Abstract
Thirteen Candida glabrata strains harboring a range of mutations in hot spot regions of FKS1 and FKS2 were studied. The mutations were linked to an echinocandin reduced susceptibility phenotype. Sequence alignments showed that 11 out of the 13 mutants harbored a mutation in FKS1 or FKS2 not previously implicated in echinocandin reduced susceptibility in C. glabrata. A detailed kinetic characterization demonstrated that amino acid substitutions in Fks1p and Fks2p reduced drug sensitivity in mutant 1,3-β-d-glucan synthase by 2 to 3 log orders relative to that in wild-type enzyme. These mutations were also found to reduce the catalytic efficiency of the enzyme (Vmax) and to influence the relative expression of FKS genes. In view of the association of FKS mutations and reduced susceptibility of 1,3-β-d-glucan synthase, an evaluation of the new CLSI echinocandin susceptibility breakpoint was conducted. Only 3 of 13 resistant fks mutants (23%) were considered anidulafungin or micafungin nonsusceptible (MIC > 2 μg/ml) by this criterion. In contrast, most fks mutants (92%) exceeded a MIC of >2 μg/ml with caspofungin. However, when MIC determinations were performed in the presence of 50% serum, all C. glabrata fks mutants showed MICs of ≥2 μg/ml for the three echinocandin drugs. As has been observed with Candida albicans, the kinetic inhibition parameter 50% inhibitory concentration may be a better predictor of FKS-mediated resistance. Finally, the close association between FKS1/FKS2 hot spot mutations provides a basis for understanding echinocandin resistance in C. glabrata.
Candida glabrata is the second most-common fungal species isolated from blood in the United States and one of the most common fungal pathogens worldwide. C. glabrata infections are a concern due to their high mortality rates (38) and the tendency of this microorganism to rapidly develop resistance to azole antifungal agents (6, 11, 15, 25, 31, 34). The introduction of the antifungal drug caspofungin (CSF) in 2001 was a significant advance in the treatment of these infections since these lipopeptides' mode of action is independent of existing antifungal drugs (7, 16, 36). All three echinocandin drugs, CSF, anidulafungin (ANF), and micafungin (MCF), inhibit 1,3-β-d-glucan synthase, which disrupts the structure of the growing cell wall, causing osmotic instability and death of susceptible yeast cells (8, 22). 1,3-β-d-Glucan synthase is a protein complex formed at least by catalytic subunits (Fksp) and a ubiquitous regulatory element (Rho1). Echinocandin drugs are presumed to bind to Fksp, which is encoded by three putative FKS genes in Candida spp. and Saccharomyces spp. (FKS1, FKS2, and FKS3) (8, 9, 12, 17, 21). Echinocandin reduced susceptibility in Candida spp. has been linked to mutations in two highly conserved hot spot regions of FKS genes (22). Recently, three reports showed that amino acid substitutions in Fks1p (D632E) and Fks2p (F659V) are responsible for clinical echinocandin resistance in C. glabrata (3, 17, 33). The aim of this study was to investigate the linkage between echinocandin resistance and FKS mutations in a collection of clinical C. glabrata strains exhibiting reduced susceptibility to echinocandin drugs. Moreover, a detailed kinetic analysis was performed to assess the effect of the different amino acid substitutions on 1,3-β-d-glucan synthase complex kinetic parameters and inhibition by different echinocandin drugs. Finally, the new CLSI echinocandin susceptibility breakpoint was evaluated to establish its value in identifying echinocandin-resistant C. glabrata strains with FKS mutations.
MATERIALS AND METHODS
Strains.
Fifteen clinical C. glabrata strains were used throughout this work. Two strains were susceptible to echinocandin drugs (strains 218 and 3168). The others showed CSF MICs of ≥2 μg/ml. Twelve of the 13 resistant strains were isolated from patients on CSF therapy or prophylaxis, while strain 234 was isolated after ANF therapy. Strain ATCC 90030 was used as wild-type control isolate. Strains 3168 (echinocandin susceptible) and 3169 (echinocandin resistant) are isogenic (3). The isolates were identified as C. glabrata by conventional microbiologic methods. The initial identification was confirmed by sequencing of the 5.8S RNA gene and adjacent internal transcribed spacer regions 1 and 2 (35). Molecular identification was performed in order to avoid misidentification with the C. glabrata novel anamorphic related species Candida bracarensis (5) and Candida nivariensis (1).
Antifungal susceptibility testing and compounds.
Echinocandin drug susceptibility testing was performed in triplicate in accordance with the CLSI document M27-A3 guidelines (4) in the presence or absence of 50% human serum (Sigma-Aldrich) (20). Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 were used as control strains for antifungal susceptibility testing. The drugs used were CSF (Merck & Co. Inc., Rahway, NJ), ANF (Pfizer, New York, NY), and MCF (Astellas Pharma USA, Inc., Deerfield, IL). The drugs were obtained as standard powders from their manufacturers. CSF and MCF were dissolved in sterile distilled water, and ANF was dissolved in 100% dimethyl sulfoxide (Sigma-Aldrich). Stock solutions of each drug were kept at −86°C.
FKS gene sequence analysis.
C. glabrata genomic DNA was extracted from yeast cells grown overnight in YPD (2% yeast extract, 4% Bacto peptone, 4% dextrose) broth medium with a Q-Biogene (Irvine, CA) FastDNA kit. PCR and sequencing primers were designed based on the C. glabrata FKS1, FKS2, and FKS3 gene sequences (GenBank accession no. XM_446406, XM_448401, and XM_449945, respectively). DNA sequencing was performed with a CEQ dye terminator cycle sequencing quick start kit (Beckman Coulter, Fullerton, CA) according to the manufacturer's recommendations. Sequence analysis was performed with CEQ 8000 genetic analysis system software (Beckman Coulter, Fullerton, CA) and BioEdit sequence alignment editor (Ibis Therapeutics, Carlsbad, CA).
1,3-β-d-Glucan synthase isolation and assay.
All isolates used in this work were grown with vigorous shaking at 35°C to early stationary phase in YPD broth, and cells were collected by centrifugation. Cell disruption, membrane protein extraction, and partial 1,3-β-d-glucan synthase purification by-product entrapment were performed, as described previously (21). Sensitivity to echinocandin drugs was measured in a polymerization assay, using a 96-well multiscreen high-throughput screen filtration system (Millipore corporation, Bedford, MA) with a final volume of 100 μl (12, 21). Serial dilutions of echinocandin drugs (0.01 to 10,000 ng/ml) were used to determine 50% inhibitory concentration (IC50) values. Control reactions were performed in the presence of 1% dimethyl sulfoxide when ANF was used. The reactions were initiated by addition of the purified 1,3-β-d-glucan synthase enzyme. Inhibition profiles and IC50 values were determined using a sigmoidal response (variable-slope) curve fitting algorithm with GraphPad Prism software (version 4.0; Prism Software, Irvine, CA).
Characterization of 1,3-β-d-glucan product.
The product of the reaction mixtures was verified as 1,3-β-d-glucan by using a Glucatell kit (Associates of Cape Cod, Inc., Falmouth, MA), following the procedure previously described (12).
Kinetic analyses.
All reactions were run in a 96-well multiscreen high-throughput screen filtration system (Millipore) with a final volume of 100 μl as previously described (12, 14). [3H]uridine diphosphoglucose (UDPG) was used as the substrate in concentrations ranging from 0.015 to 2 mM to determine the different kinetic parameters, which were analyzed by linear regressions to obtain slopes in dpm/min. This value was then converted to nM of glucose incorporated per minute. The maximum velocity (Vmax) and the Michaelis-Menten constant (Km) were determined for trapped 1,3-β-d-glucan synthase complex by varying the amount of UDPG (between 0.015 and 2 mM) using Lineweaver-Burke plots.
RNA isolation and expression profiling.
C. glabrata strains were grown in YPD at 37°C with shaking (150 rpm) for 16 h. Total RNA was extracted using an RNeasy mini kit (Qiagen), and gene expression profiles were performed using a one-step SYBR green quantitative reverse transcription-PCR kit (Stratagene, La Jolla, CA) with the Stratagene Mx3005P multiplex quantitative PCR system. Differential expression was analyzed for the three C. glabrata FKS genes. The relative expression levels were evaluated using the Pfaffl method (23). The C. glabrata URA3 gene (GenBank accession no. AY771209) was used for normalization. The primers used for gene expression profiling were described previously (12).
Statistical analysis.
The kinetic data are the result of experiments performed in triplicate. Arithmetic means and standard deviations were used to statistically analyze all the continuous variables (IC50, Km, and Vmax). Geometric means were used to statistically compare MIC results. The significance levels of MIC differences and kinetic parameters were determined by Student's t test (unpaired, unequal variance); a P value of 0.05 was considered significant. In order to approximate a normal distribution, the MICs were transformed to log2 values to establish susceptibility differences between strains. Both on-scale and off-scale results were included in the analysis. The off-scale MICs were converted to the next concentration up or down. MICs were compared with IC50s by using plots of the log2 MIC versus log10 IC50. The MIC-IC50 breakpoint was defined as the MIC for which ≥90% of the strains had IC50s for 1,3-β-d-glucan synthase at least 50-fold higher than that for the wild-type enzyme. Statistical analyses were performed with the Statistical Package for the Social Sciences software (version 13.0; SPSS, Inc., Chicago, IL).
RESULTS
Genetic analysis of FKS genes from echinocandin-resistant C. glabrata isolates.
All C. glabrata strains exhibiting high echinocandin MICs showed mutations in either FKS1 (n = 4), FKS2 (n = 8), or both FKS genes (n = 1) (Table 1). Conversely, FKS3 genes harbored only silent mutations compared with wild-type strain FKS3 gene (data not shown). All of the mutations producing amino acid substitutions were found in one of the FKS mutational hot spots previously described for Candida spp. (3, 12, 14, 17, 21, 22, 33). Sequence alignments showed that 11 out of the 13 mutants in this collection harbor a mutation not previously implicated in echinocandin resistance in C. glabrata. The majority of the FKS1 mutants showed an amino acid substitution at Asp632 (D632G, D632E, and D632Y), while the FKS2 mutants harbor two different amino acid substitutions (F659V and F659S) and an amino acid deletion at Phe659 (F659del). The Phe659 deletion at Fks2p is due to a 3-nucleotide deletion (1974-CTT-1976), which maintains the same open reading frame but with a single frameshift. Strain 42971 is a double mutant with a mutation in FKS1 (G1894T) and a nonsense mutation in FKS2 producing a premature translation termination at codon Arg1377. This isolate and strain 5416 were the only two strains in the collection with an amino acid substitution at hot spot 2 (Fks2p).
TABLE 1.
In vitro whole-cell susceptibility (MIC) to and 1,3-β-d-glucan synthase inhibition profiles (IC50) of echinocandin drugs for the strains included in the study
| Strain | Genotype
|
Phenotype
|
MICc
|
IC50d
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANF
|
CSF
|
MCF
|
|||||||||||
| FKS1 | FKS2 | Fks1pa | Fks2pb | No serum | 50% Serum | No serum | 50% Serum | No serum | 50% Serum | ANF | CSF | MCF | |
| 90030 | WT | WT | WT | WT | 0.06 | 0.50 | 0.06 | 0.12 | 0.03 | 0.25 | 1.91 | 1.92 | 0.71 |
| 218 | WT | WT | WT | WT | 0.06 | 0.50 | 0.12 | 0.25 | 0.12 | 1.00 | 1.25 | 3.11 | 0.56 |
| 3168 | WT | WT | WT | WT | 0.03 | 0.50 | 0.06 | 0.12 | 0.06 | 0.50 | 1.58 | 1.79 | 1.29 |
| 42997 | T1874C | WT | F625S | WT | 2.00 | 8.00 | 8.00 | 8.00 | 0.50 | 4.00 | 110.6 | 697.8 | 126.3 |
| 5847 | T1885C | WT | S629P | WT | 4.00 | 8.00 | 8.00 | 8.00 | 2.00 | 8.00 | 3855.4 | 4987.5 | 854.5 |
| 21900 | A1895G | WT | D632G | WT | 1.00 | 8.00 | 4.00 | 16.00 | 0.50 | 8.00 | 101.8 | 756.3 | 343.4 |
| 3169 | T1896G | WT | D632E | WT | 2.00 | 8.00 | 2.52 | 8.00 | 2.52 | 8.00 | 151.5 | 844.5 | 444.7 |
| 42971 | G1894T | A4129T | D632Y | R1377STOP | 4.00 | 8.00 | 8.00 | 8.00 | 1.00 | 8.00 | 2543.5 | 5248.5 | 413.4 |
| 31498 | WT | Del1974-1976 | WT | F659del | 2.00 | 8.00 | 2.00 | 8.00 | 4.00 | 8.00 | 160 | 117.2 | 123.5 |
| 234 | WT | T1975G | WT | F659V | 1.00 | 8.00 | 4.00 | 8.00 | 1.00 | 8.00 | 86.6 | 115.3 | 113.5 |
| 41026 | WT | T1976C | WT | F659S | 4.00 | 16.00 | 16.00 | 16.00 | 4.00 | 16.00 | 858.1 | 1064.0 | 1067.0 |
| 3830 | WT | T1987C | WT | S663P | 1.00 | 8.00 | 16.00 | 16.00 | 1.00 | 8.00 | 4003.5 | 5115.5 | 2569 |
| 41400 | WT | A1997G | WT | D666G | 2.00 | 8.00 | 4.00 | 8.00 | 0.12 | 4.00 | 175.8 | 506.6 | 100.6 |
| 42031 | WT | C1998A | WT | D666E | 1.00 | 4.00 | 4.00 | 8.00 | 0.25 | 4.00 | 127.4 | 257.9 | 88.7 |
| 51916 | WT | C1999A | WT | P667T | 1.59 | 8.00 | 2.00 | 8.00 | 0.40 | 4.00 | 99.5 | 166.2 | 110.9 |
| 5416 | WT | G4125A | WT | W1375L | 0.25 | 4.00 | 4.00 | 8.00 | 0.25 | 2.00 | 85.2 | 608.4 | 50.7 |
WT, wild type at hot spots. Fks1p hot spot number one includes amino acids between 625 and 633 (625-FLILSLRDP-633).
WT, wild type at hot spots. Fks2p hot spot number one includes amino acids between 659 and 667 (659-FLILSLRDP-667). Fks2p hot spot number two comprises amino acids between 1374 and 1381 (WT, 1374- DWVRRYTL-1381).
Geometric mean values in μg ml−1 (three repetitions on three separate days).
Arithmetic mean (three repetitions on three separate days) IC50s obtained using trapped 1,3-β-d-glucan synthase enzyme expressed in ng ml−1.
Fks1p and Fks2p amino acid substitutions confer reduced echinocandin susceptibility in C. glabrata.
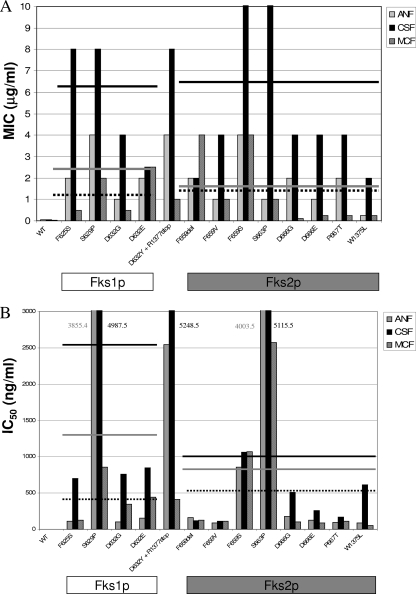
All the C. glabrata strains with Fksp amino acid substitutions in at least one of the hot spots showed statistically significant reduced in vitro echinocandin susceptibility compared with that of wild type strains (40-fold [P = 0.002], 79-fold [P = 0.005], and 19-fold [P = 0.039] for ANF, CSF, and MCF, respectively). These differences were lower when MICs were obtained in the presence of serum (16-, 60-, and 12-fold for ANF, CSF and MCF, respectively). However, when serum was used, all the mutant strains showed MICs that surpassed the MIC susceptibility breakpoint proposed by the CLSI (4). All mutant strains exhibited statistically significant higher MICs for CSF than for the other drugs in the absence of serum (P = 0.004 for CSF versus ANF, and P = 0.001 for CSF versus MCF). On the other hand, MCF was the most potent of the three echinocandin drugs against C. glabrata fks mutants. These differences between drug potencies disappear when the MICs were obtained in the presence of serum, as previously reported (19, 20, 37) (P = 0.153 and P = 0.066 when ANF and MCF, respectively, were compared with CSF) (Table 1). Echinocandin MICs varied with the specific mutations, but there was little difference in the average MICs between FKS1 and FKS2 mutants (Fig. 1). The highest MIC increases were seen with C. glabrata isolates harboring mutations F625S and S629P in Fks1p and the equivalent mutations in Fks2p, F659S and S663P (Table 1).
FIG. 1.
Summary of echinocandin MIC (A) and IC50 (B) values for the C. glabrata strains harboring various Fks1p and Fks2p amino acid substitutions. Lines represent average MICs and IC50s for ANF, CSF, and MCF for the group of mutants harboring amino acid substitutions in each Fks protein. MICs were obtained using the CLSI document M27-A3 guidelines, and IC50s were obtained using product entrapped partially purified 1,3-β-d-glucan synthase enzyme complexes. WT, wild type.
Effect of Fksp substitutions on 1,3-β-d-glucan synthase inhibition.
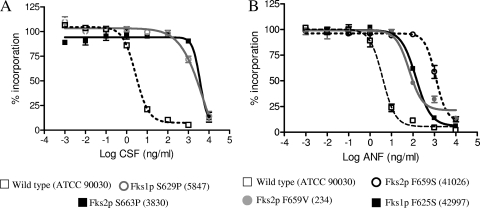
The kinetic inhibition parameter IC50 was determined for 1,3-β-d-glucan synthase complexes isolated from fks mutant C. glabrata strains. Overall, the fks mutant enzymes showed significantly higher IC50s than did corresponding enzymes isolated from wild-type strains (54-, 267-, and 60-fold on average for ANF, CSF, and MCF, respectively [P < 0.001 for all the drugs studied]). The CSF IC50s were the highest of the three drugs (693-, 366-, and 577-fold increases for CSF, ANF and MCF, respectively) (Table 1). However, the pattern of 1,3-β-d-glucan synthase echinocandin susceptibility depends on the amino acid substitution and its localization (Table 1). Among the mutant 1,3-β-d-glucan synthase enzyme complexes studied, mutations at Ser629 (Fks1p) and Ser663 (Fks2p) showed the highest IC50s for all echinocandin drugs (P = 0.011), followed by the F659S (Fks2p) (Table 1 and Fig. 2). Special consideration was required for the enzyme complex isolated from the strain 42971 (D632Y substitution in Fks1p coupled with an FKS2 onsense mutation at nucleotide A4129T). This complex showed higher IC50s for ANF and CSF than those obtained for the Fks1p D632G and D632E mutants (20- and sevenfold, respectively). However, there were no differences in MCF IC50s between these three mutants suggesting that the Fks2p lack of function does not affect the echinocandin drug interaction with the complex. The Phe659 mutants (Fks2p) showed different IC50s depending on which amino acid was replaced. The 1,3-β-d-glucan synthase complex harboring the F659S substitution in Fks2p showed echinocandin IC50s 10-fold higher than those of the F659V and the F659del mutant enzymes (Table 1 and Fig. 2). Finally, amino acid substitutions in the C termini of hot spot 1 (strain 51916) and hot spot 2 (strain 5416) showed the lowest IC50s among the collection, indicating that these enzymes were the most sensitive to drug, which was also reflected in MICs.
FIG. 2.
Echinocandin inhibition profiles for product-entrapped 1,3-β-d-glucan synthase enzyme complexes assessed by the incorporation of [3H]glucose into radiolabeled product. (A) CSF titration curves for enzymes isolated from strains ATCC 90030 (wild type), 5847 (Fks1p-S629P), and 3830 (Fks2p-S663P). (B) ANF inhibition curves for 1,3-β-d-glucan synthase enzyme complexes extracted from strains ATCC 90030 (wild type), 42997 (Fks1p-S625S), 234 (Fks2p-S659V), and 41026 (Fks2p-F659S).
C. glabrata 1,3-β-d-glucan synthase enzyme kinetics.
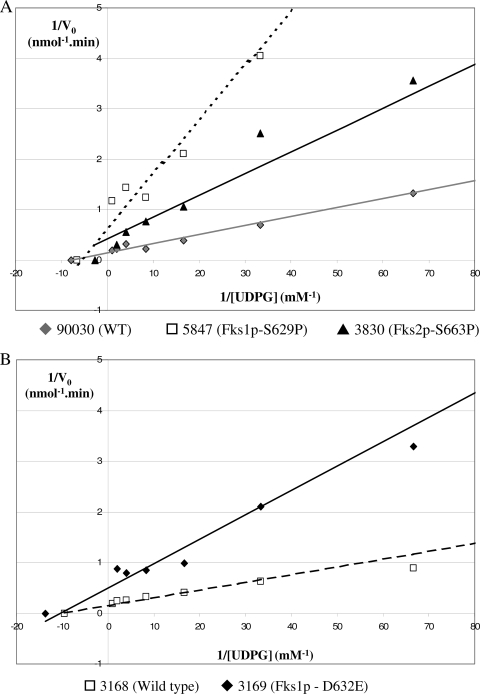
The kinetic parameters Vmax and Km for wild-type enzymes (n = 3) showed average values of 6.812 ± 0.246 nmol/min and 0.133 ± 0.015 mM, respectively (Table 2). Among the mutant enzyme complexes, amino acid substitutions in the hot spot regions generally decreased the catalytic capacity of the mutant enzyme Vmax (on average, 2.380 ± 0.375 nmol/min; P < 0.001 relative to wild-type enzyme). On the other hand, Km values for mutant enzymes were not statistically different than wild-type enzymes (P = 0.660). Amino acid substitutions that alter echinocandin sensitivity decrease enzyme velocity, but they do not change the binding affinity (Km) of the enzyme for echinocandin drugs. These data are consistent with the noncompetitive kinetic behavior of echinocandin drugs on substrate, as described previously (14, 32). The extent of the Vmax reduction depends on the amino acid substitution and its localization. The Fks1p mutant enzymes showed Vmax values lower than those of Fks2p mutant enzymes (arithmetic means ± standard deviations were 1.530 ± 0.208 nmol/min for Fks1p mutants and 2.883 ± 1.455 nmol/min for Fks2p mutants; P = 0.008) (Table 2 and Fig. 3). Fks1p mutant enzymes showed a uniform Vmax reduction with respect to wild-type strains (4.55-fold lower on average). On the other hand, Fks2p mutants showed a wider range of Vmax reduction with the highest Vmax values obtained from strains 3830 and 41400, Fks2p-S663P, and Fks2-D666G, respectively, and the lowest Vmax from strain 51916, Fks2p-P667T. These data demonstrate that a single FKS mutation at one hot spot locus is sufficient to reduce the Vmax of the C. glabrata 1,3-β-d-glucan synthase complex. This view was further supported when the 1,3-β-d-glucan synthase complexes from isogenic strains 3168 and 3169 were shown to have a Vmax reduction of more than threefold (6.55 ± 0.38 nmol/min and 1.70 ± 0.32 nmol/min, respectively) (Table 2 and Fig. 3).
TABLE 2.
Summary of kinetic properties of the C. glabrata 1,3-β-d-glucan synthase complexes included in the study
| Strain | Genotype
|
Phenotype
|
Vmax (nmol min−1)c | K m (mM)c | ||
|---|---|---|---|---|---|---|
| FKS1 | FKS2 | Fks1pa | Fks2pb | |||
| 90030 | WT | WT | WT | WT | 7.04 ± 0.81 | 0.13 ± 0.02 |
| 218 | WT | WT | WT | WT | 6.85 ± 0.52 | 0.12 ± 0.04 |
| 3168 | WT | WT | WT | WT | 6.67 ± 0.18 | 0.10 ± 0.07 |
| 42997 | T1874C | WT | F625S | WT | 1.83 ± 0.59 | 0.17 ± 0.11 |
| 5847 | T1885C | WT | S629P | WT | 1.44 ± 0.95 | 0.15 ± 0.05 |
| 21900 | A1895G | WT | D632G | WT | 1.55 ± 0.41 | 0.10 ± 0.05 |
| 3169 | T1896G | WT | D632E | WT | 1.57 ± 0.38 | 0.07 ± 0.05 |
| 42971 | G1894T | A4129T | D632Y | R1377STOP | 1.26 ± 0.24 | 0.06 ± 0.01 |
| 31498 | WT | Del1975-1977 | WT | F659del | 2.75 ± 0.22 | 0.10 ± 0.04 |
| 234 | WT | T1975G | WT | F659V | 2.87 ± 0.85 | 0.17 ± 0.04 |
| 41026 | WT | T1976C | WT | F659S | 3.05 ± 0.51 | 0.08 ± 0.01 |
| 3830 | WT | T1987C | WT | S663P | 4.45 ± 0.73 | 0.16 ± 0.05 |
| 41400 | WT | A1997G | WT | D666G | 5.05 ± 0.41 | 0.11 ± 0.03 |
| 42031 | WT | C1998A | WT | D666E | 1.85 ± 0.44 | 0.08 ± 0.01 |
| 51916 | WT | C1999A | WT | P667T | 0.33 ± 0.05 | 0.04 ± 0.02 |
| 5416 | WT | G4125A | WT | W1375L | 2.71 ± 0.09 | 0.04 ± 0.01 |
WT, wild type at hot spots. Fks1p hot spot number one includes amino acids between 625 and 633 (625-FLILSLRDP-633).
WT, wild type at hot spots. Fks2p hot spot number one includes amino acids between 659 and 667 (659-FLILSLRDP-667).
Arithmetic mean ± standard deviation (three repetitions).
FIG. 3.
Lineweaver-Burke double-reciprocal plots used to determine the maximum velocity (Vmax) and the Michaelis-Menten constant (Km) for product-entrapped 1,3-β-d-glucan synthase complexes by varying the amount of [3H]UDPG. (A) Comparison of the kinetic properties of the 1,3-β-d-glucan synthase enzymes isolated from strains ATCC 90030 (wild type [WT]), 5847 (Fks1p-S629P), and 3830 (Fks2p-S663P). (B) Evaluation of the kinetic properties of the 1,3-β-d-glucan synthase enzyme obtained from the isogenic strains 3168 (WT) and 3169 (Fks1p-D632G).
C. glabrata FKS gene expression varies with genotype.
SYBR green real-time PCR was used to evaluate the transcription levels of the three different C. glabrata FKS genes relative to C. glabrata URA3 gene. The FKS1/FKS2 ratios suggested that FKS2 is expressed in a proportion ranging between 2:1 to 3:1 with respect to FKS1 in the wild type (n = 3) and in the majority of the FKS2 mutants (n = 6). FKS3 was expressed at a very low level (750-fold lower expression on average) compared to both FKS1 and FKS2 in the complete collection (Table 3). All of the FKS1 mutants (n = 4), one FKS2 mutant (strain 51916; Fks2p P667T), and the double mutant (strain 42971; Fks1p D632Y and truncated Fks2p at the amino acid 1377) showed FKS1/FKS2 ratios of >1. Despite the similar increases in expression ratios in these strains, there were differences found for FKS1 and FKS2 expression values relative to those of the wild type (strain 90030). The FKS1 expression values were significantly higher in the FKS1 mutants than in the rest of the collection (P = 0.01) (data not shown), demonstrating that the higher FKS1/FKS2 ratios in FKS1 mutants are due to changes in the expression of FKS1. On the other hand, strains 51916 and 42971 were the only isolates showing a significant FKS2 reduction relative to FKS2 expression of wild-type strains. Collectively, these data support the notion that the expression of the FKS genes is regulated by the relative functionality of each of the components of the 1,3-β-d-glucan synthase complex.
TABLE 3.
Relative expression ratios between FKS genes
| Strain | Genotype
|
Expression ratioc
|
||
|---|---|---|---|---|
| FKS1a | FKS2b | FKS1/FKS2 | FKS3/FKS2 | |
| 90030 | WT | WT | 0.65 | 0.01 |
| 218 | WT | WT | 0.49 | 0.04 |
| 3168 | WT | WT | 0.67 | 0.06 |
| 42997 | F625S | WT | 1.79 | 0.07 |
| 5847 | S629P | WT | 1.95 | 0.01 |
| 21900 | D632G | WT | 1.30 | 0.13 |
| 3169 | D632E | WT | 1.20 | 0.07 |
| 42971 | D632Y | R1377STOP | 7.84 | 0.05 |
| 31498 | WT | F659del | 0.49 | 0.06 |
| 234 | WT | F659V | 0.95 | 0.07 |
| 41026 | WT | F659S | 0.29 | 0.01 |
| 3830 | WT | S663P | 0.58 | 0.05 |
| 41400 | WT | D666G | 0.85 | 0.06 |
| 42031 | WT | D666E | 0.42 | 0.05 |
| 51916 | WT | P667T | 1.23 | 0.01 |
| 5416 | WT | W1375L | 0.68 | 0.06 |
WT, wild type at hot spots. Fks1p hot spot number one includes amino acids between 625 and 633 (625-FLILSLRDP-633).
WT, wild type at hot spots. Fks2p hot spot number one includes amino acids between 659 and 667 (659-FLILSLRDP-667).
Values are arithmetic means (three repetitions).
DISCUSSION
Candida glabrata FKS1 and FKS2 mutations are linked to echinocandin resistance.
Large-scale surveillance studies have documented the outstanding potency of echinocandin drugs against clinical isolates of C. glabrata, using the CLSI reference method (10, 24, 25, 27). Clinical failures are uncommon, although reports of echinocandin resistance with C. glabrata are emerging (3, 17, 33). In these studies, amino acid substitutions in both Fks proteins (Fks1p-D632E and Fks2p-F659V) have been linked with echinocandin reduced susceptibilities. The data presented here from a collection of C. glabrata strains with reduced echinocandin susceptibility, including some isolated after therapeutic failure, confirm that mutations in both FKS genes are associated with echinocandin resistance. Moreover, the treatment with ANF (strain 234) or CSF (all FKS mutants, excluding strain 234) were shown to have no influence on the MICs and IC50s obtained (Table 1).
We also demonstrate that a range of mutations in FKS1 and FKS2 are responsible for echinocandin reduced susceptibility and clinical failures in C. glabrata, including 11 new FKS mutations. In contrast to FKS1 and FKS2, FKS3 appears to have no influence on the echinocandin susceptibility/resistance phenotype in C. glabrata. All FKS mutations described in this work were shown to be linked with increases in echinocandin MIC and IC50, supporting the contention that changes in FKS is a universal echinocandin resistance mechanism among Candida spp. (12-14, 17, 22). It has to be noted that the FKS mutants studied in this work do not represent the frequency of the mutations in C. glabrata FKS. These mutants were selected to represent a variety of FKS mutations and were pulled out from a larger collection of isolates with high echinocandin MICs. The real frequency of each of the mutations should be obtained using a prospective population-based surveillance study.
It should be noted that 1,3-β-d-glucan synthase complexes isolated from C. glabrata have two Fksp isoenzymes. In the majority of the cases, only one of the Fksp proteins showed an amino acid substitution. However, the IC50 and kinetic profiles did not reveal the presence of mixed enzyme species (Fig. 2 and 3), suggesting that the Fksp proteins may form a multimeric catalytic functional subunit that behaves as a single kinetic entity. Thus, a mutation in one of the Fksp subunits alters the kinetic profiles of the complete 1,3-β-d-glucan synthase complex.
CLSI echinocandin susceptibility breakpoints underrepresent echinocandin-resistant C. glabrata harboring FKS mutations.
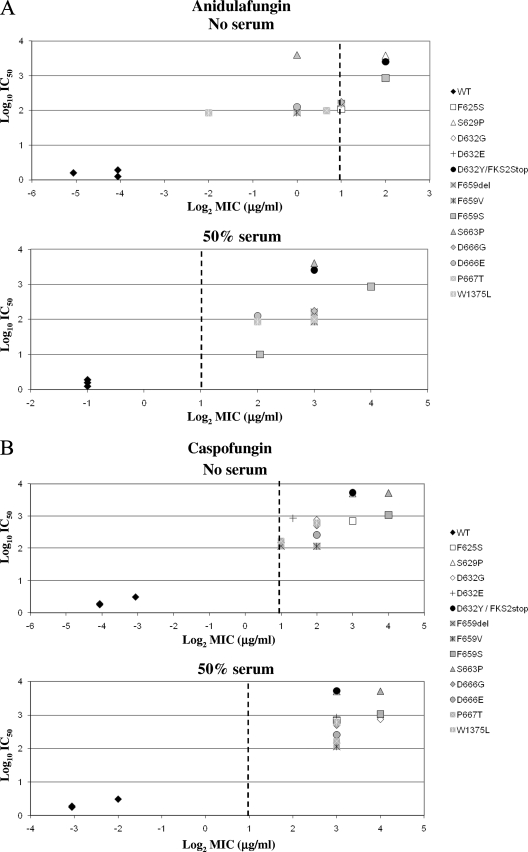
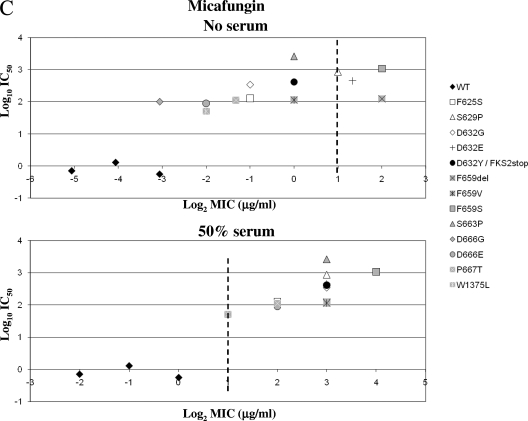
The CLSI has established an echinocandin MIC breakpoint for susceptibility of ≤2 μg/ml for Candida spp. to all three echinocandin drugs. This proposed breakpoint was based on microbiologic in vitro susceptibility data obtained from large-scale global surveillance, in vivo outcomes, and pharmacokinetic and pharmacodynamic studies (4, 26). In this study, all but one of the isolates were obtained as a result of CSF clinical failure; only 3 out of 13 C. glabrata fks clinical isolates (23%) could be considered ANF or MCF nonsusceptible using the CLSI breakpoint. In contrast, most of the fks mutants (92%) were CSF nonsusceptible. Yet, the biochemical sensitivity (IC50) of all the mutant 1,3-β-d-glucan synthase enzymes (n = 13) was consistent with a less susceptible enzyme, which showed at least 50-fold higher IC50s for all drugs than those with wild-type enzyme (Fig. 4). These IC50s were comparable with previous data obtained for other echinocandin-resistant Candida species strains with FKS mutations (13, 14, 21, 22).
FIG. 4.
MIC/IC50 correlation as a function of Fksp amino acid substitutions. The echinocandin drug ANF (A), CSF (B), or MCF (C) was used to obtain IC50 values and MICs in the presence of RPMI 1640 medium (CLSI document M27-A3) (4) or RPMI 1640 medium with 50% serum. Vertical dashed lines represent the CLSI susceptibility breakpoint (2 μg/ml). WT, wild type.
The MIC susceptibility breakpoint appears to overstate the susceptibility endpoint for C. glabrata strains with FKS mutations. This is largely because of the low number of C. glabrata clinical isolates with MICs of >2 μg/ml that were available at the time the breakpoint was established (only 1 out of 747 C. glabrata strains used in the study) (26). Moreover, the molecular mechanism of echinocandin resistance was not strictly considered. Given these limitations, it was appropriately reasoned that there was insufficient data to assign a resistance breakpoint (4, 26). While all three echinocandin drugs share the same target, in vivo potency, mechanism of resistance, and spectrum, they vary in antifungal properties in the CLSI testing medium. In the absence of serum, ANF and MCF appear to be more potent than CSF. However, these relative differences in potency are effectively minimized in the presence of 50% serum and confirmed in in vivo efficacy models (19, 20, 37). In the presence of serum, all of the C. glabrata fks mutant strains show significant shifts in MIC, which place them outside the susceptibility breakpoint, while all wild-type strains are maintained within the breakpoint MIC (Table 1 and Fig. 4).
There is an unambiguous linkage between increased echinocandin IC50s, FKS mutations, and treatment failure. Yet, there is no clear linkage between ANF and MCF MICs and Fksp amino acid substitutions. The data presented here suggest several approaches to improve the relationship between MIC breakpoint, FKS mutations, and the potential for successful clinical outcome. One is to reduce the ANF and MCF breakpoints (>0.25 μg/ml) in order to distinguish between C. glabrata FKS mutants and wild-type strains. The second is the modification of the CLSI protocol to include 50% serum to the MIC plates. However, this latter idea has some disadvantages regarding safety and standardization difficulties. A third is to use CSF as a surrogate marker to predict the C. glabrata susceptibility to the other echinocandin drugs, as previously suggested for fluconazole and azole susceptibility (28-30). Finally, the fourth is to use advances in molecular detection to identify specific fks mutations as resistance markers for Candida spp.
FKS mutations influence 1,3-β-d-glucan synthase kinetics and FKS gene expression.
Specific hot spot Fks amino acid substitutions were shown to decrease the Vmax of the enzyme complex (Table 2). This lower catalytic capacity may have a potential fitness cost to the cell, as suggested by our group for other Candida spp. (12-14), which may contribute to the relatively low frequency of echinocandin resistance. In the case of C. albicans and C. parapsilosis, the reduction in Vmax could be compensated for by an increase in FKS2 expression. The data presented here confirm that modulation of the relative expression of FKS genes is a common compensatory response when the overall catalytic potential of the 1,3-β-d-glucan synthase complex is compromised. However, in C. glabrata, the expression of level of FKS1 is mostly modulated in response to Fks1p amino acid substitutions. For most isolates, the level of FKS2 transcript remained constant relative to that of URA3, irrespective of whether mutations occurred in FKS1 or FKS2. The modulation of FKS1 was most apparent in the behavior of isogenic wild-type and fks1 mutant strains 3168 and 3169 in which a Vmax decrease (Table 2) was manifested as a relative increase in FKS1 expression (Table 3). The third gene, FKS3, is expressed at a trace amount and does not appear to play a role in 1,3-β-d-glucan synthase action; it is unaffected by mutations in FKS1 and FKS2.
In our collection, there were two strains with mutations in FKS2 showing FKS1/FKS2 ratios of >1: strains 51916 (Fks2p-P667T) and 42971 (Fks1p-D632Y and Fks2p early termination). In both cases, a decrease in FKS2 expression was observed. Strain 51916 showed one of the lowest IC50 and Vmax values in this collection. These kinetic parameters are consistent with those obtained for C. parapsilosis, Candida metapsilosis, and Candida orthopsilosis, which harbor a naturally occurring amino acid substitution in the equivalent position of Fks1p (12). However, this strain showed an FKS1/FKS2 ratio comparable to those obtained for FKS1 mutants, suggesting that this particular strain has an unknown alteration in FKS2 regulation. With respect to the strain with the Fks2p early termination codon (42971), the significant decrease in FKS2 expression may be produced by a nonsense-mediated mRNA decay. This is a natural cell mechanism to ensure the elimination of incomplete processed mRNA and has been described for various eukaryotic cells, including Saccharomyces cerevisiae (2, 18, 39).
In summary, mutations in FKS genes of C. glabrata that yield 1,3-β-d-glucan synthase enzymes with highly reduced sensitivity to echinocandin drugs result in elevated MICs with a strong potential for clinical failure. The existing CLSI breakpoint does not adequately cover all three echinocandin drugs and may require modification for suitable capture. Finally, the cell senses changes in the catalytic behavior of 1,3-β-d-glucan synthase, which influences the relative expression of FKS genes.
Acknowledgments
This work was supported by grants to D.S.P. from the NIH (AI069397) and Pfizer.
We thank the Fungus Testing Lab (Pathology Department, University of Texas San Antonio) for providing some of the strains used in this work.
Footnotes
Published ahead of print on 22 June 2009.
REFERENCES
- 1.Alcoba-Florez, J., S. Mendez-Alvarez, J. Cano, J. Guarro, E. Perez-Roth, and A. M. del Pilar. 2005. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol. 43:4107-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amrani, N., M. S. Sachs, and A. Jacobson. 2006. Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol. 7:415-425. [DOI] [PubMed] [Google Scholar]
- 3.Cleary, J. D., G. Garcia-Effron, S. W. Chapman, and D. S. Perlin. 2008. Reduced Candida glabrata susceptibility secondary to an FKS1 mutation developed during candidemia treatment. Antimicrob. Agents Chemother. 52:2263-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A3, 3rd ed. Clinical Laboratory Standards Institute, Wayne, PA.
- 5.Correia, A., P. Sampaio, S. James, and C. Pais. 2006. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int. J. Syst. Evol. Microbiol. 56:313-317. [DOI] [PubMed] [Google Scholar]
- 6.Cuenca-Estrella, M., A. Gomez-Lopez, E. Mellado, G. Garcia-Effron, and J. L. Rodriguez-Tudela. 2004. In vitro activities of ravuconazole and four other antifungal agents against fluconazole-resistant or -susceptible clinical yeast isolates. Antimicrob. Agents Chemother. 48:3107-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denning, D. W. 2003. Echinocandin antifungal drugs. Lancet 362:1142-1151. [DOI] [PubMed] [Google Scholar]
- 8.Douglas, C. M. 2001. Fungal β(1,3)-d-glucan synthesis. Med. Mycol. 39(Suppl. 1):55-66. [DOI] [PubMed] [Google Scholar]
- 9.Douglas, C. M., F. Foor, J. A. Marrinan, N. Morin, J. B. Nielsen, A. M. Dahl, P. Mazur, W. Baginsky, W. Li, and M. el Sherbeini. 1994. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-d-glucan synthase. Proc. Natl. Acad. Sci. USA 91:12907-12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinel-Ingroff, A. 2003. In vitro antifungal activities of anidulafungin and micafungin, licensed agents and the investigational triazole posaconazole as determined by NCCLS methods for 12,052 fungal isolates: review of the literature. Rev. Iberoam. Micol. 20:121-136. [PubMed] [Google Scholar]
- 11.Fidel, P. L., Jr., J. A. Vazquez, and J. D. Sobel. 1999. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12:80-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Effron, G., S. K. Katiyar, S. Park, T. D. Edlind, and D. S. Perlin. 2008. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 52:2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Effron, G., D. P. Kontoyiannis, R. E. Lewis, and D. S. Perlin. 2008. Caspofungin-resistant Candida tropicalis strains causing breakthrough fungemia in patients at high risk for hematologic malignancies. Antimicrob. Agents Chemother. 52:4181-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Effron, G., S. Park, and D. S. Perlin. 2009. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for C. albicans: implications for interpretive breakpoints. Antimicrob. Agents Chemother. 53:112-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hachem, R., H. Hanna, D. Kontoyiannis, Y. Jiang, and I. Raad. 2008. The changing epidemiology of invasive candidiasis: Candida glabrata and Candida krusei as the leading causes of candidemia in hematologic malignancy. Cancer 112:2493-2499. [DOI] [PubMed] [Google Scholar]
- 16.Kartsonis, N. A., J. Nielsen, and C. M. Douglas. 2003. Caspofungin: the first in a new class of antifungal agents. Drug Resist. Updat. 6:197-218. [DOI] [PubMed] [Google Scholar]
- 17.Katiyar, S., M. Pfaller, and T. Edlind. 2006. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 50:2892-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerenyi, Z., Z. Merai, L. Hiripi, A. Benkovics, P. Gyula, C. Lacomme, E. Barta, F. Nagy, and D. Silhavy. 2008. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 27:1585-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odabasi, Z., V. Paetznick, J. H. Rex, and L. Ostrosky-Zeichner. 2007. Effects of serum on in vitro susceptibility testing of echinocandins. Antimicrob. Agents Chemother. 51:4214-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paderu, P., G. Garcia-Effron, S. Balashov, G. Delmas, S. Park, and D. S. Perlin. 2007. Serum differentially alters the antifungal properties of echinocandin drugs. Antimicrob. Agents Chemother. 51:2253-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park, S., R. Kelly, J. N. Kahn, J. Robles, M. J. Hsu, E. Register, W. Li, V. Vyas, H. Fan, G. Abruzzo, A. Flattery, C. Gill, G. Chrebet, S. A. Parent, M. Kurtz, H. Teppler, C. M. Douglas, and D. S. Perlin. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 49:3264-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perlin, D. S. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaller, M. A., L. Boyken, R. J. Hollis, S. A. Messer, S. Tendolkar, and D. J. Diekema. 2006. Global surveillance of in vitro activity of micafungin against Candida: a comparison with caspofungin by CLSI-recommended methods. J. Clin. Microbiol. 44:3533-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfaller, M. A., and D. J. Diekema. 2002. Role of sentinel surveillance of candidemia: trends in species distribution and antifungal susceptibility. J. Clin. Microbiol. 40:3551-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaller, M. A., D. J. Diekema, L. Ostrosky-Zeichner, J. H. Rex, B. D. Alexander, D. Andes, S. D. Brown, V. Chaturvedi, M. A. Ghannoum, C. C. Knapp, D. J. Sheehan, and T. J. Walsh. 2008. Correlation of MIC with outcome for Candida species tested against caspofungin, anidulafungin, and micafungin: analysis and proposal for interpretive MIC breakpoints. J. Clin. Microbiol. 46:2620-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaller, M. A., S. A. Messer, L. Boyken, C. Rice, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2003. Caspofungin activity against clinical isolates of fluconazole-resistant Candida. J. Clin. Microbiol. 41:5729-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaller, M. A., S. A. Messer, L. Boyken, C. Rice, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2004. Cross-resistance between fluconazole and ravuconazole and the use of fluconazole as a surrogate marker to predict susceptibility and resistance to ravuconazole among 12,796 clinical isolates of Candida spp. J. Clin. Microbiol. 42:3137-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfaller, M. A., S. A. Messer, L. Boyken, C. Rice, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2007. Use of fluconazole as a surrogate marker to predict susceptibility and resistance to voriconazole among 13,338 clinical isolates of Candida spp. tested by Clinical and Laboratory Standards Institute-recommended broth microdilution methods. J. Clin. Microbiol. 45:70-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaller, M. A., S. A. Messer, L. Boyken, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2008. Selection of a surrogate agent (fluconazole or voriconazole) for initial susceptibility testing of posaconazole against Candida spp.: results from a global antifungal surveillance program. J. Clin. Microbiol. 46:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Safdar, A., V. Chaturvedi, B. S. Koll, D. H. Larone, D. S. Perlin, and D. Armstrong. 2002. Prospective, multicenter surveillance study of Candida glabrata: fluconazole and itraconazole susceptibility profiles in bloodstream, invasive, and colonizing strains and differences between isolates from three urban teaching hospitals in New York City (Candida Susceptibility Trends Study, 1998 to 1999). Antimicrob. Agents Chemother. 46:3268-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang, J., and T. R. Parr, Jr. 1991. W-1 solubilization and kinetics of inhibition by cilofungin of Candida albicans (1,3)-β-d-glucan synthase. Antimicrob. Agents Chemother. 35:99-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, G. R., III, N. P. Wiederhold, A. C. Vallor, N. C. Villareal, J. S. Lewis, and T. F. Patterson. 2008. Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob. Agents Chemother. 52:3783-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trick, W. E., S. K. Fridkin, J. R. Edwards, R. A. Hajjeh, and R. P. Gaynes. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35:627-630. [DOI] [PubMed] [Google Scholar]
- 35.White, T. J., T. D. Bruns, S. B. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA.
- 36.Wiederhold, N. P., and R. E. Lewis. 2003. The echinocandin antifungals: an overview of the pharmacology, spectrum and clinical efficacy. Expert Opin. Investig. Drugs 12:1313-1333. [DOI] [PubMed] [Google Scholar]
- 37.Wiederhold, N. P., L. K. Najvar, R. Bocanegra, D. Molina, M. Olivo, and J. R. Graybill. 2007. In vivo efficacy of anidulafungin and caspofungin against Candida glabrata and association with in vitro potency in the presence of sera. Antimicrob. Agents Chemother. 51:1616-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]
- 39.Zheng, W., J. S. Finkel, S. M. Landers, R. M. Long, and M. R. Culbertson. 2008. Nonsense-mediated decay of ash1 nonsense transcripts in Saccharomyces cerevisiae. Genetics 180:1391-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]