Abstract
A total of 905 enterohemorrhagic Escherichia coli (EHEC) O157:H7 isolates that were recovered from experimentally infected cattle, in addition to the inoculated strain, were analyzed by pulsed-field gel electrophoresis (PFGE). Twelve PFGE profiles other than that of the inoculated strain were observed. We successfully identified five distinct chromosomal deletions that affected the PFGE profiles using whole-genome PCR scanning and DNA sequencing analysis. The changes in PFGE profiles of EHEC O157:H7 isolates after passage through the intestinal tract of cattle were partially generated by deletion of chromosomal regions.
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 causes hemorrhagic colitis and hemolytic-uremic syndrome in humans worldwide (18). Cattle are considered the primary reservoir for this pathogen and play a central role in transmission to humans (6). Healthy cattle transiently carry EHEC O157:H7 and shed the bacteria in their feces (5, 7). Human infections have been associated with the consumption of contaminated meat and milk, direct contact with cattle, and the consumption of vegetables, fruits, and water contaminated with cattle manure (6).
Because of its high discriminatory power, pulsed-field gel electrophoresis (PFGE) has been widely employed as a molecular typing method in many epidemiological investigations to identify various outbreaks and routes of transmission of EHEC O157:H7 (1, 12, 15, 17). Simpson's index of diversity (9) was reported to be >0.985 in previous studies (1, 15), supporting the identification of richness (the number of types among isolates) and evenness (the relative distribution of individual strains among the different types) of molecular typing using PFGE.
Instability of the PFGE patterns of EHEC O157:H7 isolates has been reported. Changes in PFGE patterns were observed among strains after repeated subculturing and prolonged storage at room temperature (11). Loss of Shiga toxin genes and a large-scale inversion within the genome were identified as genetic events generating changes in PFGE patterns in vitro (10, 13). Shifts in the genotypes of EHEC O157:H7 clinical isolates from patients and cattle have been reported (3, 14). This phenomenon was also observed in EHEC O157:H7 experimental infections of cattle. Spontaneous curing of a 90-kb plasmid resulted in the loss of two restricted fragments from the PFGE profiles of EHEC O157:H7 isolates obtained from experimentally infected cattle (2). The purpose of the present study was to identify the genetic events affecting the PFGE patterns of EHEC O157:H7 after passage through the intestinal tract of cattle, especially for restriction fragments that are >90 kb long.
Four 5-month-old Holstein steers were housed individually in climate-controlled biosafety level 2 containment barns in accordance with the guidelines for animal experimentation defined by the National Institute of Animal Health of Japan. The pens had individual floor drains and were cleaned twice daily with water and disinfectant. All animals were healthy and culture negative for EHEC O157:H7 strains, as determined by a previously described technique (2), prior to inoculation.
EHEC O157:H7 strain Sakai-215 (12, 23), which was isolated from an outbreak in Sakai, Osaka Prefecture, in 1996 was used for inoculation. This strain harbors the genes encoding Stx1 and Stx2. A spontaneous resistant strain was selected with nalidixic acid in order to facilitate the recovery of this strain from fecal samples. All calves were inoculated using a stomach tube with an exponential-phase culture (109 CFU) of the nalidixic acid-resistant Sakai-215 strain. Fecal samples were collected from the four calves daily for 45 days. Fecal culturing was performed as described previously (2). Eight non-sorbitol-fermenting colonies were selected daily from each animal and identified as EHEC O157:H7 colonies by routine diagnostic methods (25).
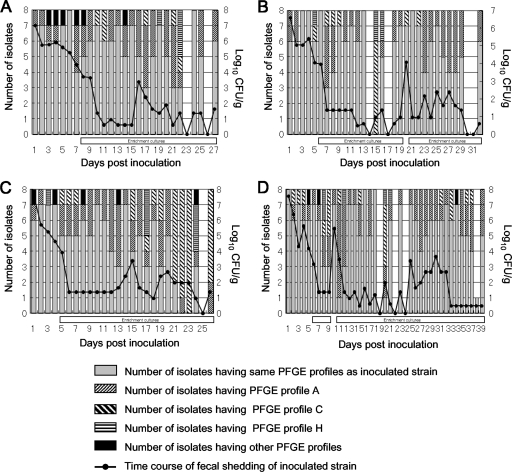
All animals were clinically normal throughout the experimental period. The EHEC O157:H7-inoculated calves (calves 1 to 4) were culture positive for the organism 24 h after inoculation. Intermittent fecal shedding by the calves was observed until 27, 32, 26, and 39 days postinoculation for calves 1, 2, 3, and 4, respectively (Fig. 1). The numbers of EHEC O157:H7 isolates recovered from calves 1, 2, 3, and 4 were 200, 224, 200, and 281, respectively.
FIG. 1.
Changes in PFGE profiles of EHEC O157:H7 isolates recovered from calves 1 (A), 2 (B), 3 (C), and 4 (D). The absence of a bar indicates that no EHEC O157:H7 was detected. The open horizontal bars under the vertical bars indicate that the eight isolates obtained on a day were obtained from the enrichment culture.
A total of 905 recovered isolates in addition to the inoculated strain were used for PFGE analysis. Genomic DNA from each EHEC O157:H7 isolate was prepared using the method of Persing (Mayo Clinic, Rochester, MN) described by Rice et al. (20). Agarose-embedded chromosomal DNA was cleaved with XbaI by following the manufacturer's instructions. PFGE was performed in a 0.85% megabase agarose gel, using a CHEF DR III apparatus (Bio-Rad Laboratories). The pulse time was increased from 12 to 35 s for 18 h. The PFGE profiles of all of the EHEC O157:H7 isolates recovered from the four calves were compared with that of the inoculated strain. The number of band differences was determined by enumerating the loss and addition of fragments (22).
Two hundred eighty-nine isolates had PFGE profiles different from that of the inoculated strain, and 12 distinct PFGE profiles were identified for these isolates (Table 1). The fact that only one to three band differences were observed for the 12 profiles suggested that these isolates were closely related (22) and were variants of the inoculated strain. In addition, the pens had individual floor drains and were cleaned twice daily with water and disinfectant, which reduced the likelihood of introduction of novel EHEC O157:H7 strains. We designated the PFGE profiles A to L. PFGE profiles A, C, and H were obtained for all four calves and accounted for 30.4% of the 905 isolates recovered. Different PFGE profiles were obtained for all animals at least 2 days postinoculation (Fig. 1). All eight isolates from calf 2 collected on day 15 postinoculation and from calf 3 collected on days 22 and 23 postinoculation had PFGE profiles different from that of the original isolate (Fig. 1). The isolates that had the same PFGE profile as the inoculated strain were detected again later.
TABLE 1.
Temporal distribution of PFGE profiles of EHEC O157:H7 isolates recovered from experimentally infected cattle
| PFGE profile | No. of isolates recovered at different times postinoculation from:
|
Total no. of isolates (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calf 1
|
Calf 2
|
Calf 3
|
Calf 4
|
||||||||||||
| 1 to 10 days | 11 to 20 days | 21 to 27 days | 1 to 10 days | 11 to 20 days | 21 to 30 days | 31 to 32 days | 1 to 10 days | 11 to 20 days | 21 to 36 days | 1 to 10 days | 11 to 20 days | 21 to 30 days | 31 to 39 days | ||
| Ina | 63 | 56 | 25 | 62 | 46 | 50 | 6 | 60 | 46 | 5 | 59 | 45 | 47 | 46 | 616 (68.1) |
| A | 11 | 19 | 10 | 13 | 7 | 21 | 2 | 13 | 24 | 4 | 10 | 19 | 16 | 12 | 181 (20.0) |
| B | 1 | 1 (0.1) | |||||||||||||
| C | 1 | 3 | 2 | 4 | 7 | 1 | 3 | 6 | 27 | 7 | 6 | 5 | 72 (8.0) | ||
| D | 1 | 1 (0.1) | |||||||||||||
| E | 1 | 1 (0.1) | |||||||||||||
| F | 1 | 2 | 3 (0.3) | ||||||||||||
| G | 1 | 1 (0.1) | |||||||||||||
| H | 1 | 3 | 1 | 4 | 1 | 3 | 3 | 2 | 2 | 1 | 1 | 22 (2.4) | |||
| I | 1 | 1 | 1 | 3 (0.3) | |||||||||||
| J | 1 | 1 (0.1) | |||||||||||||
| K | 1 | 1 | 2 (0.2) | ||||||||||||
| L | 1 | 1 (0.1) | |||||||||||||
| Total no. of variants (%)b | 17 (21.3) | 24 (30.0) | 15 (37.5) | 18 (22.5) | 18 (25.0) | 22 (30.6) | 2 (25.0) | 20 (25.0) | 34 (42.5) | 35 (87.5) | 21 (26.3) | 27 (37.5) | 17 (26.6) | 19 (29.2) | 289 (31.9) |
PFGE profile of inoculated strain Sakai-215.
Total numbers of isolates having PFGE profiles A to L.
Kudva et al. (16) demonstrated that the difference in PFGE profiles between EHEC O157:H7 strains was due to distinct insertions or deletions that contained XbaI sites rather than to single-nucleotide polymorphisms in the XbaI sites themselves. To identify the locations of insertions or deletions in the genome of the EHEC O157:H7 isolates recovered from experimentally infected cattle, whole-genome PCR scanning (WGP scanning) was performed as described previously (19). Briefly, 549 pairs of PCR primers were used to amplify 549 segments covering the whole chromosome of EHEC O157:H7 strain RIMD 0509952, with overlaps of a certain length at every segment end. The inoculated strain (strain Sakai-215) and the strain whose genome was sequenced (RIMD 0509952) were isolated from the same outbreak in Japan in 1996 (23) and had same PFGE profile after XbaI digestion. All primer sequences are available at http://genome.gen-info.osaka-u.ac.jp/bacteria/o157/pcrscan.html. PCR were performed using genomic DNA as the template and long accurate PCR (LA-PCR) kits. The cycling conditions for the LA-PCR included an initial incubation at 96°C for 100 s, followed by 30 cycles of 96°C for 20 s and 69°C for 10 min.
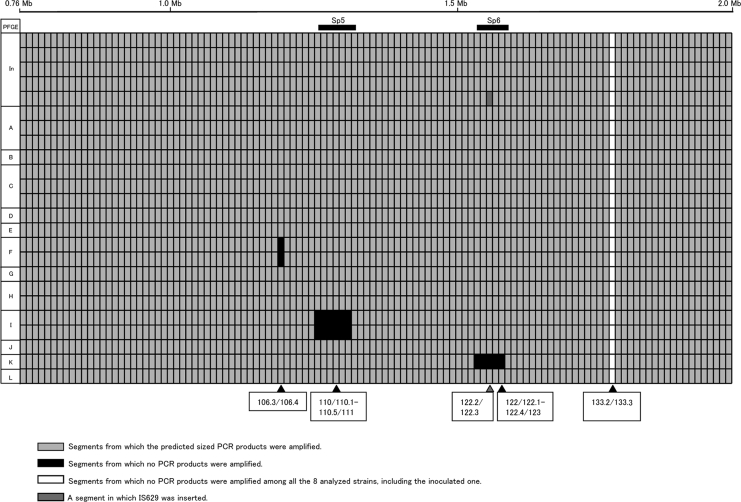
Prior to the WGP scanning of the isolates, we scanned an approximately 1.2-Mb region covered by 116 segments (71/72 to 146/147) of the EHEC O157:H7 genome using 24 strains, including inoculated strain Sakai-215, 4 isolates with the same PFGE profile as the inoculated strain, 3 isolates with PFGE profile A, 3 isolates with PFGE profile C, 2 isolates with PFGE profile F, 2 isolates with PFGE profile H, 2 isolates with PFGE profile I, and one isolate each with PFGE profiles B, D, E, G, J, K, and L. The main purpose of this preliminary scanning was to determine the extent of variation in the data for isolates having the same PFGE profiles.
As shown in Fig. 2, we successfully amplified products that were the expected sizes for 103 of the 116 segments for the 24 strains tested. No amplification in a segment was observed for the 24 strains. Polymorphism (expected amplification was observed in some but not all strains) was observed in 12 segments. Eleven of the 12 polymorphic segments consisted of two different sequentially unamplified regions. An IS629 insertion was also observed in a polymorphic segment in one strain. In other words, variation in the data for isolates with the same PFGE profile was not observed except for the isolates having the same PFGE profile as the inoculated strain. Hence, we performed WGP scanning using one isolate with each of the selected PFGE profiles.
FIG. 2.
Summary of the results of PCR scanning analysis of part of the EHEC O157:H7 genome using 24 strains recovered from experimentally infected cattle. The line at the top indicates data for the inoculated strain. The positions of Sp5 and Sp6 are indicated above the data lines. Segments showing polymorphism (expected amplification was observed in some strains but not in all strains) are indicated below the data lines. In, inoculated strain.
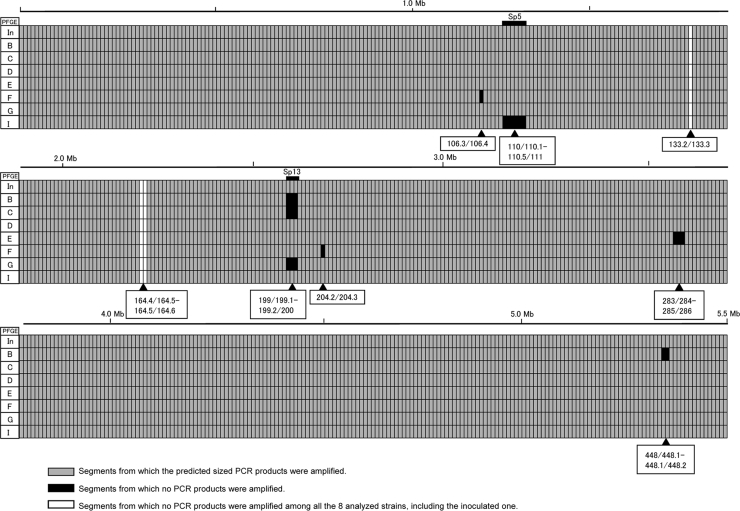
The results of WGP scanning of the seven isolates with different PFGE profiles in addition to inoculated strain Sakai-215 are summarized in Fig. 3. We successfully amplified products of the expected sizes for 530 of 549 segments for the eight strains tested. No amplification was observed for any of the eight strains for three segments (133.2/133.3, 164.4/164.5, and 164.5/164.6). Polymorphism was observed in 16 segments. Fourteen of the 16 polymorphic segments were located in four different regions.
FIG. 3.
Summary of the results of WGP scanning analysis of the EHEC O157:H7 isolates recovered from experimentally infected cattle and the inoculated strain. The positions of Sp5 and Sp13 are indicated above the data lines. Segments showing polymorphism (expected amplification was observed in some strains but not in all strains) are indicated below the data lines. In, inoculated strain.
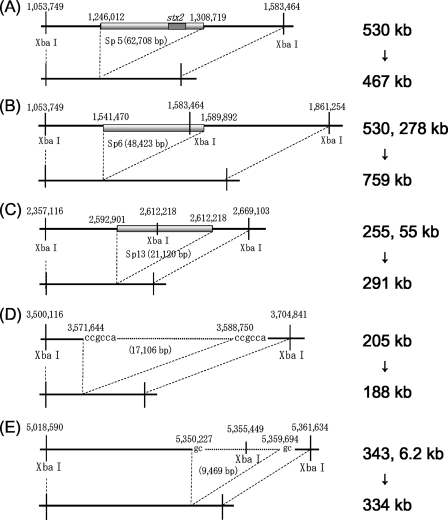
The 110/110.1-to-110.5/111 region in PFGE profile I, the 122/122.1-to-122.4/123 region in PFGE profile K, and the 199/199.1-to-199.2/200 region in PFGE profiles B, C, and G corresponded to prophages Sp5, Sp6, and Sp13, respectively. The 283/284-to-285/286 region in PFGE profile E and the 448/448.1-to-448.1/448.2 region in PFGE profile B corresponded to nonprophage regions on the chromosome. The sizes of the deletion sites of nonprophage regions 283/284 to 285/286 and 448/448.1 to 448.1/448.2 were 17 kb and 9.5 kb, respectively. We synthesized new primer pairs upstream and downstream of these five regions and performed LA-PCR (data not shown). The results of the sequencing analysis of the products indicated that the three prophage genomes were cured at their integration sites (Fig. 4A to C). It is not clear from this study whether deletion of the three prophages represented phage excisions or simple deletions. We identified short direct CCGCCA and GC repeats at both ends of the 17-kb and 9.5-kb deletion sites, respectively, compared with the sequence data for the Sakai-215 strain, although the deleted regions included one side of the direct repeats (Fig. 5D and E).
FIG. 4.
Schematic diagrams showing the relationships between deletions of chromosomal regions and changes in the sizes of restricted fragments. (A) The 467-kb restricted fragment of PFGE profile I was generated by deletion of prophage Sp5 located in the 530-kb fragments of the inoculated strain. (B) The 759-kb restricted fragment of PFGE profile K was generated by deletion of Sp6 located in the adjacent 530-kb and 278-kb fragments of the inoculated strain. (C) The 291-kb restricted fragment of PFGE profiles C, G, and H was generated by deletion of prophage Sp13 located in the adjacent 255-kb and 55-kb fragments of the inoculated strain. (D) The 188-kb restricted fragment of PFGE profile E was generated by deletion of the 17-kb chromosomal region in the 205-kb fragment of the inoculated strain. (E) The 334-kb restricted fragment of PFGE profile B was generated by deletion of the 9.5-kb region located in the adjacent 343-kb and 6.2-kb fragments of the inoculated strain.
FIG. 5.
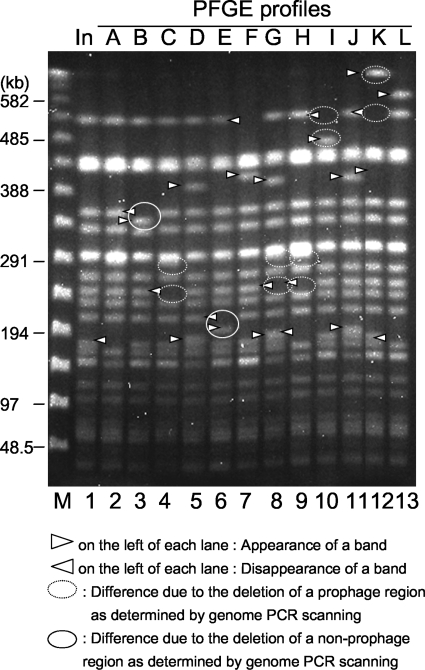
Comparison of the PFGE profiles of the EHEC O157:H7 isolates recovered from experimentally infected cattle and the inoculated strain. Lane M, λ ladder used as a size marker; lane 1, inoculated strain; lanes 2 to 13, isolates with PFGE profiles A to L, respectively.
The deleted 17-kb region contains 16 open reading frames, including formate hydrogenase-related genes (4), mutS (21), and rpoS (8), suggesting that the strain with PFGE profile E is more susceptible to environmental stresses than the inoculated strain. In fact, the isolate with PFGE profile E was more susceptible to low-pH, high-temperature, and high-osmolarity conditions or to the presence of deoxycholate in vitro than the other isolates obtained in this study (data not shown). The fact that this isolate was obtained 4 days after inoculation from calf 1 and could not be detected after that time suggested that the isolate with PFGE profile E could not survive in the intestine of the calf due to the loss of genes related to stress resistance. The deleted 9.5-kb region contains nine open reading frames whose functions are unknown. The strain with this deletion was isolated 1 day after inoculation from calf 3 and could not be detected after that time.
Sp5 is one of the prophages in EHEC O157:H7 RIMD 0509952 carrying the stx2 gene. Deletion of this prophage affected the PFGE profile of inoculated strain Sakai-215. The loss of a 530-kb fragment and the gain of a 467-kb fragment due to deletion of the 63-kb prophage Sp5 were identified in PFGE profile I (Fig. 4A and 5).
Sp6 is one of the lambda-like phages and has a single XbaI site in its genome. The loss of 530-kb and 278-kb fragments and the gain of a 759-kb fragment due to deletion of this phage were identified in PFGE profile K (Fig. 4B and 5).
Sp13 is one of the P2-like phages that have a single XbaI site in the genome. The loss of 255-kb and 55-kb fragments and the gain of a 291-kb fragment due to deletion of this prophage were identified in PFGE profiles C, G, and H (Fig. 4C and 5). The same changes in PFGE profile B were not observed, although we found a sequentially unamplified region in which Sp13 was located in the genomes of isolates with PFGE profile B (Fig. 3). We detected part of the Sp13 sequence by Southern blot analysis; however, this part of the sequence was not detected in isolates with PFGE profiles C, G, and H (data not shown). One possible explanation for this phenomenon is that deletion of part of the Sp13 sequence included deletion of primer annealing sites. However, the details of mutation in this region for the isolates with PFGE profile B are not clear.
Deletion of the two nonprophage regions also affected the PFGE profiles. The loss of a 205-kb fragment and the gain of a 188-kb fragment due to deletion of a 17-kb region were identified in PFGE profile E (Fig. 4D and 5). The loss of a 343-kb fragment and the gain of a 334-kb fragment due to deletion of a 9.5-kb region were identified in PFGE profile B (Fig. 4E and 5).
Two single unamplified segments were both observed in the strain with PFGE profile F (106.3/106.4 and 204.2/204.3). We could not amplify these regions using additional primer pairs (data not shown). Insertion of DNA or large-scale inversion might have occurred in these regions. The other unamplified segments all corresponded to deletion of chromosomal regions. Recombination successfully occurred and cured three prophages and two other chromosomal regions. These data suggest that the changes in PFGE profiles after passage through the intestinal tract of cattle are generated in part by deletion of chromosomal regions. Obviously, deletion of five chromosomal regions does not explain the other changes in the PFGE profiles, including profiles A, D, F, J, and L. The genetic events behind such changes are not clear.
Prior to drawing a conclusion, we need to consider the use of nalidixic acid, a potent inducer of bacteriophage induction (24), for selection of the isolates. In addition, most of the EHEC O157:H7 isolates obtained on day 8 postinoculation and later were isolated from enrichment cultures (Fig. 1). The possibility that the culturing process itself affected the deletion events affecting the PFGE profiles cannot be ruled out. Taken together, the results suggest that deletions can cause a single strain to mutate into several variants while it is passing through the gastrointestinal tract of a host, provided that the culture technique used does not contribute to this process. Hence, this study may explain why EHEC O157:H7 isolates with various PFGE profiles can be isolated from a single animal. What causes the deletion mutations and why the PFGE profiles show such patterns after passage through cattle are subjects for future studies.
Acknowledgments
This work was supported by a grant-in-aid from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Zoonoses Control Project).
Footnotes
Published ahead of print on 6 July 2009.
REFERENCES
- 1.Akiba, M., T. Masuda, T. Sameshima, K. Katsuda, and M. Nakazawa. 1999. Molecular typing of Escherichia coli O157:H7 (H-) isolates from cattle in Japan. Epidemiol. Infect. 122:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiba, M., T. Sameshima, and M. Nakazawa. 2000. Clonal turnover of enterohemorrhagic Escherichia coli O157:H7 in experimentally infected cattle. FEMS Microbiol. Lett. 184:79-83. [DOI] [PubMed] [Google Scholar]
- 3.Akiba, M., T. Sameshima, and M. Nakazawa. 1999. The shift of genetic subtypes of Escherichia coli O157:H7 isolates from cattle. Epidemiol. Infect. 122:343-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock, A., and G. Sawer. 1996. Fermentation, p. 262-282. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 5.Cray, W. C., Jr., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gyles, C. L. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85:E45-E62. [DOI] [PubMed] [Google Scholar]
- 7.Hancock, D. D., T. E. Besser, and D. H. Rice. 1998. Ecology of Escherichia coli O157:H7 in cattle and impact of management practices, p. 85-91. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, DC.
- 8.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iguchi, A., S. Iyoda, J. Terajima, H. Watanabe, and R. Osawa. 2006. Spontaneous recombination between homologous prophage regions causes large-scale inversions within the Escherichia coli O157:H7 chromosome. Gene 372:199-207. [DOI] [PubMed] [Google Scholar]
- 11.Iguchi, A., R. Osawa, J. Kawano, A. Shimizu, J. Terajima, and H. Watanabe. 2002. Effects of repeated subculturing and prolonged storage at room temperature of enterohemorrhagic Escherichia coli O157:H7 on pulsed-field gel electrophoresis profiles. J. Clin. Microbiol. 40:3079-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izumiya, H., J. Terajima, A. Wada, Y. Inagaki, K. I. Itoh, K. Tamura, and H. Watanabe. 1997. Molecular typing of enterohemorrhagic Escherichia coli O157:H7 isolates in Japan by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 35:1675-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karch, H., T. Meyer, H. Russmann, and J. Heesemann. 1992. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect. Immun. 60:3464-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karch, H., H. Russmann, H. Schmidt, A. Schwarzkopf, and J. Heesemann. 1995. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J. Clin. Microbiol. 33:1602-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krause, U., F. M. Thomson-Carter, and T. H. Pennington. 1996. Molecular epidemiology of Escherichia coli O157:H7 by pulsed-field gel electrophoresis and comparison with that by bacteriophage typing. J. Clin. Microbiol. 34:959-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kudva, I. T., P. S. Evans, N. T. Perna, T. J. Barrett, F. M. Ausubel, F. R. Blattner, and S. B. Calderwood. 2002. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J. Bacteriol. 184:1873-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng, J., S. Zhao, T. Zhao, and M. P. Doyle. 1995. Molecular characterisation of Escherichia coli O157:H7 isolates by pulsed-field gel electrophoresis and plasmid DNA analysis. J. Med. Microbiol. 42:258-263. [DOI] [PubMed] [Google Scholar]
- 18.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA 99:17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice, D. H., K. M. McMenamin, L. C. Pritchett, D. D. Hancock, and T. E. Besser. 1999. Genetic subtyping of Escherichia coli O157 isolates from 41 Pacific Northwest USA cattle farms. Epidemiol. Infect. 122:479-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupp, W. D. 1996. DNA repair mechanisms, p. 2277-2294. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 22.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe, H., A. Wada, Y. Inagaki, K. Itoh, and K. Tamura. 1996. Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotype strains in Japan, 1996. Lancet 348:831-832. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. K. Acheson. 2000. Quinolone antibiotics induce shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]
- 25.Zhao, T., M. P. Doyle, J. Shere, and L. Garber. 1995. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ. Microbiol. 61:1290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]