Abstract
VanA-type Staphylococcus aureus strain VRSA-7 was partially dependent on glycopeptides for growth. The vanA gene cluster, together with the erm(A) and the ant(9)-Ia resistance genes, was carried by the ca. 35- to 40-kb conjugative plasmid pIP848 present at five copies per cell. The chromosomal ddl gene had a mutation that led to a N308K substitution in the d-Ala:d-Ala ligase that resulted in a 1,000-fold decrease in activity relative to that of strain VRSA-6. Strain VRSA-7 grown in the absence or in the presence of vancomycin mainly synthesized precursors ending in d-Ala-d-Lac, indicating that the strain relied on the vancomycin resistance pathway for peptidoglycan synthesis. Greatly enhanced growth in the presence of glycopeptides and the absence of mutations in the genes for VanR and VanS indicated the inducible expression of resistance. Thus, a combination of loose regulation of the vanA operon by the two-component system and a gene dosage effect accounts for the partial glycopeptide dependence of VRSA-7. Since peptidoglycan precursors ending in d-Ala-d-Lac are not processed by PBP 2′, the strain was fully susceptible to oxacillin, despite the production of a wild-type PBP 2′.
Methicillin (meticillin)-resistant Staphylococcus aureus (MRSA) strains that have acquired the vanA operon from glycopeptide-resistant enterococci are designated vancomycin-resistant S. aureus (VRSA). The first two VRSA isolates were recovered in the United States in 2002 (25, 36). Since then, an additional seven MRSA isolates carrying the vanA gene cluster have been detected in the United States (one in New York and six in Michigan), and two tentative identifications of VRSA have been reported in India (32) and Iran (1). Vancomycin acts by binding to the C-terminal acyl-d-alanyl-d-alanine (acyl-d-Ala-d-Ala) of pentapeptide peptidoglycan precursors and inhibits transglycosylation and transpeptidation reactions, thus preventing cell wall formation (6) and with the acyl-d-Ala-d-Ala residues being incorporated into peptidoglycan precursors as dipeptides synthesized by the host d-Ala:d-Ala ligase (Ddl). VanA-type resistance is characterized by high-level inducible resistance to vancomycin and teicoplanin due to synthesis of peptidoglycan pentadepsipeptide precursors ending in d-Ala-d-lactate (d-Ala-d-Lac). This alteration is responsible for the diminished binding affinity of glycopeptides for their target. VanA-type resistance is mediated by a gene cluster often carried by Tn1546 encoding nine proteins. Two are implicated in the movement of the element: open reading frame 1 (ORF1), a transposase, and ORF2, a resolvase. Two are essential for the ultimate production of pentadepsipeptides: VanH, a dehydrogenase that reduces pyruvate to d-Lac, and VanA, a ligase that catalyzes the formation of an ester bond between d-Ala and d-Lac. Two other proteins are responsible for the elimination of pentapeptide precursors: VanX, a d,d-dipeptidase that cleaves the d-Ala-d-Ala dipeptide synthesized by the host Ddl, and VanY, a d,d-carboxypeptidase that removes the ultimate d-Ala of the pentapeptide precursors. Expression of the vanA gene cluster is regulated by the two-component regulatory system VanR-VanS, which is responsible for recognition of the presence of glycopeptides in the culture medium and transcriptional activation of the resistance and regulatory genes. The remaining protein, VanZ, whose function is unknown, is implicated in teicoplanin resistance (2).
Enterococci that require the presence of vancomycin in the culture medium for growth have been isolated from patients treated with this antibiotic (14, 39). Although many of these vancomycin-dependent strains are VanB-type Enterococcus faecium, vancomycin-dependent Enterococcus faecalis and Enterococcus avium containing the vanA operon have also been isolated (16, 38). To the best of our knowledge, no VRSA strain with this phenotype has been reported. Here we describe the mechanism of vancomycin dependence in VanA-type S. aureus strain VRSA-7, isolated from a patient treated with vancomycin for prolonged periods of time. The activity of the host Ddl and the expression of the vanA gene cluster of VRSA-7 were compared with those of strain VRSA-6, another highly vancomycin-resistant S. aureus of the VanA type (37, 41).
(An initial report of this work was presented at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy, abstr. C1-4176.)
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The origins and characteristics of the bacterial strains and plasmids used in this study are described in Table 1. S. aureus strains were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus. S. aureus strains VRSA-6 and VRSA-7 were isolated in Michigan in 2005 from a surgical site wound and a nonhealing plantar ulcer, respectively (37). Escherichia coli Top10 was used as the host in cloning experiments. S. aureus BM4602 (from a laboratory collection), E. faecalis JH2-2 (19), and E. faecium 64/3 (40) were used as recipient strains in the transfer experiments. MRSA COL (15) and methicillin-susceptible S. aureus RN4220 (21) were used as positive and negative controls, respectively, for detection of penicillin-binding protein (PBP) PBP 2′. E. coli BL21(DE3)pLys (Novagen, Madison, WI) was used with the pET28a(+) expression vector (Novagen). All strains were grown in brain heart infusion (BHI) broth or on BHI agar (Difco Laboratories, Detroit, MI) at 37°C. Kanamycin (50 μg/ml) was used as a selective agent for cloning of the PCR products into the pCR-Blunt vector (Invitrogen, Leek, The Netherlands). Spectinomycin (60 μg/ml) was used to prevent the loss of plasmids derived from pAT79 (4).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| Top10 | F−mcrA Δ(mrr hsdRMS mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| Bl21 | BL21(DE3)pLys | Novagen |
| S. aureus | ||
| VRSA-6 | vanRSHAXYZ, erm(B), ant(9)-Ia, ant(4′)-Ia, mecA | 37 |
| VRSA-7 | vanRSHAXYZ, erm(A), erm(C), ant(9)-Ia, ant(4′)-Ia, mecA | 37 |
| COL | mecA Vms Tes | 15 |
| BM4602 | Rifr Fur | Laboratory collection |
| RN4220 | Oxas | 21 |
| E. faecium | ||
| BM4339 | vanRDSDHDDXDYD | 30 |
| 64/3 | Rifr Fur | 40 |
| E. faecalis JH2-2 | Rifr Fur | 19 |
| Plasmid | ||
| pCR-Blunt | Kmr ZeocinroriR from ColE1 lacZα ccdB | Invitrogen |
| pET28a | Kmr, vector for expression of recombinant proteins | Novagen |
| pAT79 | oriR from pAMβ1 oriR from pUC oriT from RK2 SprlacZα P2cat | 4 |
| pAT515 | 1,128-bp SacI-XbaI PCR fragment (ddlVRSA-6) of S. aureus VRSA-6 cloned into pAT79 | This study |
| pAT516 | 1,128-bp SacI-XbaI PCR fragment (ddlVRSA-7) of S. aureus VRSA-7 cloned into pAT79 | This study |
| pAT518 | 1,087-bp BsaI-XhoI PCR fragment (ddlCOL) of S. aureus COL cloned into pET28a(+) | This study |
| pAT519 | 1,087-bp BsaI-XhoI PCR fragment (ddlVRSA-7) of S. aureus VRSA-7 cloned into pET28a(+) | This study |
Fu, fusidic acid; Km, kanamycin; Oxa, oxacillin; Rif, rifampin; r, resistant; s, susceptible; Sp, spectinomycin.
Susceptibility testing.
Antibiotic susceptibility was tested by disk diffusion on Mueller-Hinton (MH) agar, according to the recommendations of the Comité de l'Antibiogramme de la Société Française de Microbiologie (9). The MICs of antimicrobial agents were determined by Etest (AB Biodisk, Combourg, France) on MH agar.
Growth rate studies.
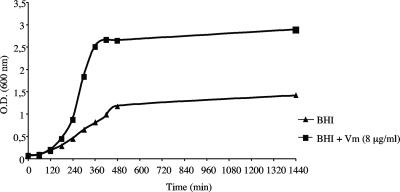
The induction of resistance by vancomycin in strain VRSA-7 was studied by the determination of growth rates under various conditions. The isolate was grown overnight at 37°C in BHI broth without or with vancomycin (8 μg/ml). The cultures were diluted 1:20 into 20 ml of BHI without or with vancomycin (8 μg/ml) and were then grown at 37°C with shaking, and the optical density at 600 nm was monitored.
PCR analysis and sequencing.
Glycopeptide resistance genotyping and species identification of VRSA-6 and VRSA-7 were performed by a multiplex PCR assay, as described previously (13). PCR mapping of the vanA operon was performed by using primers specific for every gene of the cluster (5). The strains were screened by PCR for the spectinomycin resistance gene ant(9)-Ia (20); the kanamycin and tobramycin resistance gene ant(4′)-Ia (34); and the macrolide-lincosamide-streptogramin B (MLSB) resistance genes erm(A), erm(B), and erm(C) with the following primers: ermA(+) (5′-CTTCGATAGTTTATTAATATTAGT-3′), ermA(−) (5′-TCTAAAAAGCATGTAAAAGAA-3′), ermB(+) (5′-GAAAAGGTACTCAACCAAATA-3′), ermB(−) (5′-AGTAACGGTACTTAAATTGTTTAC-3′), ermC(+) (5′-TCAAAACATAATATAGATAAA-3′), and ermC(−) (5′-GCTAATATTGTTTAAATCGTCAAT-3′). Primers MecAStaph1 (5′-TAACGTGGAGACGAGCAC-3′) and MecAStaph2 (5′-AACAGTGAAGCAACCATCGT-3′) were used to amplify the mecA gene and its promoter region. PCRs were performed with a 50-μl volume containing 20 pmol of each pair of oligodeoxynucleotide primers, 5 nmol of each 2′-deoxynucleoside 5′-triphosphate, reaction buffer, 100 ng of DNA, and 5 U of DNA Pfu polymerase (Stratagene, La Jolla, CA). PCR elongation times and temperatures were adjusted according to the expected size of the PCR product and the nucleotide sequences of the primers, respectively. Amplification was in a model 9700 thermal cycler (Perkin Elmer Cetus, Norwalk, CT). The sequences of the ddl genes from VRSA-6 and VRSA-7 and of the vanR-vanS genes from VRSA-7 were determined after the amplification of total DNA with oligodeoxynucleotides MurSA1 (5′-CCCCAGCACCATCTTGTAAT-3′) and FtsSA1 (5′-GGCCAAACGTGTACCAACTT-3′) and with oligodeoxynucleotides P9/P10 and P11/P12, respectively (5). The PCR fragments, which had expected sizes of 1,930 bp, 1,284 bp, and 1,671 bp, respectively, were purified with the PCR purification kit (Qiagen, Inc., Chatsworth, CA) and cloned into PCR-Blunt. Plasmid DNA was labeled by using a dye-labeled dideoxynucleoside triphosphate Terminator cycle sequencing kit (Beckman Coulter, Fullerton, CA) and sequenced with a model CEQ 2000 automated sequencer (Beckman Coulter). The sequences were analyzed with the GCG sequence analysis software package (version 10.1; Genetics Computer Group, Madison, WI).
Contour-clamped homogeneous electric field gel electrophoresis.
Pulsed-field gel electrophoresis of genomic DNA embedded in agarose plugs digested with I-CeuI or SmaI was performed as described previously (12). The fragments generated by I-CeuI were hybridized successively to a α-32P-labeled 16S rRNA (rrs) probe obtained by amplification of an internal portion of the rrs gene (18) and to a vanA-specific probe obtained by PCR with primers EA1 and EA2 (13).
Recombinant DNA techniques.
Plasmid DNA isolation, digestion with restriction endonucleases (Amersham Pharmacia Biotech, Uppsala, Sweden), ligation with T4 DNA ligase (Amersham), and transformation of E. coli Top10 with recombinant plasmid DNA were performed by standard methods (33). For the isolation of plasmid DNA from S. aureus, lysostaphin (Sigma-Aldrich, St. Louis, MO) was added to a Tris-HCl (10 mM)-EDTA (1 mM)-glucose (9 g/liter) buffer at a final concentration of 2 mg/ml, and the bacteria were incubated in the buffer at 37°C for 1 h before the lysis step.
Plasmid analysis.
The attempted transfer of vancomycin resistance from strain VRSA-7 to S. aureus BM4602, E. faecalis JH2-2, and E. faecium 64/3 was performed by filter mating. Plasmid DNA extracted from VRSA-7 was electrotransformed into S. aureus BM4602. Transformants were selected on agar containing rifampin (rifampicin; 20 μg/ml), fusidic acid (10 μg/ml), and vancomycin (10 μg/ml) or erythromycin (8 μg/ml). Plasmid DNA that had been digested with EcoRI was transferred to a nylon membrane and hybridized separately to α-32P-labeled probes for vanA, erm(A), erm(C), and ant(9)-Ia.
qPCR.
Quantitative PCR (qPCR) was performed in a LightCycler instrument with version 4.1 of the LightCycler software (Roche Diagnostics) and a Fast Start DNA Master Plus SYBR green I kit (Roche Diagnostics GmbH, Mannheim, Germany), according to the manufacturer's instructions. The ddl, rpoB, and rrs chromosomal genes, which are present in S. aureus in one, one, and five copies, respectively, were selected as controls. The copy numbers of the vanA and vanH genes relative to those of ddl, rpoB, and rrs were estimated from three independent experiments with five sets of primers specific for each gene: primers qPCRrrs1 (5′AGGTAACGGCTTACCAAGGCA3′) and qPCRrrs2 (5′ACGATCCGAAGACCTTCA3′), primers qPCRddl1 (5′CCCATTATTACATGGTCCTA3′) and qPCRddl2 (5′ GGTAACTGTGGTAACCCT3′), primers qPCRrpoB1 (5′GTACAGTGCTTGATCGTCGT3′) and qPCRrpoB2 (5′CAGTTGTCGTACGACCTTCA3′), primers qPCRvanA1 (5′TTCAGCTTTGCATGGCAAGT3′) and qPCRvanA2 (5′ACCCAAAAGGCGGGAGTA3′), and primers qPCRvanH1 (5′CGGATAGCGTTGCCGATTAT3′) and qPCRvanH2 (5′GCTCAATAACCGCTTTGCCT3′). The data were analyzed by the method of Peirson et al. (28).
Plasmid construction.
The chromosomal ddl genes from S. aureus VRSA-6 (ddlVRSA-6) and VRSA-7 (ddlVRSA-7) with their ribosome-binding sites were amplified by PCR from total DNA with oligodeoxynucleotides DdlSASacI (5′-CGCTGCAGAGCTCATATATCATTGGAGGAA-3′) and DdlSAXbaI (5′-TTTGGGATCTAGATTTAATGTAACATTAAT-3′) containing a SacI site and a XbaI site (underlined), respectively. The PCR fragments were cloned under the control of the constitutive P2 promoter and upstream from the cat reporter gene of the shuttle vector pAT79, leading to plasmids pAT515 (pAT79ΩP2ddlVRSA-6cat) and pAT516 (pAT79ΩP2ddlVRSA-7cat), which were electrotransformed into VanD-type E. faecium BM4339 with selection on spectinomycin (120 μg/ml), followed by screening for resistance to chloramphenicol. The nucleotide sequence of the amplified fragments was redetermined.
Production and purification of Ddls of S. aureus VRSA-7 and VRSA-6 strains.
Oligodeoxynucleotides DdlSABsaI (5′-CGTGGTCTCCCATGACAAAAGAAAATATTT-3′) and DdlSAXhoI (5′-CTCCTCGAGGTCAATTTTGTATTTATTTTT-3′) containing a BsaI and a XhoI restriction site (underlined), respectively, were used to amplify the ddl gene from strains VRSA-6 and VRSA-7 with Pfu DNA polymerase. The PCR products were cloned in the PCR-Blunt vector, sequenced, and subcloned under the control of the T7 promoter in pET28a(+) previously digested with NcoI and XhoI, producing pAT518 [pET28a(+)ΩddlVRSA-6] and pAT519 [pET28a(+)ΩddlVRSA-7]. Protein production and purification were performed as described previously (22), with slight modifications. Briefly, Ddl enzymes engineered to have a C-terminal His6 tag were produced from E. coli BL21(DE3)pLys harboring plasmid pAT518 or pAT519 in 1 liter of LB medium containing 25 μg/ml kanamycin and 100 μg/ml ampicillin. Extraction buffer was supplemented with 10% (vol/vol) BugBuster 10× protein extraction reagent and Benzonase (25 U; Sigma-Aldrich). Recombinant His6 proteins were purified by using 5 ml HisTrap fast-flow columns (GE Healthcare, Uppsala, Sweden). The purified proteins were stored at −80°C in buffer containing 50 mM HEPES, pH 7.5, 150 mM KCl, and 1 mM EDTA.
Size-exclusion chromatography.
Size-exclusion chromatography was performed with a Hiload 16/60 Superdex 200 column (GE Healthcare) equilibrated with 50 mM HEPES, pH 7.5, 150 mM KCl, and 1 mM EDTA and eluted in the same buffer at a flow rate of 0.8 ml/min. The column was calibrated according to the manufacturer's instructions.
Enzyme kinetics.
Steady-state kinetic constants for the Ddl activities were determined by a spectrophotometric assay at 340 nm, in which the production of ADP is coupled to the oxidation of NADH through pyruvate kinase and l-lactate dehydrogenase (10). The assays were performed at 37°C in 100 mM HEPES, pH 7.5, 10 mM KCl, 10 mM MgCl2, 10 mM ATP, 2.5 mM phosphoenolpyruvate, 0.2 mM NADH, 50 U/ml l-lactate dehydrogenase, 50 U/ml pyruvate kinase, and d-Ala in a total volume of 0.5 ml. The initial rates of hydrolysis were determined with a Uvikon UV931 spectrophotometer (Kontron Instruments, Saint-Quentin-en-Yvelines, France). The steady-state kinetic parameters were determined by using the equations described previously (26). To obtain the kcat and Km2 values for the Ddl activities, the enzymes were incubated in the presence of increasing concentrations of d-Ala (lowest concentration, 20 mM). Km1 was calculated from the activity measured in the presence of low concentrations of d-Ala (0.5-3 mM). The Km for ATP was measured with fixed concentrations of d-Ala of 10 and 80 mM for the VRSA-6 Ddl and the VRSA-7 Ddl, respectively.
Analysis of peptidoglycan precursors.
Extraction and analysis of peptidoglycan precursors were performed with strains VRSA-6 and VRSA-7 after growth without or with 8 μg/ml of vancomycin, as described previously (24). The results were expressed as the percentages of the total late peptidoglycan precursors, represented by tetrapeptides, pentapeptides, and pentadepsipeptides, that were determined from the integrated peak areas.
Detection of PBP 2′.
Membrane preparations of strains VRSA-7, RN4220, and COL were obtained as described previously (27). Briefly, exponentially growing bacteria were harvested, washed in 50 mM Tris-HCl-145 mM NaCl, pH 7.5, and resuspended at 30 mg (dry weight)/ml in the same buffer containing lysostaphin (20 mg/liter). The suspension was incubated at 37°C for 15 min, and after addition of 5 mM MgCl2 and DNase (30 mg/liter) the suspension was incubated for an additional 5 min. Membranes were collected by centrifugation at 45,000 × g for 20 min at 4°C. The proteins were solubilized by heating the suspension in buffer containing 10% sodium dodecyl sulfate (SDS), separated by SDS-polyacrylamide gel electrophoresis (PAGE), and transferred electrophoretically to a Hybond-P (polyvinylidene difluoride) membrane (Amersham Biosciences, Buckinghamshire, United Kingdom) by blotting at 80 mA for 45 min. The detection of PBP 2′ was performed with a primary mouse anti-PBP 2′ monoclonal antibody (monoclonal antibody 14A9C9-C6), which was used at a 1:20,000 dilution, and sheep anti-mouse immunoglobulin G antibodies conjugated to horseradish peroxidase (ECL Plus Western blotting; Amersham Biosciences) was used as the secondary reagent at a 1:20,000 dilution.
RESULTS AND DISCUSSION
Characterization of strains VRSA-6 and VRSA-7.
Both VRSA-6 and VRSA-7 were resistant to vancomycin (MICs > 256 μg/ml) and teicoplanin (MICs = 16 μg/ml). Multiplex PCR for identification of the van genotype and of staphylococci at the species level (13) confirmed that VRSA-6 and VRSA-7 were S. aureus and that the glycopeptide resistance was of the VanA type. SmaI-digested genomic DNA of the two strains separated by pulsed-field gel electrophoresis revealed two patterns indicating that the isolates were distinct (data not shown). The stability of the vancomycin resistance was tested by replica plating. In three independent experiments, no derivatives susceptible to vancomycin were obtained. The organization of the vanA gene cluster was determined by PCR mapping; and the fragments had sizes indicating that the vanR, vanS, vanH, vanA, vanX, vanY, and vanZ genes constituting the vanA operon were present and were present in the same order as they are in Tn1546. Both strains were constitutively resistant to MLSB antibiotics, and PCR with specific primers indicated the presence of erm(B) in VRSA-6 and of erm(A) along with erm(C) in VRSA-7. The two strains were also resistant to spectinomycin due to the presence of the ant(9)-Ia gene and to kanamycin and tobramycin due to the ant(4′)-Ia gene.
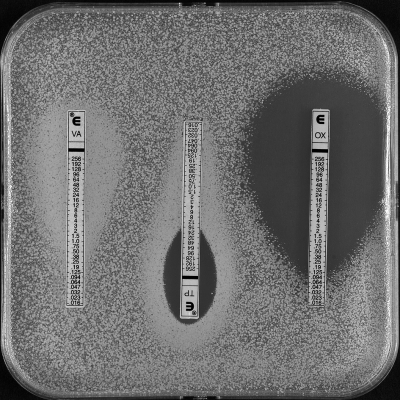
Strain VRSA-7 grew slowly on plates without antibiotic as small colonies with various morphologies but grew normally on plates supplemented with vancomycin (8 μg/ml). Determination of the antimicrobial susceptibility of VRSA-7 by Etest showed a high-density culture surrounding the vancomycin strip or the edge of the teicoplanin inhibition zone (Fig. 1), suggesting that the strain was partially dependent on a glycopeptide for growth. The effect of exposure to vancomycin (8 μg/ml) on the growth of VRSA-7 was determined in liquid medium, which better revealed the growth of the strain in the presence of vancomycin (Fig. 2).
FIG. 1.
Determination of MICs for VRSA-7 by Etest. VA, vancomycin; TP, teicoplanin; OX, oxacillin.
FIG. 2.
Effect of exposure to vancomycin (Vm; 8 μg/ml) on growth of VRSA-7. O.D., optical density.
Transfer of antibiotic resistance from VRSA-7.
Attempts to transfer VanA-type resistance from strain VRSA-7 to E. faecalis JH2-2 and E. faecium 64/3 by conjugation were unsuccessful. However, vancomycin, erythromycin, and spectinomycin resistance was cotransferred to S. aureus BM4602 at a frequency of ca. 1. 10−5 per donor cell. The transconjugants acquired, in addition to the vanA gene cluster, the erm(A) and ant(9)-Ia genes but not erm(C) or ant(4′)-Ia. The SmaI-generated patterns of four randomly selected transconjugants were indistinguishable and differed from that of the recipient by a fragment of ca. 35-40 kb that was also present in the donor and that hybridized with the vanA probe (data not shown).
Plasmid analysis and localization of the resistance genes in VRSA-7.
The vanA gene cluster in strain VRSA-7 was assigned to plasmid pIP848 by contour-clamped homogeneous electric field gel electrophoresis of total DNA digested with I-CeuI, followed by successive hybridization with 16S rRNA (rrs) and vanA probes (data not shown). The copy numbers of the vanA and vanH genes relative to those of the ddl, rpoB, and rrs chromosomal genes were estimated by qPCR from three independent experiments to be 4.6 ± 1.2 and 5.2 ± 1.9, respectively. Electrotransformation of the VRSA-7 plasmid DNA content into BM4602 with selection on erythromycin (8 μg/ml) gave rise to transformants that were resistant only to MLSB antibiotics. The plasmid DNA EcoRI restriction patterns of the transconjugants and of the transformants differed from each other: the former harbored only pIP848, whereas two smaller plasmids were present in the transformants. Hybridization experiments indicated that the vanA, erm(A), and ant(9)-Ia genes were located on the ca. 35- to 40-kb pIP848 conjugative plasmid present at five copies per cell and that erm(C) was carried by one of the other plasmids in VRSA-7 (data not shown).
Sequences of the ddl genes of VRSA-6 and VRSA-7.
Vancomycin dependence results from mutations that impair the host Ddl (39). The sequences of the ddl gene of strain VRSA-7 and the ddl gene of strain VRSA-6, which was used as a control, were determined; and the deduced amino acid sequences were compared (Fig. 3). The ddl sequence of VRSA-6 was identical to that of vancomycin-susceptible S. aureus COL (GenBank accession no. NC_002951), whereas the Ddl of VRSA-7 had a single point mutation at position 308 that led to an asparagine-to-lysine (N308K) substitution. The asparagine at position 308 corresponds to one of the highly conserved residues of the active site of Ddls, and its substitution is therefore likely to affect the activity of the enzyme (Fig. 3).
FIG. 3.
Alignment of the amino acid sequences of VRSA-6 Ddl, VRSA-7 Ddl, and E. coli DdlB. The amino acid substitution N308K is indicated by an arrow. Numbers refer to the positions of the amino acids in DdlB. Conserved residues are highlighted in dark gray. The active-site residues conserved in all Ddls are underlined.
Functionality of Ddl of VRSA-7.
To the best of our knowledge, mutation of the N308 residue in the Ddl has not been described previously. It has been demonstrated that the introduction of a ddl gene coding for a functional ligase into VanD-type glycopeptide-resistant E. faecium BM4339 (vancomycin MIC = 64 μg/ml), which has an impaired Ddl, restores its susceptibility to glycopeptides (8). To determine if the N308K mutation was responsible for the loss of activity of the Ddl, the ddl genes from strains VRSA-7 and VRSA-6 were cloned under the control of the P2 promoter in pAT79 (4); and the recombinant plasmids obtained, pAT516 (pAT79ΩP2ddlVRSA-7) and pAT515 (pAT79ΩP2ddlVRSA-6), respectively, were electrotransformed into BM4339. Transformant BM4339/pAT516 (P2ddlVRSA-7) had the same level of vancomycin resistance as BM4339 (vancomycin MIC = 64 μg/ml), whereas BM4339/pAT515 (P2ddlVRSA-6) was less resistant (MIC = 16 μg/ml).
The ca. 41-kDa C-terminal His6-tagged VRSA-6 Ddl and VRSA-7 Ddl overproduced from E. coli BL21(DE3)pLys harboring plasmid pAT518 [pET28a(+)ΩddlVRSA-6] or pAT519 [pET28a(+)ΩddlVRSA-7], respectively, were obtained as soluble proteins and were >99% pure, as determined by SDS-PAGE analysis (data not shown). The Mr of Ddl VRSA-7, determined by size-exclusion chromatography, was estimated to be 82,000, suggesting that the dimeric structure was preserved. Kinetic analysis indicated that the VRSA-6 Ddl had Kms of 1.2 mM and 17 mM for d-Ala subsite 1 (Km1) and d-Ala subsite2 (Km2), respectively, and kcats of 1,905 min−1 (Table 2). The kcat of the N308K mutant Ddl of VRSA-7 was found to be 250-fold lower than that of the VRSA-6 Ddl. The N308K mutation also affected the binding affinity of d-Ala at subsite 2, where the Km2 was 4.5-fold higher than that of the VRSA-6 Ddl. Overall, the VRSA-7 Ddl was approximately 1,000-fold less efficient than the VRSA-6 Ddl, as indicated by the relative kcat/Km2 catalytic efficiency values (Table 2). The Km values for ATP were similar, indicating that the N308K mutation did not affect the ATP affinity binding site. The crystal structure of the Ddl of S. aureus COL (StaDDl) in complex with ADP has been reported previously (23). However, the crystal structure of substrate analog-bound StaDDl is not available, and the role of the N308 residue is not clearly identified. However, modeling of the homology with other ligases suggests that the side chain nitrogen of N308, the side chain of R291, and the main chain amide of G312 form an oxyanion hole that probably interacts with the phosphate of the d-alanyl-phosphate intermediate. The N308K mutation thus presumably affects the binding of the transition-state intermediate, leading to the loss of activity.
TABLE 2.
Kinetic parameters of Ddl VRSA-6 and Ddl VRSA-7 ligases for d-Ala-d-Ala synthesis
| Ddl source | Km1a (mM) | Km2b (mM) | kcat(min−1) | kcat/Km2(min−1/mM) | Relative kcat/Km2 | ATP Kmc(mM) |
|---|---|---|---|---|---|---|
| VRSA-6 | 1.2 | 17 | 1,925 | 113 | 1 | 0.70 |
| VRSA-7 | NDd | 76.5e | 7.6f | 0.01 | 0.9 × 10−3 | 0.86 |
Km1, Km at subsite 1.
Km2, Km at subsite 2.
Determined with 10 mM d-Ala for the VRSA-6 Ddl and 80 mM d-Ala for the VRSA-7 Ddl.
ND, not detectable.
The highest substrate concentration tested.
Activity was measured with 50 μg (1.25 μM) of enzyme.
Expression of vanA gene cluster.
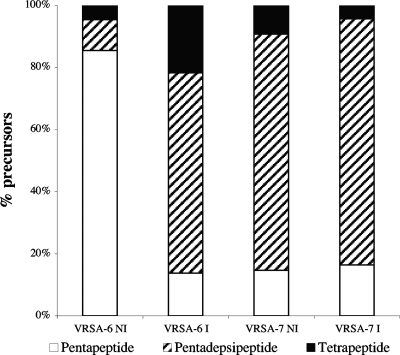
The level of expression of the vanA operon in strains VRSA-7 and VRSA-6 was determined by comparative analysis of the nature and relative amounts of cytoplasmic peptidoglycan precursors (Fig. 4). In the absence of induction by vancomycin, pentapeptides were the main late precursors synthesized by VRSA-6 (86%), whereas induction by vancomycin resulted in the presence of pentapeptides (22%), pentadepsipeptides (64%), and tetrapeptides (14%). Strain VRSA-7 synthesized mainly pentadepsipeptide precursors (ca. 80%) without or with induction. The synthesis of very low levels of pentapeptide precursors (15%) in VRSA-7 could be due to the residual activity of the host ligase or to that of the VanA d-Ala:d-Lac ligase, which can also synthesize d-Ala-d-Ala (7). The loose regulation of the vanA operon by the VanR-VanS system, which allows low-level expression of the vanA operon in the absence of induction (3), combined with a gene dosage effect due to the location of the vanA gene cluster on a multicopy plasmid, could account for the significant expression of the vanA operon in the absence of an inducer in VRSA-7. These data, combined with the facts that VRSA-7 grows more efficiently in the presence of glycopeptides and that the sequences of vanR and vanS in this strain had no mutations, indicate that the expression of the vanA operon is inducible.
FIG. 4.
Cytoplasmic peptidoglycan precursors in VRSA-6 and VRSA-7. NI, not induced; I, induced by growth in the presence of 8 μg/ml of vancomycin.
Methicillin resistance in VRSA-7.
To date, all S. aureus isolates that have acquired the vanA operon also possess the mecA gene. Methicillin resistance is due to the synthesis of an additional PBP, PBP 2′, encoded by the mecA gene, which exhibits a low affinity for β-lactams (31). PBP 2′ is the only transpeptidase that remains active in the presence of β-lactams in the medium (11). It has been demonstrated that pentadepsipeptide peptidoglycan precursors ending in d-Ala-d-Lac are not substrates for PBP 2′ (35), resulting in a strong synergism between glycopeptides and ß-lactams against VRSA isolates (17, 29). Strain VRSA-7 was susceptible to oxacillin (MIC = 0.125 μg/ml), even in the absence of glycopeptides in the culture medium. Determination of the sequence of a 2,291-bp PCR fragment containing the mecA gene and its promoter revealed no mutations. The PBP 2′ protein, which has an expected size of 76 kDa, was detected by Western blotting with an anti-PBP 2′ monoclonal antibody (data not shown). It therefore appears that inactivation of the VRSA-7 Ddl and acquisition of the vanA gene cluster, which are responsible for a lack of peptidoglycan precursors ending in d-Ala-d-Ala and the synthesis of precursors mainly ending in d-Ala-d-Lac, respectively, result in the loss of resistance to ß-lactams, despite the presence of a functional mecA gene.
In summary, the N308K mutation in the Ddl of strain VRSA-7 has impaired the activity of the enzyme, and thus, the strain must rely on the vancomycin-inducible activity of the acquired VanA d-Ala:d-Lac ligase for growth. The basal level of expression of the vanA operon due to a combination of loose regulation by the two-component regulatory system and a gene dosage effect accounts for the slow growth of VRSA-7 in the absence of vancomycin, whereas following induction, full expression of the operon allows the normal growth of the strain. The synthesis of pentadepsipeptide peptidoglycan precursors by VRSA-7, even in the absence of vancomycin in the medium, that cannot be processed by PBP 2′ is responsible for susceptibility to ß-lactams. Thus, it is conceivable that patients infected with this type of strain could be treated with ß-lactams, despite the presence of a functional mecA gene, a hypothesis that remains to be critically tested in an animal model. The prevalence of clinical isolates of VRSA partially dependent on vancomycin for growth may be underestimated, since these strains can be missed in routine laboratory practice because of their particular nutritional requirement.
Acknowledgments
We thank M. Page for the gift of the anti-PBP 2′ monoclonal antibody and P. E. Reynolds for critical review of the manuscript. S. aureus strains were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus.
This work was supported in part by the Institut National de la Veille Sanitaire.
Footnotes
Published ahead of print on 15 June 2009.
REFERENCES
- 1.Aligholi, M., M. Emaneini, F. Jabalameli, S. Shahsavan, H. Dabiri, and H. Sedaght. 2008. Emergence of high-level vancomycin-resistant Staphylococcus aureus in the Imam Khomeini Hospital in Tehran. Med. Princ. Pract. 17:432-434. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, M., F. Depardieu, C. Molinas, P. Reynolds, and P. Courvalin. 1995. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 154:87-92. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, M., F. Depardieu, P. Reynolds, and P. Courvalin. 1996. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol. Microbiol. 21:33-44. [DOI] [PubMed] [Google Scholar]
- 4.Arthur, M., C. Molinas, and P. Courvalin. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arthur, M., P. E. Reynolds, and P. Courvalin. 1996. Glycopeptide resistance in enterococci. Trends Microbiol. 4:401-407. [DOI] [PubMed] [Google Scholar]
- 7.Bugg, T. D., S. Dutka-Malen, M. Arthur, P. Courvalin, and C. T. Walsh. 1991. Identification of vancomycin resistance protein VanA as a d-alanine:d-alanine ligase of altered substrate specificity. Biochemistry 30:2017-2021. [DOI] [PubMed] [Google Scholar]
- 8.Casadewall, B., and P. Courvalin. 1999. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J. Bacteriol. 181:3644-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comité de l' A ntibiogramme de la S ociété F rançaise de M icrobiologie. 2009. Communiqué 2009. Comité de l'Antibiogramme de la Société Française de Microbiologie, Paris, France. http://www.sfm.asso.fr.
- 10.Daub, E., L. E. Zawadzke, D. Botstein, and C. T. Walsh. 1988. Isolation, cloning, and sequencing of the Salmonella typhimurium ddlA gene with purification and characterization of its product, d-alanine:d-alanine ligase (ADP forming). Biochemistry 27:3701-3708. [DOI] [PubMed] [Google Scholar]
- 11.Dejonge, B. L. M., Y. S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. J. Biol. Chem. 267:11248-11254. [PubMed] [Google Scholar]
- 12.Depardieu, F., M. G. Bonora, P. E. Reynolds, and P. Courvalin. 2003. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 50:931-948. [DOI] [PubMed] [Google Scholar]
- 13.Depardieu, F., B. Perichon, and P. Courvalin. 2004. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J. Clin. Microbiol. 42:5857-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dever, L. L., S. M. Smith, S. Handwerger, and R. H. K. Eng. 1995. Vancomycin-dependent Enterococcus faecium isolated from stool following oral vancomycin therapy. J. Clin. Microbiol. 33:2770-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyke, K. G., M. P. Jevons, and M. T. Parker. 1966. Penicillinase production and intrinsic resistance to penicillins in Staphylococcus aureus. Lancet i:835-838. [DOI] [PubMed] [Google Scholar]
- 16.Farrag, N., I. Eltringham, and H. Liddy. 1996. Vancomycin-dependent Enterococcus faecalis. Lancet 348:1581-1582. [DOI] [PubMed] [Google Scholar]
- 17.Fox, P. M., R. J. Lampert, K. S. Stumpf, G. L. Archer, and M. W. Climo. 2006. Successful therapy of experimental endocarditis caused by vancomycin-resistant Staphylococcus aureus with a combination of vancomycin and ß-lactam antibiotics. Antimicrob. Agents Chemother. 50:2951-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greisen, K., M. Loeffelholz, A. Purohit, and D. Leong. 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32:335-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi, N., M. Alam, Y. Nishimoto, S. Urasawa, N. Uehara, and N. Watanabe. 2001. Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium. Epidemiol. Infect. 126:197-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 22.Lessard, I. A., V. L. Healy, I. S. Park, and C. T. Walsh. 1999. Determinants for differential effects on d-Ala-d-lactate vs d-Ala-d-Ala formation by the VanA ligase from vancomycin-resistant enterococci. Biochemistry 38:14006-14022. [DOI] [PubMed] [Google Scholar]
- 23.Liu, S., J. S. Chang, J. T. Herberg, M. M. Horng, P. K. Tomich, A. H. Lin, and K. R. Marotti. 2006. Allosteric inhibition of Staphylococcus aureus d-alanine:d-alanine ligase revealed by crystallographic studies. Proc. Natl. Acad. Sci. USA 103:15178-15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messer, J., and P. E. Reynolds. 1992. Modified peptidoglycan precursors produced by glycopeptide-resistant enterococci. FEMS Microbiol. Lett. 94:195-200. [DOI] [PubMed] [Google Scholar]
- 25.Miller, D., V. Urdaneta, A. Weltman, and S. Park. 2002. Vancomycin-resistant Staphylococcus aureus. MMWR Morb. Mortal. Wkly. Rep. 51:902. [PubMed] [Google Scholar]
- 26.Neuhaus, F. C. 1962. The enzymatic synthesis of d-alanyl-d-alanine. II. Kinetic studies on d-alanyl-d-alanine synthase. J. Biol. Chem. 237:3128-3135. [PubMed] [Google Scholar]
- 27.O'Hara, D. M., and P. E. Reynolds. 1987. Antibody used to identify penicillin-binding protein 2′ in methicillin-resistant strains of Staphylococcus aureus (MRSA). FEBS Lett. 212:237-241. [DOI] [PubMed] [Google Scholar]
- 28.Peirson, S. N., J. N. Butler, and R. G. Foster. 2003. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 31:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Périchon, B., and P. Courvalin. 2006. Synergism between beta-lactams and glycopeptides against VanA-type methicillin-resistant Staphylococcus aureus and heterologous expression of the vanA operon. Antimicrob. Agents Chemother. 50:3622-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Périchon, B., P. Reynolds, and P. Courvalin. 1997. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob. Agents Chemother. 41:2016-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds, P. E., and C. Fuller. 1986. Methicillin-resistant strains of Staphylococcus aureus; presence of identical additional penicillin-binding protein in all strains examined. FEMS Microbiol. Lett. 33:251-254. [Google Scholar]
- 32.Saha, B., A. K. Singh, A. Ghosh, and M. Bal. 2008. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). J. Med. Microbiol. 57:72-79. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Schmitz, F. J., A. C. Fluit, M. Gondolf, R. Beyrau, E. Lindenlauf, J. Verhoef, H. P. Heinz, and M. E. Jones. 1999. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 43:253-259. [PubMed] [Google Scholar]
- 35.Severin, A., K. Tabei, F. Tenover, M. Chung, N. Clarke, and A. Tomasz. 2004. High level oxacillin and vancomycin resistance and altered cell wall composition in Staphylococcus aureus carrying the staphylococcal mecA and the enterococcal vanA gene complex. J. Biol. Chem. 279:3398-3407. [DOI] [PubMed] [Google Scholar]
- 36.Sievert, D. M., M. L. Boulton, G. Stolman, D. Johnson, M. G. Stobierski, F. P. Downes, P. A. Somsel, J. T. Rudrik, W. Brown, W. Hafeez, T. Lundstrom, E. Flanagan, R. Johnson, J. Mitchell, and S. Chang. 2002. Staphylococcus aureus resistant to vancomycin. MMWR Morb. Mortal. Wkly. Rep. 51:565-567.12139181 [Google Scholar]
- 37.Sievert, D. M., J. T. Rudrik, J. B. Patel, L. C. McDonald, M. J. Wilkins, and J. C. Hageman. 2008. Vancomycin-resistant Staphylococcus aureus in the United States, 2002-2006. Clin. Infect. Dis. 46:668-674. [DOI] [PubMed] [Google Scholar]
- 38.Sifaoui, F., and L. Gutmann. 1997. Vancomycin dependence in a VanA-producing Enterococcus avium strain with a nonsense mutation in the natural d-Ala-d-Ala ligase gene. Antimicrob. Agents Chemother. 41:1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Bambeke, F., M. Chauvel, P. E. Reynolds, H. S. Fraimow, and P. Courvalin. 1999. Vancomycin-dependent Enterococcus faecalis clinical isolates and revertant mutants. Antimicrob. Agents Chemother. 43:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werner, G., R. J. Willems, B. Hildebrandt, I. Klare, and W. Witte. 2003. Influence of transferable genetic determinants on the outcome of typing methods commonly used for Enterococcus faecium. J. Clin. Microbiol. 41:1499-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, W., N. C. Clark, L. K. McDougal, J. Hageman, L. C. McDonald, and J. B. Patel. 2008. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 52:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]