Abstract
Escherichia coli produces H2 from formate via the formate hydrogenlyase (FHL) complex during mixed acid fermentation; the FHL complex consists of formate dehydrogenase H (encoded by fdhF) for forming 2H+, 2e−, and CO2 from formate and hydrogenase 3 (encoded by hycGE) for synthesizing H2 from 2H+ and 2e−. FHL protein production is activated by the σ54 transcriptional activator FhlA, which activates transcription of fdhF and the hyc, hyp, and hydN-hypF operons. Here, through random mutagenesis using error-prone PCR over the whole gene, as well as over the fhlA region encoding the first 388 amino acids of the 692-amino-acid protein, we evolved FhlA to increase H2 production. The amino acid replacements in FhlA133 (Q11H, L14V, Y177F, K245R, M288K, and I342F) increased hydrogen production ninefold, and the replacements in FhlA1157 (M6T, S35T, L113P, S146C, and E363K) increased hydrogen production fourfold. Saturation mutagenesis at the codons corresponding to the amino acid replacements in FhlA133 and at position E363 identified the importance of position L14 and of E363 for the increased activity; FhlA with replacements L14G and E363G increased hydrogen production (fourfold and sixfold, respectively) compared to FhlA. Whole-transcriptome and promoter reporter constructs revealed that the mechanism by which the FhlA133 changes increase hydrogen production is by increasing transcription of all of the genes activated by FhlA (the FHL complex). With FhlA133, transcription of PfdhF and Phyc is less sensitive to formate regulation, and with FhlA363 (E363G), Phyc transcription increases but Phyp transcription decreases and hydrogen production is less affected by the repressor HycA.
Hydrogen is a promising fuel, since it can be produced from renewable sources (16) and its combustion does not produce pollutants, such as CO, CO2, and SO2, like conventional fossil fuels (34). To create a sustainable energy system based on hydrogen, improvements in hydrogen production are required to make it competitive with fossil fuels (34). It is important to note that the cost of new infrastructure to transport hydrogen may be avoided if hydrogen can be generated at the end user's location rather than at a central production facility (65).
Microbial fermentation is a potential method for large-scale hydrogen production (11), and there are two primary means of microbial hydrogen production: photosynthesis and fermentation. Fermentative reactors have the advantage that waste biomass (20) may be used as a feedstock. In addition, reactors with fermentative bacteria are considered more practical than those with photosynthetic bacteria, as photosynthetic systems require reactors with large surface areas (7) and have hydrogen production rates orders of magnitude lower than those of fermentative bacteria (25).
The hydrogen required to power a home using a 1-kW hydrogen fuel cell is 24 mol H2/h (25). If hydrogen is produced by fermentation of glucose, the annual cost of the glucose is approximately $6,400 (59). To decrease the cost, it is necessary to increase the yield or use less expensive feedstocks (59). The hydrogen yield may be increased by utilizing additional fermentation end products, such as acetate, succinate, and lactate, to produce hydrogen. To power a home, the required size of the reactor for hydrogen production by fermentation of glucose or formate is approximately 500 liters. This size may be reduced by increasing the hydrogen production rate (59).
Escherichia coli is an attractive fermentative microorganism to engineer for hydrogen production because the majority of enzymes and genes related to hydrogen production are known (9) and it is easy to manipulate genetically (13). Under anaerobic conditions, E. coli produces hydrogen from formate through the reaction HCOO− + H2O ↔ H2 + HCO3−, which is catalyzed by the formate hydrogenlyase (FHL) complex (63). The structural components of the FHL complex are formate dehydrogenase H, encoded by fdhF (3), which converts formate to 2H+, 2e−, and CO2; hydrogenase 3 (Hyd-3), encoded by hycE (large subunit) and hycG (small subunit), which is reported to be a NiFe hydrogenase (49) that synthesizes molecular hydrogen from 2H+ and 2e− (48); and the electron transfer proteins encoded by hycBCDF, which are thought to shuttle electrons between formate dehydrogenase H and Hyd-3 (49). An active FHL complex also requires the protease HycI (43), the putative electron carrier HydN (33), and the maturation proteins HycH (48), HypF (33), and HypABCDE (19).
The FHL complex has at least two regulators, FhlA and HycA. FhlA, the product of the last gene of the hyp operon (50) (Fig. 1), is the transcriptional activator of the fdhF gene and the hyc, hyp, and hydN-hypF operons, which form the formate regulon (22). In addition to the FhlA-dependent promoter Phyp, fhlA is also transcribed by an FNR-dependent promoter located within hypA and by its own weak constitutive promoter (44). Translation of FhlA is inhibited by the RNA regulator OxyS, which forms a stable antisense-target complex with fhlA mRNA overlapping the ribosome binding site (2). HycA, the product of the first gene of the hyc operon, represses hyc and hyp, possibly by binding FhlA (48).
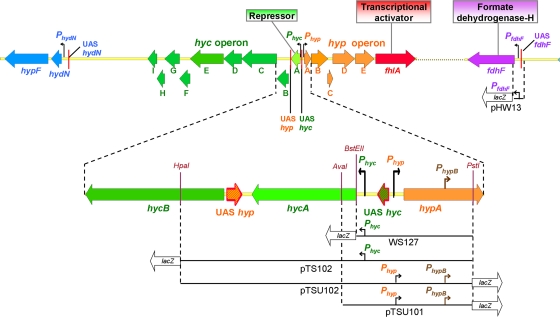
FIG. 1.
Physical map of the transcriptional units activated by FhlA. Coding regions are represented by block arrows, −12/−24 promoters are indicated by black right-angled arrows, the FNR-dependent promoter PhypB is indicated by brown right-angled arrows, and the UAS where FhlA binds (8, 33, 51) are shown by a red-hatched arrow and red boxes. The fragments present in the lacZ reporter fusions used for the transcriptional studies are indicated in black (the lacZ gene is not drawn to scale).
FhlA requires formate (44) to activate transcription from −12/−24 promoters by the σ54-RNA polymerase complex. FhlA with bound formate (18) binds to the upstream activating sequences (UAS) located about 100 bp upstream of the transcriptional start site of fdhF (8) and to hydN-hypF (33), in the region between the divergently transcribed hyp and hyc operons for activation of hyc, and in the intergenic region between hycA and hycB for the activation of hyp (51) (Fig. 1). Intracellular molybdate is required for transcription of fdhF and hyc (42). Also, for maximum transcription of hyc, the integration host factor must bind between the UAS and the promoter of the hyc operon (17). FhlA, as a member of the σ54 family, has a structure composed of three domains (35, 55). The N domain (amino acids 1 to 381) (35) is responsible for binding formate and oligomerization as a tetramer (23); it is very large, and its sequence does not show similarity to other σ54 regulators (50). The central domain (amino acids 388 to 617) (35) is responsible for ATP hydrolysis once formate is bound to the N domain; this reaction is essential for the formation of the open complex of RNA polymerase with DNA, which leads to transcription initiation (18). This region is not influenced by formate and is thought to interact with the RNA polymerase-σ54 complex (23). The C-terminal domain (amino acids 618 to 692) (35) contains a helix-turn-helix motif responsible for DNA binding (50).
Most of the previous studies to enhance E. coli hydrogen production have focused on metabolic engineering (36, 37, 64); for example, we achieved a 141-fold enhancement with the hyaB, hybC, hycA, and fdoG mutations coupled with overexpression of fhlA+ using formate as the substrate (30), and a 4.6-fold enhancement was achieved with the hyaB, hybC, hycA, fdoG, frdC, ldhA, and aceE mutations using glucose as the substrate (28). In contrast, protein engineering has not been used extensively to increase hydrogen production, although we recently reported that hydrogen production may be increased 30-fold by using error-prone PCR (epPCR), DNA shuffling, and saturation mutagenesis of hycE (the large subunit of Hyd-3) (31). In this work, we sought to increase hydrogen production by E. coli through epPCR and saturation mutagenesis of fhlA.
MATERIALS AND METHODS
Bacterial strains, growth, and total protein.
The E. coli strains and plasmids used in this study are listed in Table S1 in the supplemental material; all strains were grown at 37°C. Overnight cultures were made from fresh, single colonies using either Luria-Bertani medium (46), modified complex medium (30) without formate, or modified complex-formate medium with 20 mM formate. Antibiotics were used to maintain plasmids, as well as to select the host, and were used at the following concentrations: ampicillin at 100 μg/ml, chloramphenicol (Cm) at 30 μg/ml, kanamycin (Km) at 100 μg/ml, and spectinomycin at 50 μg/ml. The total protein concentration was 0.22 mg ml−1 (turbidity at 600 nm)−1 (58). JW0098 (ΔoxyS) was constructed via P1 transduction (57) by selecting for Cm resistance that was transferred along with the oxyS deletion from E. coli K-12 GSO35 (1). For each strain from the Keio collection, the deletion of the target gene was verified by two PCRs (see Table S2 in the supplemental material). First, to determine if the wild-type allele was deleted, a PCR using a primer upstream of the target gene and a primer inside the coding region of the target gene was performed. Second, to verify that the Km resistance gene was inserted at the target locus, a PCR using a primer upstream of the target gene and a primer inside the coding region of the Km resistance gene was performed. The deletions in strains JW0098 and MW1002 were also verified by PCR.
Random mutagenesis of fhlA.
The fhlA gene from plasmid pASKA2701 (21) under the control of the pT5-lac promoter was mutated by epPCR as described previously (14) using 50 pmol of each primer (FhlAfront and FhlArev) (see Table S2 in the supplemental material), 0.5 mM MnCl2, and a 3-min extension time. The epPCR product was cloned into pASKA2701 using the MfeI and HindIII restriction enzymes with Antarctic phosphatase (New England Biolabs, Beverly, MA) treatment of the vector; the ligation mixture was electroporated (24) into strain JW2701-1 (ΔfhlA) (4) (complementation of the fhlA deletion by pASKA2701 was reported by us previously [29]). For epPCR of the fhlA region encoding the N domain of FhlA, the conditions were the same as described above, but primers FhlAfront and FhlAN (see Table S2 in the supplemental material) were used with a 2-min extension time. The enzymes used for the cloning were MfeI and BsrGI.
Saturation mutagenesis.
A QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to perform saturation mutagenesis of fhlA at all of the mutated codons of fhlA133 (Table 1) and at the codon corresponding to E363, which is mutated in fhlA1157 (Table 1), using pASKA2701 as a template. The DNA primers (see Table S2 in the supplemental material) contained NNS at the target codon (N is A, G, C, or T, and S is G or C) to allow the substitution of all 20 amino acids using the 32 possible codons (10); the constructed plasmids were electroporated (24) into JW2701-1, and at least 300 colonies (45) were screened for enhanced hydrogen production using chemochromic membranes.
TABLE 1.
Hydrogen production by JW2701-1 expressing the fhlA alleles via pCA24N after 0.5 h of anaerobic incubation in modified complex 20 mM formate medium
| Strain | fhlA allele | FhlA amino acid change(s) | na | H2 production rate (μmol mg protein−1 h−1)e | Relative H2 production rate |
|---|---|---|---|---|---|
| JW2701-1(pASKA2701) | fhlA | 24 | 0.8 ± 0.3 | 1 | |
| JW2701-1(pVSC133) | fhlA133b | Q11H, L14V, Y177F, K245R, M288K, I342F | 5 | 7 ± 2 | 9 |
| JW2701-1(pVSC14) | fhlA14c | L14G | 4 | 3.5 ± 0.5 | 4 |
| JW2701-1(pVSC1157) | fhlA1157d | M6T, S35T, L113P, S146C, E363K | 4 | 2.9 ± 0.5 | 4 |
| JW2701-1(pVSC363) | fhlA363c | E363G | 5 | 5 ± 1 | 6 |
n, number of independent cultures.
Obtained via epPCR of whole fhlA.
Obtained via saturation mutagenesis.
Obtained via epPCR of the fhlA region encoding the N domain of FhlA.
The values are averages ± standard deviations.
Hydrogen screening.
Chemochromic membranes (GVD Corp., Cambridge, MA) formed by a thin film of WO3 covered with a catalytic layer of palladium, were used to detect hydrogen gas, by a colorimetric response, from colonies grown anaerobically (52). These membranes were used to identify mutants with enhanced hydrogen production due to mutations in fhlA generated by epPCR and saturation mutagenesis as described previously (31). Modified complex (30) agar plates containing 20 mM formate and Cm were used for screening; isopropyl β-d-1-thiogalactopyranoside (IPTG) was not added because overexpression of fhlA by adding IPTG is not beneficial for hydrogen production (28).
Hydrogen assay.
For all of the mutants with enhanced hydrogen production that were identified with the chemochromic membranes, hydrogen production was quantified using anaerobic cells. Overnight aerobic cultures (25 ml) in modified complex medium (30) supplemented with 20 mM formate and Cm, as well as uninoculated modified complex medium supplemented with 20 mM formate and Cm, were sparged for 5 min with nitrogen to remove oxygen. Sealed crimped-top vials (27 ml) were also sparged for 2 min with nitrogen. Inside an anaerobic glove box, 9 ml of sparged uninoculated modified complex medium and 1 ml of sparged overnight culture were added to each vial. The amount of hydrogen generated in the headspace was measured after 0.5 h of anaerobic incubation at 37°C by gas chromatography using a 6890N gas chromatograph (Agilent Technologies Inc., Santa Clara, CA) as described previously (32). The work in the anaerobic glove box took about 36 min; therefore, at the time of analysis (listed as 0.5 h of incubation), the cells had been anaerobic for over 1 h.
Cloning of fhlA alleles.
To study the impacts of the beneficial mutations on the transcription of the FhlA-controlled loci using compatible plasmids, the fhlA, fhlA133, and fhlA363 alleles were cloned from plasmids pASKA2701, pVSC133, and pVSC363 into plasmid pVLT35 (12). The plasmids harboring the fhlA alleles were digested with XhoI and HindIII, and pVLT35 was digested with SalI and HindIII (New England Biolabs, Beverly, MA). The DNA fragments were ligated after the digested pVLT35 was treated with Antarctic phosphatase (New England Biolabs, Beverly, MA) and were electroporated into JW2701-1 (4).
Hydrogen production with overexpression of hycA and with isogenic mutants.
The hydrogen production of the JW2701-1 strains harboring the pVLT35-derived plasmids pVSV2701, pVSV133, and pVSV363 (carrying the fhlA, fhlA133, and fhlA363 alleles, respectively) with and without pASKA2695 (hycA+) was evaluated with a hydrogen assay (hycA and fhlA were expressed constitutively). Hydrogen production by JW0833-1 (ΔgrxA), JW0599-1 (ΔahpF), and JW0098 (ΔoxyS) (see Table S1 in the supplemental material) was also evaluated with a hydrogen assay. At least three independent cultures of each strain were assayed.
Hydrogen uptake assay.
pVSC133 and pASKA2701 were electroporated into MW1002, a strain that lacks activity of the uptake hydrogenases Hyd-1 and Hyd-2, as well as chromosomal fhlA. Hydrogen uptake activity by Hyd-3 was assayed in modified complex medium with 20 mM formate, as described previously (32), by measuring the increase in absorbance that results from reducing colorless, oxidized methyl viologen to a purple product (MV2+ + 1/2H2 → MV+ + H+). Two independent cultures of each strain were evaluated.
Transcription of the fdhF gene and hyc and hyp operons.
To explore the mechanism by which the FhlA variants enhance hydrogen production, transcription of the hyc, hyp, and fdhF promoters was evaluated using a β-galactosidase assay in strains lacking fhlA in the chromosome. For the hyc promoter (Phyc) and the hyp promoter (Phyp), two lacZ fusion systems were studied: one set included the hyc UAS (strain WS127 [54] for Phyc and pTSU101 [51] for Phyp) (see Table S1 in the supplemental material), and the other set included the hyc UAS, hycA, and the hyp UAS (pTS102 [51] for Phyc and pTSU102 [51] for Phyp) (see Table S1 in the supplemental material). Thus, the transcriptional activation levels of hyp in the presence of one or two FhlA binding regions could be compared. pHW13 (60), which harbors a PfdhF::lacZ fusion, was used for the fdhF promoter (PfdhF). The DNA fragments in these lacZ fusion systems are shown in Fig. 1.
Plasmids pASKA2701, pVSC133, and pVSC363 (harboring the fhlA alleles) were electroporated into strain WS127, which lacks fhlA and contains the chromosomal lacZ reporter harboring the hyc UAS and Phyc, whereas the lacZ reporter plasmids to study the Phyc, Phyp, and PfdhF promoters were electroporated into JW2701-1 strains harboring plasmids pVSV2701, pVSV133, and pVSV363. For the β-galactosidase assay, cells were prepared in the same manner as for the hydrogen assay using appropriate antibiotics, and enzyme activity was conducted as described previously (62).
Whole-transcriptome analysis.
To investigate why strains with FhlA133 produce more hydrogen, whole-transcriptome analysis was performed. JW2701-1(pVSC133) and JW2701-1(pASKA2701) were cultured as for the hydrogen assay, and total RNA was isolated with the RNeasy kit (Qiagen, Inc., Valencia, CA) as described previously (41) using a bead beater. E. coli GeneChip Genome 2.0 arrays (part no. 511302; Affymetrix, Inc., Santa Clara, CA) were used; they contained 10,208 probe sets for open reading frames, rRNA, tRNA, and 1,350 intergenic regions for four E. coli strains (MG1655, CFT073, O157:H7-Sakai, and O157:H7-EDL933). cDNA synthesis, fragmentation, end terminus biotin labeling, and hybridization were performed as described previously (15). Background values, noise values, and scaling factors for the two arrays were comparable, and the intensities of polyadenosine RNA controls were used to monitor the labeling process. For each binary microarray comparison of differential gene expression, if the gene with the higher transcription rate did not have a consistent transcription rate based on the 11 probe pairs (a detection P value of less than 0.05), the genes were discarded. A gene was considered differentially expressed when the P value for comparing two chips was less than 0.05 (to ensure that the change in gene expression was statistically significant and that false positives arose at less than 5%) and when the expression ratio was higher (1.2-fold) than the standard deviation for all K-12 genes of the microarrays (1.2-fold) (40).
qRT-PCR.
To validate the whole-transcriptome analysis data, the transcription of grxA, ahpF, hycE, hypD, fdhF, and hydN was quantified using quantitative real-time reverse transcription PCR (qRT-PCR) (5) with the RNA samples used for the whole-transcriptome analysis. The housekeeping gene rrsG (16S rRNA) was used to normalize the expression data. Three technical replicates were performed for each gene using the StepOne Real-Time PCR system (Applied Biosystems, Foster City, CA) and the Power SYBR green RNA-to-CT 1-Step Kit (Applied Biosystems, Foster City, CA). The primers for qRT-PCR are given in Table S2 in the supplemental material. The expression ratios for the genes analyzed were calculated according to the 2−ΔΔCT method (26).
Plasmid isolation, SDS-PAGE, and DNA sequencing.
Plasmids were isolated using the QIAprep Spin Miniprep kit (Qiagen, Inc., Valencia, CA). The formation of recombinant proteins under the conditions used for the hydrogen assay was analyzed with standard Laemmli discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12%) (46). A dideoxy chain termination method (47) with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer, Wellesley, MA) was used to determine the nucleotide changes in the fhlA alleles; the primers used for sequencing are given in Table S2 in the supplemental material.
Microarray data accession number.
The expression data for the whole-transcriptome analysis of JW2701-1(pVSC133) and JW2701-1(pASKA2701) have been deposited in the NCBI Gene Expression Omnibus (GEO) (6) and are accessible as GSE13902.
RESULTS
Isolation of mutants with enhanced H2 production.
To increase hydrogen production and to better understand the transcription activation of the genes of FHL by FhlA, epPCR was used to construct a random-mutagenesis library of fhlA. We screened 2,200 colonies using the chemochromic membranes in a host that lacks fhlA in the chromosome (JW2701-1). Using the hydrogen assay, we identified variant FhlA133, which allows JW2701-1(pVSC133) to have a ninefold-higher hydrogen production rate than JW2701-1(pASKA2701) (Table 1).
The FhlA133 variant has six amino acid changes (Q11H, L14V, Y177F, K245R, M288K, and I342F) in the N-terminal domain, which motivated us to focus on mutagenesis of fhlA only in the N-terminal domain (the fhlA region coding for the first 388 amino acids of FhlA). Thus, a second epPCR library was constructed targeting only this region, and an additional 4,400 colonies were screened from this new library. From this screening, variant FhlA1157 (expressed via pVSC1157), which has five amino acid changes (M6T, S35T, L113P, S146C, and E363K), was identified as causing a fourfold increase in hydrogen production (Table 1).
Saturation mutagenesis.
To identify which amino acid replacements in FhlA133 are important for enhanced hydrogen production, saturation mutagenesis of fhlA was performed on each of the six mutated codons in fhlA133 that corresponded to Q11, L14, Y177, K245, M288, and I342. For each position, at least 300 colonies were screened to ensure, with a probability of 99.99%, that all possible codons were utilized (45). Only the mutation encoding L14G (expressed in pVSC14) resulted in an increase in the hydrogen production rate (fourfold) (Table 1); therefore, position L14 of FhlA is important for controlling hydrogen production.
Saturation mutagenesis was also performed at the codon corresponding to E363 of FhlA, since the replacement E363K was identified in FhlA1157 and because the E363K amino acid replacement increases Phyc transcription approximately 3-fold in the presence of 30 mM formate (34-fold without formate) and decreases the impact of formate (22). FhlA363 (E363G) was identified from the hydrogen screen with JW2701-1, and this replacement caused hydrogen production rates sixfold higher than that of the strain with FhlA.
Plasmids harboring each of four mutated fhlA alleles found through epPCR and saturation mutagenesis of fhlA (fhlA133, fhlA1157, fhlA14, and fhlA363) were isolated and reelectroporated into JW2701-1; hydrogen production was assayed to confirm that the mutations in the plasmid were responsible for the higher hydrogen production rates (Table 1). In addition, enhanced hydrogen production by the mutants harboring fhlA133 and fhlA363 was confirmed for a third time with the hydrogen assay after the fhlA, fhlA133, and fhlA363 alleles were cloned into pVLT35 (data not shown). Hence, consistent data were obtained demonstrating that the beneficial mutations in fhlA were directly related to enhanced hydrogen production. SDS-PAGE of the cell lysates from JW2701-1 expressing the fhlA alleles from pASKA2701, pVSC133, pVSC1157, pVSC14, and pVSC363 indicated that the higher hydrogen production rates were not due to changes in the amount of FhlA (data not shown).
To confirm that the hydrogen produced by the strains studied came from the formate added to the medium (rather than from other medium components), we compared hydrogen production by JW2701-1(pVSC133) and JW2701-1(pASKA2701) with and without formate. Hydrogen production in the absence of formate was 0.5 ± 0.1 μmol H2 mg protein−1 h−1 for JW2701-1(pVSC133) and 0.290 ± 0.005 μmol H2 mg protein−1 h−1 for JW2701-1(pASKA2701). The hydrogen production of JW2701-1(pVSC133) in modified complex medium with 20 mM formate was 14- ± 5-fold higher than the hydrogen production without formate; for JW2701-1(pASKA2701), hydrogen production with 20 mM formate was 3- ± 1-fold higher than without formate. Since the amount of hydrogen produced in the medium without formate was very small relative to the amount produced with 20 mM formate, we concluded that the hydrogen produced by the strains studied came predominantly from the formate added to the medium.
Transcription of fdhF, the hyc operon, and the hyp operon.
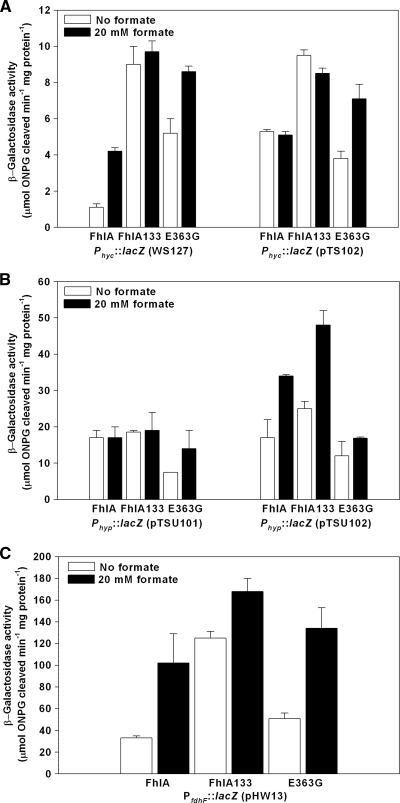
Since FhlA is the transcriptional activator of the genes of the FHL complex, the impacts of formate (20 mM) on the transcription of fdhF (which encodes formate dehydrogenase H), the hyc operon (which encodes the structural proteins of the FHL complex), and the hyp operon (which encodes maturation proteins) (Fig. 1) were evaluated by the β-galactosidase assay. With fhlA133 expressed from pVSC133, Phyc transcription in strain WS127 (including the hyc UAS and Phyc) was increased 2.3-fold with 20 mM formate and 8-fold in the absence of formate (Fig. 2A). Using plasmid pTS102, which contains the hyc UAS, hycA, and the hyp UAS, Phyc transcription in the strain harboring fhlA133 was 1.7-fold higher than in the strain with fhlA with and without formate (Fig. 2A). Using pTSU101 (including the hyc UAS and Phyp), transcription rates of Phyp in the strains with fhlA133 and fhlA were very similar (Fig. 2B); for pTSU102 (including the hyp UAS), Phyp transcription increased 1.5-fold in the strain with fhlA133 relative to the strain with fhlA (Fig. 2B). Increased transcription due to the mutations in fhlA133 was also observed for PfdhF; transcription of this promoter was 1.7-fold higher with 20 mM formate and 3.8-fold higher without formate (Fig. 2C). In addition, Phyc and PfdhF transcriptions were less affected by formate addition in the strains with fhlA133 than in the strains with fhlA (Fig. 2A and C). Hence, the ninefold-higher hydrogen production attained by JW2701-1(pVSC133) appears to be due to an increase in transcription of all three promoters of hyc, hyp, and fdhF.
FIG. 2.
Transcriptional activation of hyc, hyp, and fdhF by FhlA variants. Cells were cultured anaerobically in modified complex medium without formate or with 20 mM formate for 40 min. The results are the averages of two independent cultures. (A) Transcription of Phyc::lacZ using E. coli WS127 (the fhlA alleles were expressed via pCA24N) and E. coli JW2701-1 (ΔfhlA) harboring plasmid pTS102 (the fhlA alleles were expressed via pVLT35). (B) Transcription of Phyp::lacZ using plasmids pTSU101 and pTSU102 with JW2701-1 (the fhlA alleles were expressed via pVLT35). (C) Transcription of PfdhF::lacZ using plasmid pHW13 with JW2701-1 (the fhlA alleles were expressed via pVLT35). ONPG, o-nitrophenyl-β-d-galactopyranoside.
In the presence of 20 mM formate, the replacement E363G in FhlA increased Phyc transcription; WS127(pVSC363) had 2.1-fold-higher Phyc transcription than WS127(pASKA2701), and using the pTS102 reporter, the strain with fhlA363 had 1.4-fold-higher Phyc transcription than the wild-type strain. A higher Phyc transcription rate was also observed in the absence of formate for WS127(pVSC363) (fivefold) (Fig. 2A). The transcription of PfdhF was slightly higher in the strain with fhlA363 than in the strain with fhlA (1.5-fold without formate and 1.3-fold with 20 mM formate) (Fig. 2C), while the transcription of Phyp decreased (Fig. 2B). Therefore, the E363G replacement confers a higher transcription rate for hyc and fdhF that appears to lead to increased hydrogen production.
Whole-transcriptome analysis.
To investigate why the mutant JW2701-1(pVSC133) produced more hydrogen than the wild-type strain, we performed a whole-transcriptome analysis. The amino acid changes in FhlA133 induced all four of the transcriptional units regulated by FhlA: the hyc operon (1.6-fold), the hyp operon (1.5-fold), fdhF (1.7-fold), and hydN-hypF (1.7-fold) (see Table S3 in the supplemental material). These results corroborate those found using the promoter reporters (1.7-fold for the hyc operon, 1.5-fold for the hyp operon, and 1.6-fold for fdhF) and demonstrate that the hydN-hypF operon is also induced by this protein variant.
Surprisingly, the genes of the FHL complex were not the most induced genes. Instead, the highest induction was observed for genes activated by OxyR under conditions of oxidative stress (66): grxA (2.8-fold), ahpF (2.8-fold), and ahpC (2.5-fold) (see Table S4 in the supplemental material). Other stress-related genes were also induced, such as the psp operon, which is transcribed by σ54-RNA polymerase (61).
qRT-PCR.
qRT-PCR was used to verify the expression of the most induced genes (grxA and ahpF) and of some genes activated by FhlA (hycE, hypD, fdhF, and hydN) (see Table S5 in the supplemental material). The differential changes in expression were comparable to those in the whole-transcriptome analysis: grxA, 3.0-fold versus 2.8-fold; ahpF, 3.9-fold versus 2.8-fold; hycE, 1.2-fold versus 1.5-fold; hypD, 1.4-fold versus 1.5-fold; fdhF, 1.2-fold versus 1.7-fold; and hydN, 1.5-fold versus 1.7-fold.
grxA, ahpF, and oxyS mutations and hydrogen production.
To explore whether the oxidative-stress genes induced in the whole-transcriptome analysis were related to hydrogen production, we analyzed the effects of deleting grxA, ahpF, and oxyS on hydrogen production by BW25113. The rate of hydrogen production by BW25113 was slightly reduced upon deletion of grxA (1.6- ± 0.2-fold) and ahpF (1.4- ± 0.4-fold); however, the oxyS deletion increased hydrogen production by 1.7- ± 0.4-fold.
Hydrogen uptake assay.
Hydrogen uptake was assayed directly to determine if the increase in hydrogen production by JW2701-1(pVSC133) was due to a decrease in Hyd-3-mediated hydrogen uptake. MW1002 was used because it lacks fhlA and the large subunits of uptake hydrogenases Hyd-1 and Hyd-2; thus, only uptake by Hyd-3, a reversible enzyme capable of hydrogen uptake (29) that is activated by FhlA, is possible. There was no significant difference in the hydrogen uptake activity of MW1002(pVSC133) (0.53 nmol min−1 mg protein−1) relative to that of MW1002(pASKA2701) (0.55 nmol min−1 mg protein−1); therefore, the increase in hydrogen production by JW2701-1(pVSC133) was not due to a change in hydrogen uptake.
Hydrogen production with overexpression of hycA.
To ascertain if the mutations in fhlA133 and fhlA363 alter the HycA-mediated repression of genes encoding FHL (48), we evaluated the effect of hycA overexpression on hydrogen production. pASKA2695 (expressing hycA) was electroporated into JW2701-1(pVSV2701) and the derived strains harboring fhlA133 and fhlA363. As expected, overexpression of hycA reduced hydrogen production; however, the strain with fhlA363 was repressed less by HycA (1.4- ± 0.7-fold) than the strain with fhlA (3.7- ± 0.9-fold) and the strain with fhlA133 (5- ± 1-fold). These results indicate that E363 of FhlA may be involved in the HycA-mediated inhibition of transcription of the genes encoding FHL.
DISCUSSION
Random mutagenesis of fhlA had been conducted previously by Korsa and Böck (22) to find FhlA variants that activate Phyc transcription independently of formate; however, mutagenesis was not conducted to increase hydrogen production, as these variants were used to study the kinetics of ATP hydrolysis in the presence and absence of formate. They found that FhlA with amino acid replacements E358K and E363K activated hyc transcription with reduced dependence on formate, whereas E183K conferred a constitutive phenotype (22). Similarly, Self and Shanmugam (54) found several FhlA variants that activated hyc transcription without molybdate. Here, using direct screening for hydrogen production, we identified four mutants with increased hydrogen production obtained through epPCR and saturation mutagenesis (Table 1) of fhlA and discovered the importance of position L14 of FhlA.
The N-terminal domains of some σ54 regulators (e.g., DmpR [56] and XylR [38]) inhibit transcription activation in the absence of their corresponding effectors. FhlA-C, an N-terminally truncated FhlA protein lacking the first 378 amino acids, is active independently of formate and is not affected by the repressor HycA (23). Similarly, FhlA165, which has a deletion from amino acids 5 to 374, activates hyc transcription independently of formate, but unlike FhlA-C, its activity was reduced by HycA (53). FhlA-N, a C-terminally and central-domain-truncated protein lacking the last 314 amino acids, repressed transcriptional activation of the hyc operon by FhlA in the presence and absence of formate (23). Hence, the N domain of FhlA inhibits FhlA transcriptional activation and is influenced by formate and HycA (53).
Even though the entire fhlA gene was mutated here, all six amino acid replacements in FhlA133 were in the N domain. This suggests that these replacements may decrease the repressive effect of the N domain. Saturation mutagenesis at each codon affected by the mutations in fhlA133 led only to the discovery of replacement L14G, which stimulates hydrogen production fourfold in a medium supplemented with 20 mM formate. L14 is in the region between amino acids 7 and 37 of FhlA, and Self et al. (53) showed that a truncation of this region abolishes transcriptional activation of the hyc operon; therefore, this region is important for hydrogen production. Since FhlA is a transcriptional activator of four loci (33, 50, 51), the increase in hydrogen production due to the mutations in fhlA should be related to changes in the transcription of the units activated by FhlA; therefore, we studied the transcriptional activation of the fdhF gene and the hyc and hyp operons by strains harboring fhlA, fhlA133, and fhlA363.
The results of the β-galactosidase transcription assay for strains with fhlA (Fig. 2) agree with the data reported previously in which formate induced an increase in PfdhF transcription (60) and an increase in Phyp when the hyc UAS and hyp UAS were present (using pTSU102) (51). Also, as reported by Schlensog et al. (51), Phyc transcription was not induced by formate using the pTS102 reporter plasmid (Fig. 2A) and Phyp transcription was not induced by formate using pTSU101. From these transcription reporter results, the replacements in FhlA133 led to increased transcription of all three of the promoters studied (Phyc, Phyp, and PfdhF) with and without formate (Fig. 2). Moreover, transcription from Phyc and PfdhF in strains with FhlA133 was less dependent on formate. This is reflected in the hydrogen production rate in the absence of formate [JW2701-1(pVSC133) had 1.7- ± 0.3-fold-higher hydrogen production than JW2701-1(pASKA2701)]. Since the intracellular level of formate determines the transcription rate of the FHL genes by FhlA (44), FhlA133 may be able to activate transcription with a smaller internal concentration of formate than FhlA.
Strain WS127, which was used to measure Phyc transcription, has a deletion of all of the genes of the formate regulon except fdhF (54). Thus, we studied Phyc transcription in the absence of the repressor HycA and with only the hyc UAS and the fdhF UAS present, since the other UAS were deleted. Using this strain, replacements in FhlA133 led to an 8-fold increase in Phyc transcription in the absence of formate and a 2.3-fold increase in Phyc transcription in the presence of 20 mM formate.
Along with indicating that all four of the known FhlA-controlled operons are induced in JW2701-1(pVSC133) versus JW2701-1(pASKA2701), the whole-transcriptome analysis indicated that the replacements in FhlA133 also induced eight genes related to oxidative stress (see Table S4 in the supplemental material). A role for oxidative-stress proteins during anaerobic fermentations is surprising. However, removal of OxyS inhibition of FhlA translation by deleting oxyS from BW25113 was expected to provide a small beneficial effect on hydrogen production, and a nearly twofold increase was measured. OxyS RNA forms a stable antisense-target complex with fhlA mRNA by binding to a sequence overlapping the ribosome binding site and to a sequence located in the fhlA coding region; mutations at either site decrease the stability of the complex (2). For the JW2701-1(pASKA2701) derivatives, deletion of oxyS should have less impact on hydrogen production, since the ribosome binding sequence from plasmid pASKA2701 differs from the native sequence where OxyS binds to fhlA mRNA. In addition, for fhlA133 and fhlA14, the replacement at position L14 is located in one of the OxyS binding regions (2). Another 12 stress-related genes were also induced (see Table S4 in the supplemental material), which suggests that the increased hydrogen production affects cell physiology and that increases in hydrogen production may be facilitated by increasing the production of proteins that alleviate stress.
Among these stress-related genes is the psp operon; transcription of this operon, as well as that of the operons activated by FhlA, depends on σ54. Among the other 16 σ54-dependent promoters (39), the promoter with the highest similarity to the psp promoter is Phyc (66.7% identity). Therefore, FhlA133 may increase transcription of the psp operon because of its similarity to the promoters controlling the expression of the genes of the FHL complex.
Mutagenesis in the fhlA region coding for the N domain of FhlA produced variant FhlA1157 with replacements M6T, S35T, L113P, S146C, and E363K. In the absence of formate, FhlAE363K has kinetic parameters (Km and Vmax) for ATP hydrolysis similar to those of FhlA with bound formate; therefore, mutation E363K renders FhlA less sensitive to formate (22). Here, saturation mutagenesis at position E363 produced the replacement E363G, which increased hydrogen production sixfold. E363G, like E363K, increased transcription of Phyc with formate; E363G also slightly increased PfdhF transcription (50% without formate and 30% with 20 mM formate).
Transcription of Phyp in the presence of fhlA363 decreased for the two systems studied, pTSU101 (hyc UAS) and pTSU102 (hyc and hyp UAS). Transcription of hypBCDE in the presence of only the hyc UAS (pTSU101) is due to the FNR-dependent promoter located within hypA (27) and is not due to the FhlA-dependent promoter Phyp (Fig. 2B); this promoter did not show significant induction with formate (51). However, using pTSU101, the E363G mutation led to a twofold induction of Phyp transcription by formate (Fig. 2B). Hence, the mechanism for increasing hydrogen production of strains harboring fhlA133 is different than that of strains harboring fhlA363. Strains with fhlA133 have increased transcription of all of the genes of the FHL complex and have PfdhF and Phyc transcription that is less sensitive to formate regulation, whereas strains harboring fhlA363 have increased Phyc and PfdhF transcription and less Phyp transcription and have hydrogen production that is less affected by the repressor HycA.
Supplementary Material
Acknowledgments
We are grateful for plasmids sent by August Böck, Oliver Lenz, and Robert P. Gunsalus; for the strains sent by William Self; and for the chemochromic membrane sensors provided by Hilton G. Pryce Lewis of GVD Corporation.
This research was supported by the National Science Foundation (CBET-0753702).
Footnotes
Published ahead of print on 6 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altuvia, S., D. Weinstein-Fischer, A. Zhang, L. Postow, and G. Storz. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43-53. [DOI] [PubMed] [Google Scholar]
- 2.Argaman, L., and S. Altuvia. 2000. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 300:1101-1112. [DOI] [PubMed] [Google Scholar]
- 3.Axley, M. J., D. A. Grahame, and T. C. Stadtman. 1990. Escherichia coli formate-hydrogen lyase. Purification and properties of the selenium-dependent formate dehydrogenase component. J. Biol. Chem. 265:18213-18218. [PubMed] [Google Scholar]
- 4.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bansal, T., D. Englert, J. Lee, M. Hegde, T. K. Wood, and A. Jayaraman. 2007. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect. Immun. 75:4597-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett, T., T. O. Suzek, D. B. Troup, S. E. Wilhite, W.-C. Ngau, P. Ledoux, D. Rudnev, A. E. Lash, W. Fujibuchi, and R. Edgar. 2005. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 33:D562-D566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benemann, J. R. 1998. The technology of biohydrogen, p. 19-30. In O. R. Zaborsky (ed.), Biohydrogen. Plenum Press, New York, NY.
- 8.Birkmann, A., and A. Böck. 1989. Characterization of a cis regulatory DNA element necessary for formate induction of the formate dehydrogenase gene (fdhF) of Escherichia coli. Mol. Microbiol. 3:187-195. [DOI] [PubMed] [Google Scholar]
- 9.Böck, A., and G. Sawers. 1996. Fermentation, p. 262-282. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 10.Canyuk, B., P. J. Focia, and A. E. Eakin. 2001. The role for an invariant aspartic acid in hypoxanthine phosphoribosyltransferases is examined using saturation mutagenesis, functional analysis, and X-ray crystallography. Biochemistry 40:2754-2765. [DOI] [PubMed] [Google Scholar]
- 11.Das, D., and T. N. Veziroğlu. 2001. Hydrogen production by biological processes: a survey of literature. Int. J. Hydrogen Energy 26:13-28. [Google Scholar]
- 12.de Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 13.Fischbach, M. A., and C. T. Walsh. 2006. Biochemistry: directing biosynthesis. Science 314:603-605. [DOI] [PubMed] [Google Scholar]
- 14.Fishman, A., Y. Tao, W. Bentley, and T. K. Wood. 2004. Protein engineering of toluene 4-monooxygenase of Pseudomonas mendocina KR1 for synthesizing 4-nitrocatechol from nitrobenzene. Biotechnol. Bioeng. 87:779-790. [DOI] [PubMed] [Google Scholar]
- 15.González Barrios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum sensing regulator (MqsR, B3022). J. Bacteriol. 188:305-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawkes, F. R., I. Hussy, G. Kyazze, R. Dinsdale, and D. L. Hawkes. 2007. Continuous dark fermentative hydrogen production by mesophilic microflora: principles and progress. Int. J. Hydrogen Energy 32:172-184. [Google Scholar]
- 17.Hopper, S., M. Babst, V. Schlensog, H.-M. Fischer, H. Hennecke, and A. Böck. 1994. Regulated expression in vitro of genes coding for formate hydrogenlyase components of Escherichia coli. J. Biol. Chem. 269:19597-19604. [PubMed] [Google Scholar]
- 18.Hopper, S., and A. Böck. 1995. Effector-mediated stimulation of ATPase activity by the σ54-dependent transcriptional activator FHLA from Escherichia coli. J. Bacteriol. 177:2798-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobi, A., R. Rossmann, and A. Böck. 1992. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch. Microbiol. 158:444-451. [DOI] [PubMed] [Google Scholar]
- 20.Kapdan, I. K., and F. Kargi. 2006. Bio-hydrogen production from waste materials. Enzyme Microb. Technol. 38:569-582. [Google Scholar]
- 21.Kitagawa, M., T. Ara, M. Arifuzzaman, T. Ioka-Nakamichi, E. Inamoto, H. Toyonaga, and H. Mori. 2005. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): unique resources for biological research. DNA Res. 12:291-299. [DOI] [PubMed] [Google Scholar]
- 22.Korsa, I., and A. Böck. 1997. Characterization of fhlA mutations resulting in ligand-independent transcriptional activation and ATP hydrolysis. J. Bacteriol. 179:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonhartsberger, S., A. Ehrenreich, and A. Böck. 2000. Analysis of the domain structure and the DNA binding site of the transcriptional activator FhlA. Eur. J. Biochem. 267:3672-3684. [DOI] [PubMed] [Google Scholar]
- 24.Leungsakul, T., G. R. Johnson, and T. K. Wood. 2006. Protein engineering of the 4-methyl-5-nitrocatechol monooxygenase from Burkholderia sp. strain DNT for enhanced degradation of nitroaromatics. Appl. Environ. Microbiol. 72:3933-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin, D. B., L. Pitt, and M. Love. 2004. Biohydrogen production: prospects and limitations to practical application. Int. J. Hydrogen Energy 29:173-185. [Google Scholar]
- 26.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 27.Lutz, S., A. Jacobi, V. Schlensog, R. Böhm, G. Sawers, and A. Böck. 1991. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol. Microbiol. 5:123-135. [DOI] [PubMed] [Google Scholar]
- 28.Maeda, T., V. Sanchez-Torres, and T. K. Wood. 2007. Enhanced hydrogen production from glucose by metabolically-engineered Escherichia coli. Appl. Microbiol. Biotechnol. 77:879-890. [DOI] [PubMed] [Google Scholar]
- 29.Maeda, T., V. Sanchez-Torres, and T. K. Wood. 2007. Escherichia coli hydrogenase 3 is a reversible enzyme possessing hydrogen uptake and synthesis activities. Appl. Microbiol. Biotechnol. 76:1035-1042. [DOI] [PubMed] [Google Scholar]
- 30.Maeda, T., V. Sanchez-Torres, and T. K. Wood. 2008. Metabolic engineering to enhance bacterial hydrogen production. Microb. Biotechnol. 1:30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda, T., V. Sanchez-Torres, and T. K. Wood. 2008. Protein engineering of hydrogenase 3 to enhance hydrogen production. Appl. Microbiol. Biotechnol. 79:77-86. [DOI] [PubMed] [Google Scholar]
- 32.Maeda, T., G. Vardar, W. T. Self, and T. K. Wood. 2007. Inhibition of hydrogen uptake in Escherichia coli by expressing the hydrogenase from the cyanobacterium Synechocystis sp. PCC 6803. BMC Biotechnol. 7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maier, T., U. Binder, and A. Böck. 1996. Analysis of the hydA locus of Escherichia coli: two genes (hydN and hypF) involved in formate and hydrogen metabolism. Arch. Microbiol. 165:333-341. [DOI] [PubMed] [Google Scholar]
- 34.Momirlan, M., and T. N. Veziroglu. 2005. The properties of hydrogen as fuel tomorrow in sustainable energy system for a cleaner planet. Int. J. Hydrogen Energy 30:795-802. [Google Scholar]
- 35.Morett, E., and L. Segovia. 1993. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175:6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penfold, D. W., C. F. Forster, and L. E. Macaskie. 2003. Increased hydrogen production by Escherichia coli strain HD701 in comparison with the wild-type parent strain MC4100. Enzyme Microb. Technol. 33:185-189. [Google Scholar]
- 37.Penfold, D. W., F. Sargent, and L. E. Macaskie. 2006. Inactivation of the Escherichia coli K-12 twin-arginine translocation system promotes increased hydrogen production. FEMS Microbiol. Lett. 262:135-137. [DOI] [PubMed] [Google Scholar]
- 38.Pérez-Martín, J., and V. de Lorenzo. 1995. The amino-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc. Natl. Acad. Sci. USA 92:9392-9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65:422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren, D., L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Differential gene expression shows natural brominated furanones interfere with the autoinducer-2 bacterial signaling system of Escherichia coli. Biotechnol. Bioeng. 88:630-642. [DOI] [PubMed] [Google Scholar]
- 41.Ren, D., L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515-524. [DOI] [PubMed] [Google Scholar]
- 42.Rosentel, J. K., F. Healy, J. A. Maupin-Furlow, J. H. Lee, and K. T. Shanmugam. 1995. Molybdate and regulation of mod (molybdate transport), fdhF, and hyc (formate hydrogenlyase) operons in Escherichia coli. J. Bacteriol. 177:4857-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossmann, R., T. Maier, F. Lottspeich, and A. Böck. 1995. Characterisation of a protease from Escherichia coli involved in hydrogenase maturation. Eur. J. Biochem. 227:545-550. [DOI] [PubMed] [Google Scholar]
- 44.Rossmann, R., G. Sawers, and A. Bock. 1991. Mechanism of regulation of the formate-hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol. Microbiol. 5:2807-2814. [DOI] [PubMed] [Google Scholar]
- 45.Rui, L., Y. M. Kwon, A. Fishman, K. F. Reardon, and T. K. Wood. 2004. Saturation mutagenesis of toluene ortho-monooxygenase of Burkholderia cepacia G4 for enhanced 1-naphthol synthesis and chloroform degradation. Appl. Environ. Microbiol. 70:3246-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauter, M., R. Böhm, and A. Böck. 1992. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol. Microbiol. 6:1523-1532. [DOI] [PubMed] [Google Scholar]
- 49.Sawers, G. 1994. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie van Leeuwenhoek 66:57-88. [DOI] [PubMed] [Google Scholar]
- 50.Schlensog, V., and A. Böck. 1990. Identification and sequence analysis of the gene encoding the transcriptional activator of the formate hydrogenlyase system of Escherichia coli. Mol. Microbiol. 4:1319-1327. [DOI] [PubMed] [Google Scholar]
- 51.Schlensog, V., S. Lutz, and A. Böck. 1994. Purification and DNA-binding properties of FHLA, the transcriptional activator of the formate hydrogenlyase system from Escherichia coli. J. Biol. Chem. 269:19590-19596. [PubMed] [Google Scholar]
- 52.Seibert, M., T. Flynn, D. Benson, E. Tracy, and M. Ghirardi. 1998. Development of selection and screening procedures for rapid identification of H2-producing algal mutants with increased O2 tolerance, p. 227-234. In O. R. Zaborsky (ed.), Biohydrogen. Plenum Press, New York, NY.
- 53.Self, W. T., A. Hasona, and K. T. Shanmugam. 2001. N-terminal truncations in the FhlA protein result in formate- and MoeA-independent expression of the hyc (formate hydrogenlyase) operon of Escherichia coli. Microbiology 147:3093-3104. [DOI] [PubMed] [Google Scholar]
- 54.Self, W. T., and K. T. Shanmugam. 2000. Isolation and characterization of mutated FhlA proteins which activate transcription of the hyc operon (formate hydrogenlyase) of Escherichia coli in the absence of molybdate. FEMS Microbiol. Lett. 184:47-52. [DOI] [PubMed] [Google Scholar]
- 55.Shingler, V. 1996. Signal sensing by σ54-dependent regulators: derepression as a control mechanism. Mol. Microbiol. 19:409-416. [DOI] [PubMed] [Google Scholar]
- 56.Shingler, V., and H. Pavel. 1995. Direct regulation of the ATPase activity of the transcriptional activator DmpR by aromatic compounds. Mol. Microbiol. 17:505-513. [DOI] [PubMed] [Google Scholar]
- 57.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 58.Vardar, G., and T. K. Wood. 2004. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for synthesizing 4-methylresorcinol, methylhydroquinone, and pyrogallol. Appl. Environ. Microbiol. 70:3253-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vardar-Schara, G., T. Maeda, and T. K. Wood. 2008. Metabolically engineered bacteria for producing hydrogen via fermentation. Microb. Biotechnol. 1:107-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, H., and R. P. Gunsalus. 2003. Coordinate regulation of the Escherichia coli formate dehydrogenase fdnGHI and fdhF genes in response to nitrate, nitrite, and formate: roles for NarL and NarP. J. Bacteriol. 185:5076-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weiner, L., J. L. Brissette, and P. Model. 1991. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on sigma 54 and modulated by positive and negative feedback mechanisms. Genes Dev. 5:1912-1923. [DOI] [PubMed] [Google Scholar]
- 62.Wood, T. K., and S. W. Peretti. 1991. Effect of chemically-induced, cloned-gene expression on protein synthesis in E. coli. Biotechnol. Bioeng. 38:397-412. [DOI] [PubMed] [Google Scholar]
- 63.Woods, D. D. 1936. Hydrogenlyases. The synthesis of formic acid by bacteria. Biochem. J. 30:515-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshida, A., T. Nishimura, H. Kawaguchi, M. Inui, and H. Yukawa. 2005. Enhanced hydrogen production from formic acid by formate hydrogen lyase-overexpressing Escherichia coli strains. Appl. Environ. Microbiol. 71:6762-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Y. H. P., B. R. Evans, J. R. Mielenz, R. C. Hopkins, and M. W. W. Adams. 2007. High-yield hydrogen production from starch and water by a synthetic enzymatic pathway. PLoS ONE 2:e456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. Larossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.