by MET. From the sensitivity and the specificity of that overall cascade, quantified as described below, the prevalence of hVISA among the non-blood isolates of MRSA in China was estimated. The precision of the estimation method was then assessed by rescreening a sample of 120 (randomly selected by using SAS software) of the 812 non-blood isolates while also subjecting them to the reference PAP-AUC method.
Agar screening.
Agar screening was performed as described previously (8). Briefly, all isolates were screened on BHIT5 and BHIV6 by using 10-μl volumes of bacterial suspensions with densities equivalent to a 0.5 McFarland turbidity standard. Twelve isolates were inoculated onto each plate, and the screening tests were performed in duplicate. The plates were incubated for 48 h at 35°C. A strain was considered positive if one or more colonies had grown after 48 h. S. aureus Mu3 (an hVISA strain), Mu50 (a VISA strain), and ATCC 29213 (a vancomycin-susceptible S. aureus [VSSA] strain) were used as control strains.
MET.
Screening by MET with heavy inocula was performed as described by Fitzgibbon et al. (8). Briefly, a 200-μl volume of bacterial suspension with a density equivalent to a 2.0 MaFarland turbidity standard was pipetted onto a BHI agar plate and spread evenly with a swab, after which the plate was allowed to dry at room temperature. Vancomycin and teicoplanin Etest strips (AB Biodisk, Ltd., Solna, Sweden) were applied and the plates were incubated for 48 h at 35°C. Two different outcomes, designated MET A and MET B, were established by defining two different criteria of positivity (i.e., two different cutoff values). For MET A, a strain was considered positive if the readings were ≥8 mg/liter for both vancomycin and teicoplanin or ≥12 mg/liter for teicoplanin. For MET B, a strain was considered positive if the readings were simply ≥8 mg/liter for both vancomycin and teicoplanin.
PAP-AUC.
PAP-AUC was determined as described in detail elsewhere (29). Colonies grown overnight on Columbia agar containing 5% sheep blood were suspended in saline to a density equivalent to a 0.5 McFarland turbidity standard. A 2-ml sample of this bacterial suspension was added to 8 ml tryptic soy broth (Oxoid), mixed well, and incubated for 24 h at 35°C. Dilutions of 10−3 (105 CFU/ml) and 10−6 (102 CFU/ml) were then prepared in saline. Fifty-microliter volumes were inoculated onto BHI agar plates containing 0, 0.5, 1.0, 2.0, 2.5, 4.0, and 8.0 mg/liter of vancomycin by using a spiral plater (Don Whitley Scientific Limited, West Yorkshire, United Kingdom). After 48 h incubation at 35°C, the colonies were counted and the log of the number of CFU/ml was plotted against the vancomycin concentration by using Prism software (GraphPad Software, Inc., San Diego, CA). The AUC (in units of mg/liter) was measured from the graph by the construction of trapezoids. The ratio of the AUC of the test isolate to the AUC of S. aureus Mu3 was calculated and interpreted as follows: for VSSA, a ratio of <0.9; for hVISA, a ratio of 0.9 to 1.3; and for VISA, a ratio of ≥1.3. S. aureus ATCC 29213 was used as the reference VSSA strain.
Sensitivity, specificity, and the estimation of prevalence.
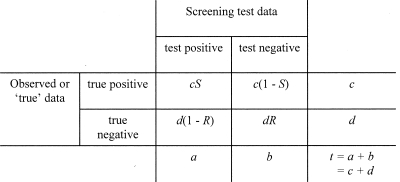
Figure 1 shows the 2 × 2 matrix used for analyzing the performance of the screening tests. It was also used to derive the expression shown below for estimating the prevalence of hVISA from the results of a screening test when the reference, PAP-AUC, result was unknown. For a given sample of isolates, the sensitivity (S in Fig. 1) was calculated as the number of hVISA isolates that were positive by the screening test divided by the number of hVISA isolates demonstrated to be present by PAP-AUC. That is, it was a quantitative measurement of the ability of the screening test to detect hVISA isolates in a sample of vancomycin-susceptible MRSA strains (9a). For the same sample of isolates, the specificity (R in Fig. 1) was calculated as the number of isolates correctly identified as negative for hVISA by the screening test divided by the number of isolates that were non-hVISA by the reference method, PAP-AUC. It was thus a quantitative expression of the ability of a screening test to screen out hVISA isolates and correctly identify non-hVISA isolates in a sample of vancomycin-susceptible MRSA isolates (9a) (Fig. 1). Sensitivity and specificity were determined using the same sample of isolates. The prevalence of hVISA among a collection of vancomycin-susceptible MRSA isolates [i.e., c/(c + d) using the symbols of Fig. 1] could be estimated from the results of a screening test of known sensitivity and specificity using the following relationship:
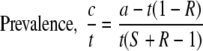 |
(1) |
The derivation of equation 1 is described in full in the supplementary material.
FIG. 1.
A 2 × 2 presentation of screening test results mapped against “true” (or reference) data for the purposes of analysis. a and b are the measured values; c and d are to be estimated.
Susceptibility testing.
The MICs of oxacillin (Sigma Chemical Co., St. Louis, MO), rifampin (rifampicin; Sigma), gentamicin (Sigma), levofloxacin (Daiichi Pharmaceutical Tokyo, Japan), erythromycin (Sigma), vancomycin (Eli Lilly), teicoplanin (Sanofi-Aventis), and tigecycline (Wyeth Pharmaceuticals, Collegeville, PA) were determined by agar dilution. Ceftobiprole and linezolid were supplied by Johnson & Johnson and Pfizer, respectively. The MICs of ceftobiprole and linezolid were determined by agar dilution. The MICs of daptomycin (AstraZeneca) were determined by broth microdilution in BBL broth (BBL, Becton Dickinson and Company) supplemented with calcium chloride to bring the final calcium concentration to 50 mg/liter, which was confirmed by biochemical analysis (6).
DNA extraction.
DNA was extracted as described by Unal et al. (26). Briefly, isolates cultured for 24 h on Columbia agar with 5% sheep blood were successively incubated with lysostaphin and proteinase K, boiled, and centrifuged. The lysate was used as a DNA template in all PCRs described below.
SCCmec typing.
The staphylococcal chromosome cassette mec (SCCmec) types were determined by a multiplex PCR strategy, as described previously (33). PCRs were carried out with an initial denaturation at 95°C for 45 s, 58°C for 45 s, and 72°C for 45 s. The reaction mixture was maintained at 72°C for a further 7 min. Nontypeable strains were defined as those showing unexpected fragments or lacking some fragments in the multiplex PCR. International clones of SCCmec types I to V were used as quality control strains.
agr typing.
agr typing was performed as previously described by Gilot et al. (10). Briefly, five primers were used to amplify the target region: Pan (5′-ATGCACATGGTGCACATGC-3′), agr 1 (5′-GTCACAAGTACTATAAGCTGCGAT-3′), agr 2 (5′-TATTACTAATTGAAAAGTGGCCATAGC-3′), agr 3 (5′-GTAATGTAATAGCTTGTATAATAATACCCAG-3′), and agr 4 (5′-CGATAATGCCGTAATACCCG-3′). The following temperature program was used: 1 cycle of 5 min at 94°C, followed by 26 cycles of 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C and, finally, 1 cycle of 10 min at 72°C.
spa typing.
spa typing was performed as described previously (17). Purified spa PCR products were sequenced, and short sequence repeats were assigned by using the spa database website (http://www.ridom.de/spaserver). The spa complex was defined by visual analysis, whereby spa types with similar short sequence repeats were clustered into the complexes previously described by Ruppitsch et al. (21).
MLST and data analysis.
Multilocus sequence typing (MLST) was performed as described previously (7). PCR fragments were purified and sequenced with an ABI 3700 sequencer. The sequences of the PCR products were compared with the existing sequences available in the MLST website for S. aureus (http://saureus.mlst.net), and the allelic number was determined for each sequence.
RESULTS
Prevalence of hVISA and VISA in blood specimens.
Among the 200 isolates of MRSA from blood specimens, PAP-AUC identified 173 isolates as VSSA, 26 isolates as hVISA, and 1 isolate as VISA. Thus, the prevalence rates of hVISA and VISA among MRSA isolates from blood specimens were 13.1% (26/199) and 0.5% (1/200), respectively. Although the numbers were small (and excluding the VISA isolate, as described above), the prevalence rates of hVISA by year were 17.9% from 2002 to 2004, 11.1% in 2005, 12.3% in 2006, and 12.8% in 2007.
Calibration of screening methods for application to higher numbers of strains.
The results of screening by BHIT5, BHIV6, and MET with the cutoff values of MET A and MET B are summarized in Table 2. The sensitivity and the specificity of BHIT5 were 88.5% and 17.3%, respectively; but those of BHIV6 were 3.8% and 98.8%, respectively. The sensitivity and the specificity of MET A were 69.2% and 48.0%, respectively. For MET B, the sensitivity was only 46.1% but the specificity was improved to 85.0%. The composite sensitivity and specificity of the cascade of BHIT5 followed by MET B were 42.3% (11/26) and 86.1% (149/173), respectively.
TABLE 2.
Sensitivities and specificities of BHIT5, BHIV6, MET, and cascade screening methods, based on screening of 199 blood isolates of MRSA of known vancomycin phenotype
| Method and result | No. of isolates with:
|
Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|
| PAP-AUC ratio ≥ 0.9 (n = 26) | PAP-AUC ratio < 0.9 (n = 173) | |||
| BHIT5 | 88.5 | 17.3 | ||
| Positive | 23 | 143 | ||
| Negative | 3 | 30 | ||
| BHIV6 | 3.8 | 98.8 | ||
| Positive | 1 | 2 | ||
| Negative | 25 | 171 | ||
| MET A | 69.2 | 48.0 | ||
| Positive | 18 | 90 | ||
| Negative | 8 | 83 | ||
| MET B | 46.1 | 85.0 | ||
| Positive | 12 | 26 | ||
| Negative | 14 | 147 | ||
| Cascadea | 42.3 | 86.1 | ||
| Positive | 11 | 24 | ||
| Negative | 15 | 149 | ||
Cascade, a composite screening method whereby the isolates positive by BHIT5 were then screened by MET B.
As pointed out by Galen and Gambino (9a), the positive predictive value of a screening test varies as the prevalence in the tested sample varies, even when the sensitivity and the specificity remain constant. Thus, the positive predictive value cannot be used to estimate prevalence (9a). However, if the sensitivity and the specificity of a screening test are known, i.e., the screening test has been calibrated against a reference method, then the screening test can be used to estimate the prevalence in a sample of uncharacterized isolates. The method of estimation is provided above. When the reference method is too cumbersome for application to large numbers of isolates, as is the case with PAP-AUC, making an estimate in this way allows a sample of isolates larger than the number that could be analyzed by the reference method itself to be analyzed. Thus, in the present study, the total collection of MRSA was 1,012 isolates, giving the opportunity to broaden the applicability of the estimate by screening the rest of the isolates by the optimal and calibrated screening test. For these reasons, we used the results of the cascade screening test for the remaining 812 isolates to estimate the prevalence of hVISA in that larger sample (Table 3). By the cascade screening method, 149 isolates tested positive for hVISA. By using that result and equation 1, with a sensitivity and a specificity of 42.3% and 86.1%, respectively, as described above, the estimated number of hVISA isolates among the remaining 812 MRSA isolates was 127.8 and the prevalence was 15.7%. As an independent check that the screening method was applicable to such samples of MRSA of unknown hVISA status, a random sample of 120 isolates from the 812 isolates described above was retested by the cascade screening method and by the reference method, PAP-AUC. The prevalence was estimated from the screening results and compared with the reference data (Table 3). Of those 120 isolates, 25 were screening test positive. The estimation method described above yielded a prevalence of hVISA of 24.5%, which was high compared with both the earlier estimate made by use of the 812 isolates and the prevalence measured among the 199 blood isolates. Indeed, the actual prevalence of hVISA in this random sample of 120 was 14.2% (17 isolates), as determined by PAP-AUC, the reference method. Finally, among the remaining 692 isolates, the number of isolates with positive results by the screening test was 127, which yielded an estimated prevalence using equation 1 of 15.7% (Table 3).
TABLE 3.
Measurement and estimate of prevalence of hVISA within various groups of clinically isolated MRSA strains
| Group | No. of isolates | No. of isolates positive by BHIT5 and MET B | Prevalence (%)
|
|
|---|---|---|---|---|
| Estimated by BHIT5- MET B | Measured by PAP-AUC analysis | |||
| Calibration seta | 199 | 11 | —e | 13.1 |
| All other isolates than the calibration setb | 812 | 149 | 15.7 | — |
| Test setc | 120 | 25 | 24.5 | 14.2 |
| Remaining isolatesd | 692 | 127 | 15.7 | — |
The blood isolates used to calibrate the screening methods.
The MRSA isolates remaining after removal of the blood isolates.
A random sample of 120 isolates from the 812 isolates in the non-calibration set; the random sample of isolates was used to assess the variability of the estimates of prevalence from the sensitivity and specificity measured by the BHIT5-MET B screening cascade.
The MRSA isolates remaining after removal of the randomly selected test set of 120 isolates from the noncalibration set of 812 isolates.
—, not applicable or not measured.
Antibiotic susceptibilities of the different MRSA phenotypes.
The MICs of 12 antimicrobial agents were determined for all 200 isolates in the blood isolate subset, as shown in Table 4. All isolates were susceptible to teicoplanin, daptomycin, tigecycline, ceftobiprole, and linezolid. The MIC range of daptomycin for isolates in both the hVISA/VISA group and the VSSA group was 0.25 to 1 mg/liter. The percentage of isolates in the hVISA group with a vancomycin MIC of ≥1.0 mg/liter was significantly higher than that in the VSSA group (96.3% and 64.2%, respectively; P = 0.001).
TABLE 4.
Activities of vancomycin, teicoplanin, daptomycin, tigecycline, ceftobiprole, and linezolid against hVISA/VISA and VSSA strains
| Group | Antimicrobial agentb | Susceptibility categorizationc
|
Cumulative % of isolates inhibited at MIC (mg/liter) of:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % R | % I | % S | MIC50 (mg/liter) | MIC90 (mg/liter) | MIC range (mg/liter) | 0.125 | 0.25 | 0.5 | 1.0 | 2.0 | 4.0 | 16 | ||
| hVISA/VISAa (n = 27) | VAN | 0 | 3.7 | 96.3 | 1 | 2 | 0.5-4 | 0.0 | 0.0 | 3.70 | 85.2 | 96.3 | 100 | 100 |
| TEC | 0 | 0 | 100 | 2 | 8 | 0.5-8 | 0.0 | 0.0 | 3.70 | 25.9 | 66.7 | 85.2 | 100 | |
| DAP | 0 | 0 | 100 | 0.5 | 1 | 0. 25-1 | 0.0 | 14.8 | 88.9 | 100 | 100 | 100 | 100 | |
| TGC | 0 | 0 | 100 | 0.25 | 0.25 | 0.032-0.5 | 44.4 | 92.6 | 100 | 100 | 100 | 100 | 100 | |
| BPR | 0 | 0 | 100 | 2 | 2 | 1-4 | 0.0 | 0.0 | 11.1 | 33.3 | 96.3 | 100 | 100 | |
| LZD | 0 | 0 | 100 | 1 | 2 | 0.5-2.0 | 0.0 | 0.0 | 33.3 | 85.2 | 100 | 100 | 100 | |
| VSSA (n = 173) | VAN | 0 | 0 | 100 | 1 | 1 | 0.5-2 | 0.0 | 0.0 | 35.8 | 99.4 | 100 | 100 | 100 |
| TEC | 0 | 0 | 100 | 2 | 4 | 0.125-4 | 0.58 | 2.89 | 12.1 | 49.1 | 79.8 | 100 | 100 | |
| DAP | 0 | 0 | 100 | 0.5 | 0.5 | 0.125-1 | 0.58 | 10.7 | 95.1 | 100 | 100 | 100 | 100 | |
| TGC | 0 | 0 | 100 | 0.125 | 0.25 | 0.032-0.5 | 54.3 | 90.2 | 100 | 100 | 100 | 100 | 100 | |
| BPR | 0 | 0 | 100 | 2 | 2 | 0.25-2 | 0.0 | 2.31 | 4.62 | 27.2 | 100 | 100 | 100 | |
| LZD | 0 | 0 | 100 | 1 | 2 | 0.125-2 | 0.58 | 1.16 | 31.2 | 82.7 | 100 | 100 | 100 | |
The MICs for the VISA strain were as follows: vancomycin, 4 mg/liter; teicoplanin, 8 mg/liter; daptomycin, 0.5 mg/liter; tigecycline, 0.25 mg/liter; ceftobiprole, 0.5 mg/liter; and linezolid, 1 mg/liter.
VAN, vancomycin; TEC, teicoplanin; DAP, daptomycin; TGC, tigecycline; BPR, ceftobiprole; LZD, linezolid.
R, resistant; I, intermediate; S, susceptible.
Comparison of SCCmec and agr types between hVISA/VISA and VSSA isolates.
The 200 isolates from blood infections were characterized by SCCmec typing and agr typing (Table 5). The predominant types among hVISA were SCCmec type III (74.1%) and agr type 1 (77.8%), followed by SCCmec type II (22.2%) and agr type 2 (22.2%). Similarly, the predominant types in the VSSA group were also SCCmec type III (81.5%) and agr type 1 (80.9%), followed by other types or nontypeable strains. The prevalence of SCCmec type II was slightly higher among the hVISA isolates than the VSSA isolates (22.2% and 9.8%, respectively), but the difference was not significant (χ2 = 2.41, P > 0.05). Similarly, the prevalence of agr type 2 was higher among the hVISA isolates than VSSA isolates, but again, the difference was not significant (χ2 = 3.24, P > 0.05).
TABLE 5.
SCCmec and agr typing of the isolates in the hVISA/VISA and VSSA group
| Molecular typing method | Type | No. (%) of isolates
|
||
|---|---|---|---|---|
| hVISA/VISA (n = 27) | VSSA (n = 173) | Total (n = 200) | ||
| SCCmec | I | 0 (0) | 0 (0) | 0 (0) |
| II | 6 (22.2) | 17 (9.8) | 23 (11.5) | |
| III | 20 (74.1) | 141 (81.5) | 161 (80.5) | |
| IV | 1 (3.7) | 1 (0.6) | 2 (1.0) | |
| NTa | 0 (0.0) | 14 (8.1) | 14 (7.0) | |
| agr | 1 | 21 (77.8) | 140 (80.9) | 161 (80.5) |
| 2 | 6 (22.2) | 15 (8.7) | 21 (10.5) | |
| 3 | 0 (0.0) | 0 (0) | 0 (0.0) | |
| 4 | 0 (0.0) | 2 (1.6) | 2 (1.0) | |
| NT | 0 (0.0) | 16 (9.2) | 16 (8.0) | |
NT, nontypeable.
Geographical sources and detailed typing results for hVISA and VISA.
Detailed data on the 1 VISA isolate and the 26 hVISA isolates are displayed in Table 6. The 26 hVISA isolates were distributed among eight cities: Beijing (n = 11), Guangzhou (n = 5), Shenyang (n = 3), Hangzhou (n = 3), Shanghai (n = 1), Changsha (n = 1), Ji'nan (n = 1), and Xi'an (n = 1). One isolate (isolate 4sy39) from Shenyang was identified as VISA because it had a PAP-AUC ratio of 1.3 and a vancomycin MIC of 4 mg/liter. This isolate was sequence type 5 (ST5)-SCCmec type II and agr type 2. Three STs were identified among the hVISA isolates: ST239 was the most common clone, accounting for 76.9% (20/26) of them. Two agr types, namely, agr type 1 and agr type 2 were found, with prevalence rates of 80.8% (21/26) and 19.2% (5/26), respectively. One isolate displayed the types ST59-SCCmec type IV, agr type 1, and spa type 437.
TABLE 6.
Characteristics of the 27 hVISA/VISA isolates evaluated in the present study
| Isolate | City in China | Region in China | MIC (mg/liter)a
|
Molecular type
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OXA | VAN | TEC | DAP | TGC | LZD | BPR | RIF | GEN | CIP | LVX | ERY | SCCmec | agr | spa | ST | |||
| 4sy39b | Shenyang | Northeast | 256 | 4 | 8 | 0.5 | 0.25 | 1 | 0.5 | 0.008 | 64 | 32 | 16 | 256 | II | 2 | t002 | 5 |
| 4sy32 | Shenyang | Northeast | 256 | 1 | 8 | 0.5 | 0.25 | 1 | 0.5 | 0.008 | 64 | 32 | 8 | 1 | II | 2 | t002 | 5 |
| 5zjsau3 | Hangzhou | Southeast | >64 | 1 | 1 | 0.5 | 0.25 | 1 | 0.5 | 0.008 | >256 | 32 | 16 | >256 | III | 1 | t037 | 239 |
| sh06024 | Shanghai | Southeast | >64 | 1 | 1 | 0.5 | 0.125 | 0.5 | 2 | >256 | 256 | 32 | 8 | >256 | II | 2 | t030 | 239 |
| gz06040 | Guangzhou | South | 32 | 1 | 2 | 0.5 | 0.125 | 1 | 1 | 0.008 | 256 | 32 | 64 | >256 | III | 1 | t037 | 239 |
| gz06051 | Guangzhou | South | >64 | 1 | 2 | 0.25 | 0.25 | 2 | 2 | >256 | 128 | 64 | 64 | 0.25 | III | 1 | t030 | 239 |
| 6zjsau7 | Hangzhou | Southeast | >64 | 2 | 2 | 0.25 | 0.25 | 1 | 4 | 0.008 | 256 | >64 | 32 | >256 | II | 2 | t002 | 5 |
| bd4768 | Beijing | North | >64 | 1 | 8 | 0.5 | 0.125 | 1 | 2 | 256 | 256 | 64 | 32 | >256 | III | 1 | t030 | 239 |
| b2k4781 | Beijing | North | >64 | 1 | 4 | 0.5 | 0.125 | 0.5 | 2 | 0.008 | >256 | 64 | 32 | >256 | II | 2 | t002 | 5 |
| b3k2345 | Beijing | North | >64 | 1 | 2 | 0.5 | 0.064 | 0.5 | 2 | 0.008 | 128 | 64 | 16 | >256 | III | 1 | t030 | 239 |
| b2k3421 | Beijing | North | >64 | 1 | 2 | 0.5 | 0.125 | 0.5 | 2 | 0.008 | 128 | 64 | 16 | >256 | III | 1 | t037 | 239 |
| b3k5923 | Beijing | North | >64 | 0.5 | 2 | 0.25 | 0.032 | 0.5 | 2 | 256 | 128 | 64 | 32 | >256 | III | 1 | t030 | 239 |
| b4k6307 | Beijing | North | >64 | 1 | 4 | 0.25 | 0.064 | 0.5 | 1 | 256 | 128 | 16 | 4 | >256 | III | 1 | t030 | 239 |
| b5k3529 | Beijing | North | >64 | 1 | 2 | 0.5 | 0.125 | 0.5 | 2 | 0.5 | >256 | 64 | 16 | >256 | III | 1 | t037 | 239 |
| b5k3578 | Beijing | North | >64 | 1 | 2 | 0.5 | 0.25 | 0.5 | 2 | 0.5 | >256 | 32 | 16 | 2 | III | 1 | t002 | 5 |
| b6k1642 | Beijing | North | >64 | 1 | 4 | 0.5 | 0.25 | 1 | 2 | 16 | >256 | 64 | 16 | >256 | III | 1 | t037 | 239 |
| b6k7974 | Beijing | North | >64 | 2 | 8 | 1 | 0.25 | 1 | 2 | 256 | 256 | 64 | 64 | >256 | III | 1 | t030 | 239 |
| 06b41 | Guangzhou | South | 8 | 1 | 1 | 1 | 0.25 | 1 | 1 | 0.016 | 0.25 | 1 | 0.25 | 128 | IV | 1 | t437 | 59 |
| 13b16 | Beijing | North | >64 | 1 | 0.5 | 0.5 | 0.125 | 2 | 1 | 0.008 | 256 | 16 | 32 | >256 | III | 1 | t037 | 239 |
| 11b091 | Changsha | South | >64 | 1 | 1 | 0.5 | 0.125 | 2 | 2 | 0.008 | 256 | >64 | 32 | >256 | III | 1 | t037 | 239 |
| 07b63 | Shenyang | Northeast | >64 | 1 | 4 | 0.5 | 0.5 | 1 | 1 | 0.008 | 64 | 64 | 8 | >256 | II | 2 | t002 | 5 |
| 15b52 | Jinan | North | >64 | 1 | 4 | 0.5 | 0.25 | 0.5 | 2 | 0.016 | 256 | 16 | 4 | 256 | III | 1 | t030 | 239 |
| zj07028 | Hangzhou | East | >64 | 2 | 2 | 0.5 | 0.5 | 1 | 2 | ≤0.002 | >256 | 32 | 8 | >256 | III | 1 | t037 | 239 |
| sh07043 | Shenyang | Northeast | >64 | 1 | 2 | 1 | 0.25 | 2 | 2 | 0.016 | 128 | >64 | 32 | >256 | III | 1 | t030 | 239 |
| xa07029 | Xi'an | Northwest | >64 | 1 | 2 | 0.5 | 0.25 | 1 | 2 | >256 | 128 | 32 | 8 | >256 | III | 1 | t037 | 239 |
| gz07030 | Guangzhou | South | >64 | 1 | 1 | 0.5 | 0.125 | 1 | 2 | 0.004 | >256 | 64 | 64 | >256 | III | 1 | t030 | 239 |
| gs07022 | Guangzhou | South | 8 | 1 | 1 | 0.5 | 0.25 | 1 | 1 | 0.008 | 8 | 64 | 32 | >256 | III | 1 | t037 | 239 |
OXA, oxacillin; VAN, vancomycin; TEC, teicoplanin; DAP, daptomycin; TGC, tigecycline; LZD, linezolid; BPR, ceftobiprole; RIF, rifampin; GEN, gentamicin; CIP, clarithromycin; LVX, levofloxacin; ERY, erythromycin.
Isolate 4sy39 was identified as a VISA strain with a vancomycin MIC of 4 mg/liter by agar dilution and a PAP-AUC ratio of 1.3.
DISCUSSION
By using a set of vancomycin-susceptible MRSA isolates (i.e., MICs ≤ 2 mg/liter) recovered from blood specimens from Chinese patients (n = 199), two agar screening methods (BHIT5 and BHIV6) and MET with different cutoff values were calibrated for the detection of hVISA on the basis of the results of PAP-AUC as the reference method. The prevalence of hVISA in these blood isolates was 13.1%. The application of the cascade of BHIT5 followed by MET B as a composite screening test to all 812 non-blood MRSA isolates recovered from Chinese patients yielded an estimate of the prevalence of hVISA of 15.7% (Table 3).
This is the first study of the prevalence of hVISA to have used a large collection of MRSA isolates from a range of cities in mainland China and to have applied PAP-AUC as the “gold standard” reference method. The prevalence of hVISA is high and is of potential concern when the treatment of patients with glycopeptides is considered. Clearly, studies of the outcome of glycopeptide treatment are needed to understand the clinical impact of hVISA in China.
The use of a screening test for the estimation of prevalence in a large number of isolates is, by definition, less accurate and precise than the use of a reference method. However, it is impractical to measure PAP-AUC ratios for large numbers of isolates. Therefore, as described in the Results and Table 3, in order to understand better the accuracy and the precision of the estimated prevalence, we further estimated the prevalence in a randomly selected set of 120 of the 812 non-blood isolates while also characterizing them by the reference PAP-AUC method. The estimated prevalence of 24.5% was rather high compared with the measured prevalence of 14.2% in the same 120-isolate subset (Table 3), revealing the extent of the variation in agreement between the robust screening assay and the current best reference assay. In comparison, an approach used by others to estimate the prevalence of hVISA among large numbers of isolates has been to apply a screening test, followed by measurement of the number of PAP-AUC-positive strains among the screening test-positive isolates (21a), the inaccuracy of which cannot be determined because the sensitivity and the specificity are unknown. We suggest that the calibration of the screening test, followed by the estimation method used here, should yield a more accurate value for the prevalence of hVISA. Moreover, a useful additional step, which was taken in the present study, is to gain an idea of the accuracy and the precision of the screening test by applying it to a blinded test set for which reference measurements are also made. In summary, we infer from our data that the prevalence of hVISA among 1,012 MRSA isolates recovered in China is 13 to 16%. In a large study such as this one, the estimated prevalence must be regarded as a rough and somewhat imprecise guide that can alert investigators to a high prevalence in hospital wards, institutions, or geographic regions; this can be followed up by more precise investigations.
The BHIV6 screening method displayed the highest specificity, but its sensitivity was very low (Table 2), in agreement with the findings of others (18, 27). The sensitivity is too low for it to be useful in the clinical laboratory. On the contrary, the sensitivity of BHIT5 was 88.5% in this study, but the specificity was low, which would also make it impractical for use in the clinical laboratory, owing to the need to investigate a large number of false-positive results by more labor-intensive methods. BHIT5 could perhaps be used as a preliminary screen in a screening cascade. Another prospective screening method is MET, despite the high cost of Etest strips. Previous investigators reported both a sensitivity and a specificity greater than 95.0% when MET was used with a cutoff value of either 8 mg/liter for vancomycin and teicoplanin or 12 mg/liter for teicoplanin alone (27). In the present study, the specificity of the test when this cutoff value was used was only 48.0%, which would again lead to an impractical number of false-positive results requiring further follow-up in the clinical laboratory. However, application of the cutoff value of 8 mg/liter for vancomycin and teicoplanin to MET resulted in an improved specificity of 85.0%, but about half of the hVISA isolates in a clinical laboratory would be missed owing to the lower sensitivity. At least one can state from the data reported here that the teicoplanin cutoff value of 12 mg/liter seems not to be appropriate for use in China. In the present study, a cascade of BHIT5 followed by MET with a cutoff value of 8 mg/liter for vancomycin and teicoplanin yielded an overall sensitivity of 42.3% and a specificity of 86.1%, and thus, the use of this cascade seems applicable for screening for hVISA in mainland China. However, the cutoff value of teicoplanin of 12 mg/liter was not sensitive enough for the detection of hVISA in China. It has been reported that a strain of MRSA might become heterogeneously resistant to teicoplanin before it becomes heterogeneously resistant to vancomycin (2, 18), a phenomenon that requires further investigation.
The points summarized above refer to surveillance studies aimed at measuring or estimating the prevalence rates of hVISA among large numbers of isolates. However, if it is important to know whether an individual patient's MRSA isolate is hVISA; i.e., for determination of the likely efficacy of glycopeptide chemotherapy, the screening test would not be adequately sensitive or specific. We propose that, currently, only a PAP-AUC measurement would give a result with enough precision for patient management.
The clinical significance of hVISA is still debated (18). Such strains might be responsible for vancomycin treatment failures, especially in infections with high bacterial loads, such as bacterial endocarditis and bone and joint infections (1, 5). Moreover, the mechanism of heterogeneous vancomycin-intermediate resistance is only partially understood (28), and heterogeneous vancomycin-intermediate resistance might be a pre-stage of vancomycin-intermediate resistance (13). In the present study, the MICs of several antibacterial agents such as daptomycin, tigecycline, ceftobiprole, and linezolid were measured against hVISA in vitro. The hVISA isolates were generally susceptible to these other antibacterial agents, and therefore, these other agents might be considered alternatives for the treatment of hVISA-infected patients (9).
SCCmec and agr typing were performed with 200 isolates of MRSA from the blood specimens. It showed that SCCmec type III and agr type 1 were the predominant types in China, which is different from the situation in South Korea and Japan but similar to the findings in Taiwan (14, 15, 16). Interestingly, the percentages of SCCmec type II and agr type 2 isolates in the hVISA group were higher than those in the VSSA group, but they were not significantly different. It was reported that when the agr function is lost, MRSA isolates of wild-type agr type 2 are more likely than MRSA isolates possessing other agr types to develop glycopeptide heteroresistance in the presence of low vancomycin concentrations (20, 25). By reducing autolysis after exposure to subinhibitory concentrations of glycopeptide, this type of strain has an intrinsic survival advantage under the selective pressure of a glycopeptide (22). Moise-Broder et al. also showed that agr type 2 was related to vancomycin treatment failure (20). Nevertheless, other published studies have suggested that glycopeptide resistance can develop in other S. aureus agr groups with sufficient selection pressure (25). Whether the higher percentage of agr type 2 in the hVISA subgroup (compared with the percentage in the VSSA subgroup) had any relationship to its low-level vancomycin resistance was not clear. However, the present study found only 26 hVISA isolates, a small number on which to base such hypotheses. The one VISA isolate found in the present investigation was the first VISA isolate to have been reported in mainland China. It belonged to ST5, while ST239 was the most frequent genotype of MRSA in China, as we have reported previously (19).
In conclusion, this study was the first study in mainland China of hVISA isolates, which were collected from 14 cities. We conclude that the prevalence of hVISA from blood is similar to the prevalence of hVISA from other specimens in China (13.1 to 15.7%). A cascade screening method consisting of BHIT5 and MET with a cutoff value of 8 mg/liter for vancomycin and teicoplanin may be a promising method for the detection of hVISA in laboratory practice in China, especially for the estimation of local prevalence rates. Further studies of the characteristics of hVISA, including susceptibility, molecular types, and the clinical outcomes of vancomycin therapy, with larger sample sizes are needed.
Supplementary Material
Acknowledgments
This study was funded by AstraZeneca and Pfizer. It was partially supported by grant 2007-2015 from the Beijing Medical Scientific Development Foundation.
We thank Xu Yingchun from Peking Union Medical College Hospital for providing some surveillance data. We thank Qiu Ning from AstraZeneca China for assistance with manuscript preparation and Zhiping You and Aaron Dane for help with statistical analyses. We also express our appreciation to Keiichi Hiramatsu from Juntendo University in Japan for providing the control strains Mu3 and Mu50. We thank all of the participants of the GPRS, SEANIR, and CARES surveillance networks for providing isolates.
Footnotes
Published ahead of print on 22 June 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Ariza, J., M. Pujol, J. Cabo, C. Pena, N. Fernandez, J. Linares, J. Ayats, and F. Gudiol. 1999. Vancomycin in surgical infections due to methicillin-resistant Staphylococcus aureus with heterogeneous resistance to vancomycin. Lancet 353:1587-1588. [DOI] [PubMed] [Google Scholar]
- 2.Brunet, F., G. Vedel, F. Dreyfus, J. F. Vaxelaire, T. Giraud, B. Schremmer, and J. F. Monsallier. 1990. Failure of teicoplanin in two neutropenic patients with staphylococcus septicemia who recovered after administration of vancomycin. Eur. J. Clin. Microbiol. Infect. Dis. 9:145-147. [DOI] [PubMed] [Google Scholar]
- 3.Cartolano, G. L., M. Cheron, D. Benabid, M. Leneveu, and A. Boisivon. 2004. Methicillin-resistant Staphylococcus aureus (MRSA) with reduced susceptibility to glycopeptides (GISA) in 63 French general hospitals. Clin. Microbiol. Infect. 10:448-451. [DOI] [PubMed] [Google Scholar]
- 4.Charles, P. G., P. B. Ward, P. D. Johnson, B. P. Howden, and M. L. Grayson. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38:448-451. [DOI] [PubMed] [Google Scholar]
- 5.Chi, C. Y., T. L. Lauderdale, S. M. Wang, J. M. Wu, Y. J. Yang, and C. C. Liu. 2008. Health care-associated endocarditis caused by Staphylococcus aureus with reduced susceptibility to vancomycin. J. Clin. Microbiol. 46:810-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement; M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgibbon, M. M., A. S. Rossney, and B. O'Connell. 2007. Investigation of reduced susceptibility to glycopeptides among methicillin-resistant Staphylococcus aureus isolates from patients in Ireland and evaluation of agar screening methods for detection of heterogeneously glycopeptide-intermediate S. aureus. J. Clin. Microbiol. 45:3263-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fowler, V. G., Jr., H. W. Boucher, G. R. Corey, E. Abrutyn, A. W. Karchmer, M. E. Rupp, D. P. Levine, H. F. Chambers, F. P. Tally, G. A. Vigliani, C. H. Cabell, A. S. Link, I. DeMeyer, S. G. Filler, M. Zervos, P. Cook, J. Parsonnet, J. M. Bernstein, C. S. Price, G. N. Forrest, G. Fatkenheuer, M. Gareca, S. J. Rehm, H. R. Brodt, A. Tice, and S. E. Cosgrove. 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 355:653-665. [DOI] [PubMed] [Google Scholar]
- 9a.Galen, R. S., and S. R. Gambino. 1975. Beyond normality: the predictive value and efficiency of medical diagnoses, p. 16-17. John Wiley & Sons, Inc., New York, NY.
- 10.Gilot, P., G. Lina, T. Cochard, and B. Poutrel. 2002. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 40:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 12.Howden, B. P., P. D. Johnson, P. B. Ward, T. P. Stinear, and J. K. Davies. 2006. Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 50:3039-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howe, R. A., M. Wootton, T. R. Walsh, P. M. Bennett, and A. P. Macgowan. 2000. Heterogeneous resistance to vancomycin in Staphylococcus aureus. J. Antimicrob. Chemother. 45:130-132. [DOI] [PubMed] [Google Scholar]
- 14.Huang, Y. C., L. H. Su, T. L. Wu, C. E. Liu, T. G. Young, P. Y. Chen, P. R. Hseuh, and T. Y. Lin. 2004. Molecular epidemiology of clinical isolates of methicillin-resistant Staphylococcus aureus in Taiwan. J. Clin. Microbiol. 42:307-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, H. B., W. B. Park, K. D. Lee, Y. J. Choi, S. W. Park, M. D. Oh, E. C. Kim, and K. W. Choe. 2003. Nationwide surveillance for Staphylococcus aureus with reduced susceptibility to vancomycin in Korea. J. Clin. Microbiol. 41:2279-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko, K. S., J. Y. Lee, J. Y. Suh, W. S. Oh, K. R. Peck, N. Y. Lee, and J. H. Song. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, C., and H. F. Chambers. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 47:3040-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Y., H. Wang, N. Du, E. Shen, H. Chen, J. Niu, H. Ye, and M. Chen. 2009. Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob. Agents Chemother. 53:512-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moise-Broder, P. A., G. Sakoulas, G. M. Eliopoulos, J. J. Schentag, A. Forrest, and R. C. Moellering, Jr. 2004. Accessory gene regulator group II polymorphism in methicillin-resistant Staphylococcus aureus is predictive of failure of vancomycin therapy. Clin. Infect. Dis. 38:1700-1705. [DOI] [PubMed] [Google Scholar]
- 21.Ruppitsch, W., A. Indra, A. Stöger, B. Mayer, S. Stadlbauer, G. Wewalka, and F. Allerberger. 2006. Classifying spa types in complexes improves interpretation of typing results for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:2442-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Rybak, M. J., S. N. Leonard, K. L. Rossi, C. M. Cheung, H. S. Sader, and R. N. Jones. 2008. Characterization of vancomycin-heteroresistant Staphylococcus aureus from the metropolitan area of Detroit, Michigan, over a 22-year period (1986 to 2007). J. Clin. Microbiol. 46:2950-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., R. P. Novick, L. Venkataraman, C. Wennersten, P. C. DeGirolami, M. J. Schwaber, and H. S. Gold. 2003. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J. Infect. Dis. 187:929-938. [DOI] [PubMed] [Google Scholar]
- 23.Song, J. H., K. Hiramatsu, J. Y. Suh, K. S. Ko, T. Ito, M. Kapi, S. Kiem, Y. S. Kim, W. S. Oh, K. R. Peck, and N. Y. Lee. 2004. Emergence in Asian countries of Staphylococcus aureus with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 48:4926-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover, F. C., and R. C. Moellering, Jr. 2007. The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin. Infect. Dis. 44:1208-1215. [DOI] [PubMed] [Google Scholar]
- 25.Tsuji, B. T., M. J. Rybak, K. L. Lau, and G. Sakoulas. 2007. Evaluation of accessory gene regulator (agr) group and function in the proclivity towards vancomycin intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:1089-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unal, S., J. Hoskins, J. E. Flokowitsch, C. Y. Wu, D. A. Preston, and P. L. Skatrud. 1992. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J. Clin. Microbiol. 30:1685-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh, T. R., A. Bolmstrom, A. Qwarnstrom, P. Ho, M. Wootton, R. A. Howe, A. P. MacGowan, and D. Diekema. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 39:2439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh, T. R., and R. A. Howe. 2002. The prevalence and mechanisms of vancomycin resistance in Staphylococcus aureus. Annu. Rev. Microbiol. 56:657-675. [DOI] [PubMed] [Google Scholar]
- 29.Wootton, M., R. A. Howe, R. Hillman, T. R. Walsh, P. M. Bennett, and A. P. MacGowan. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399-403. [DOI] [PubMed] [Google Scholar]
- 30.Wootton, M., A. P. MacGowan, T. R. Walsh, and R. A. Howe. 2007. A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 45:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wootton, M., T. R. Walsh, and A. P. MacGowan. 2005. Evidence for reduction in breakpoints used to determine vancomycin susceptibility in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3982-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao, Y. H., J. Wang, and Y. Li. 2008. Bacterial resistance surveillance in China: a report from Mohnarin 2004-2005. Eur. J. Microbiol. Infect. Dis. 27:697-708. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.