Abstract
Gene manipulation tools for an arachidonic-producing filamentous fungus, Mortierella alpina 1S-4, have not been sufficiently developed. In this study, Agrobacterium tumefaciens-mediated transformation (ATMT) was investigated for M. alpina 1S-4 transformation, using the uracil-auxotrophic mutant (ura5− strain) of M. alpina 1S-4 as a host strain and the homologous ura5 gene as a selectable marker gene. Furthermore, the gene for ω3-desaturase, catalyzing the conversion of n-6 fatty acid to n-3 fatty acid, was overexpressed in M. alpina 1S-4 by employing the ATMT system. As a result, we revealed that the frequency of transformation surpassed 400 transformants/108 spores, most of the integrated T-DNA appeared as a single copy at a random position in chromosomal DNA, and most of the transformants (60 to 80%) showed mitotic stability. Moreover, the accumulation of n-3 fatty acid in transformants was observed under the conditions of optimal ω3-desaturase gene expression. In particular, eicosapentaenoic acid (20:5n-3), an end product of n-3 fatty acids synthesized in M. alpina 1S-4, reached a maximum of 40% of total fatty acids. In conclusion, the ATMT system was found to be effective and suitable for the industrial strain Mortierella alpina 1S-4 and will be a useful tool for basic mutagenesis research and for industrial breeding of this strain.
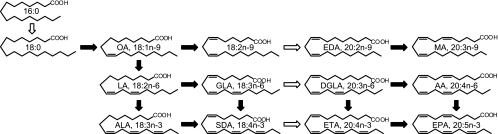
Two decades ago, a filamentous zygomycete fungus, Mortierella alpina 1S-4, was isolated from soil as a potent producer of polyunsaturated fatty acids (PUFAs) in our laboratory and was utilized for commercial production of arachidonic acid (AA) (20:4n-6) (21). Breeding of mutants derived from the wild strain led to the production of dihomo-γ-linolenic acid (20:3n-6) and Mead acid (20:3n-9) (10-12) (Fig. 1). Furthermore, we attempted to produce other PUFAs synthesized in M. alpina 1S-4, since some fatty acids (e.g., 18:2n-9, 18:4n-3, and 20:4n-3) have limited natural sources and could have promising beneficial physiological effects (9). In particular, for microbial production of n-3 PUFAs, currently prepared from fish oil, it is necessary to achieve stable productivity and quality; however, mutation treatment caused low activity of the specific enzymes involved in PUFA biosynthesis, which is unsuitable for industrial application. In addition, gene manipulation tools have not been sufficiently developed for metabolic control of the PUFA synthetic pathway. Genetic manipulation is a new means of molecularly breeding industrial strains, analyzing their physiological properties, and clarifying the biosynthetic pathway to PUFAs. A comprehensive transformation system for this fungus has been fundamentally established. It involves a uracil-auxotrophic mutant (ura5− strain) as a host strain, a homologous ura5 gene as a selectable marker gene, and transformation through the biolistic method, which is the only effective method (24).
FIG. 1.
Putative biosynthetic pathway of PUFAs in Mortierella alpina 1S-4. OA, oleic acid; LA, linoleic acid; ALA, α-linolenic acid; GLA, γ-linolenic acid; SDA, stearidonic acid; EDA, n-9 eicosadienoic acid; DGLA, dihomo-γ-linolenic acid; ETA, n-3 eicosatetraenoic acid; MA, Mead acid. Open and black arrows indicate elongase and desaturase reactions, respectively.
Agrobacterium tumefaciens-mediated transformation (ATMT) has been employed for a wide range of plants (7, 27). Recently, it was reported that A. tumefaciens is also able to transfer its DNA to various fungi, including ascomycetes, basidiomycetes, zygomycetes, and oomycetes, as well as to plants (2, 5, 16). Additionally, this bacterium can transform intact cells and spores as well as protoplasts. Under mild conditions, the ATMT system generates a large number of stable transformants, which show vigorous growth, indicating that the ATMT system can be an efficient tool for molecular manipulation of M. alpina 1S-4. Moreover, the frequency of homologous recombination was higher than that with conventional transformation methods (8). In this study, we evaluated the external gene transfer system using the ATMT system and determined the optimal conditions for M. alpina 1S-4. Furthermore, we overexpressed the ω3-desaturase gene to improve n-3 PUFA productivity in an industrial n-6-PUFA-producing strain, M. alpina 1S-4 (18, 20), using ATMT.
MATERIALS AND METHODS
Enzymes and chemicals.
Restriction enzymes and other DNA-modifying enzymes were obtained from Takara Bio Inc. (Shiga, Japan) and New England BioLabs (Beverly, MA). Nitrocellulose membranes (70-mm diameter; hardened low-ash grade 50) for cocultivation were purchased from Whatman (Maidstone, United Kingdom). Nile blue A was purchased from Sigma (St. Louis, MO). All other chemicals were of analytical grade.
Strains and growth conditions.
The Mortierella alpina 1S-4 ura5− strain derived from mutation was obtained as described previously, maintained on potato dextrose agar, and used as the recipient host strain for transformation (24). For plasmid construction, Escherichia coli strain DH5α was used and grown on LB agar plates containing 50 μg/ml ampicillin or kanamycin, when appropriate. Agrobacterium tumefaciens C58C1 and plasmid pBIG2RHPH2 were kindly provided by Yasuyuki Kubo (Kyoto Prefectural University, Japan). This strain was used as a T-DNA donor for fungal transformation. The compositions of Czapek-Dox and synthetic complete (SC) agar media were described previously (24). Czapek-Dox agar medium, supplemented with 0.05 mg/ml uracil, was used for sporulation of the ura5− strain. SC agar medium with or without uracil was used to cultivate M. alpina 1S-4 ura5− and its transformants. GY medium (2% [wt/vol] glucose and 1% yeast extract) was used for liquid culture, except for extracting genomic DNA (2% [wt/vol] glucose and 3% yeast extract). All liquid culture of transformants was performed with reciprocal shaking at 125 strokes/min at 28°C. LB-Mg agar (1% tryptone, 0.5% yeast extract, 85 mM NaCl, 0.5 mM MgSO4·7H2O, 0.5 mM NaOH, 1.5% agar, pH 7.0) was used for transformation of A. tumefaciens C58C1. Minimal medium (MM) [10 mM K2HPO4, 10 mM KH2PO4, 2.5 mM NaCl, 2 mM MgSO4·7H2O, 0.7 mM CaCl2, 9 μM FeSO4·7H2O, 4 mM (NH4)2SO4, 10 mM glucose, pH 7.0] was used for cultivation of A. tumefaciens C58C1. Induction medium (IM) [MM containing 0.5% (wt/vol) glycerol, 200 μM acetosyringone, 40 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.3] was used with A. tumefaciens C58C1 to infect M. alpina 1S-4.
Construction of T-DNA binary vector.
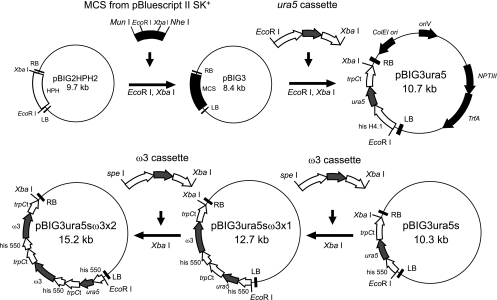
A T-DNA binary vector was constructed on the backbone of pBIG2RHPH2 (25). A fragment carrying a multiple-cloning site (MCS) was amplified with the sense primer MCS F (5′-AAACAGCTAGCACCATGATTACGCCAAGC-3′) and the antisense primer MCS R (5′-GTAATACGACTCACTATAGGGCCAATTGGG-3′) from pBluescript II SK+ (Stratagene, La Jolla, CA). The sense and antisense primers contained NheI and MunI recognition sites, respectively (underlined). The resulting fragments were digested with NheI and MunI for ligation to pBIG2RHPH2 digested with XbaI and EcoRI. A binary vector was constructed by replacing the hygromycin B resistance gene cassette in pBIG2RHPH2 with the MCS region and was designated pBIG3. The ura5 expression cassette derived from pDura5 (24) was then inserted between the left and right border repeats of T-DNA, with the resulting vector being designated pBIG3ura5 (Fig. 2). Other binary vectors, pBIG3ura5sω3x1 and pBIG3ura5sω3x2, were constructed on the base of a slightly modified pBIG3ura5 for easy handling gene manipulation, as follows. The ura5 gene expression cassette was amplified by a sense primer, hisproD1EcoRISpeIF (5′-GAGACGAATTCGCCCGTACGGCCGACTAGTTTTAGTTGATGTGAG-3′), and an antisense primer, trpCtXbaIR (5′-GTTCCTCGTCTAGACCTCTAAACAAGTGTACCTGTGCATTCTGGG-3′), with pDura5 as a template. These primers contained EcoRI, SpeI, and XbaI restriction sites, respectively (underlined). The resultant 550-bp fragment, containing a whole histone H4.1 promoter, was treated with EcoRI and XbaI and ligated to pBIG3 treated with the same enzymes to construct pBIG3ura5s. Subsequently, after the ω3-desaturase gene was amplified with the sense primer w3F2PciI (5′-GGGAATATTAAGCTTACATGTCCCC-3′) and the antisense primer w3R2BamHI (5′-GCCGGATCCAAATTGTTAATGCTTG-3′), containing PciI and BamHI restriction sites (underlined), and the Zap-cDNA library constructed previously (19) as a template, the PCR fragment was treated with PciI and BamHI and ligated to pD4 treated with NcoI and BamHI, with the resultant construct being designated pD4ω3 (14). The ω3-desaturase gene expression cassette, which was amplified by the sense primer hisproD1EcoRISpeIF and the antisense primer trpCtXbaIR with pD4ω3 as a template, was treated with SpeI and XbaI and ligated to pBIG3ura5s digested with XbaI to construct pBIG3ura5sω3x1. Finally, the same ω3-desaturase expression cassette ligated into pBIG3ura5sω3x1 was digested with XbaI and designated pBIG3ura5sω3x2.
FIG. 2.
Construction of binary vector pBIG3ura5, pBIG3ura5s, pBIG3ura5sω3x1, and pBIG3ura5sω3x2 for M. alpina 1S-4 transformation. The 1-kb M. alpina his H4.1 and his 550 promoter fragment was derived from strain CBS 528.72. The 700-bp fragment containing the trpC transcription terminator region was derived from Aspergillus nidulans. HPH, hygromycin B phosphotransferase gene; ura5, orotate phosphoribosyl transferase gene of M. alpina 1S-4; NPTIII, neomycin phosphotransferase III gene; TrfA, TrfA locus, which produces two proteins that promote replication of the plasmid; ColEI ori, ColEI origin of replication; oriV, pRK2 origin of replication; RB, right border; LB, left border; ω3, ω3-desaturase gene.
ATMT.
A spore suspension of the uracil-auxotrophic M. alpina 1S-4 mutant was freshly prepared by harvesting from cultures growing on Czapek-Dox agar medium supplemented with 0.05 mg/ml uracil and then filtering the suspension through Miracloth (Calbiochem) (24). ATMT was performed with a slight modification of the previously described protocol (5). Initially, A. tumefaciens C58C1 was transformed with each binary vector via electroporation as described previously (15). A. tumefaciens transformants were isolated on LB-Mg agar plates supplemented with kanamycin, ampicillin, and rifampin (rifampicin) (50 μg/ml), respectively. Positive A. tumefaciens transformants carrying each binary vector were confirmed by PCR and grown under good aeration conditions at 28°C for 48 h in MM supplemented with kanamycin (50 μg/ml) and ampicillin (50 μg/ml) after autoclaving. Bacterial cells were harvested by centrifugation at 5,800 × g, washed once with fresh IM, and then diluted to an optical density of 660 nm (OD660) of 0.5 to 1.5 in 10 ml of fresh IM. After preincubation for 8 to 12 h at 28°C on a rotary shaker (300 strokes/min) to an OD660 of 0.4 to 3.7, 100 μl of the bacterial cell suspension was mixed with an equal volume of a spore suspension (108 ml−1) and then spread on nitrocellulose membranes. The membranes were placed on a cocultivation medium (same as IM except containing 5 mM instead of 10 mM glucose and 1.5% agar) and incubated at 20 to 28°C for 0 to 5 days. After cocultivation, the membranes were transferred to uracil-free SC agar plates that contained 50 μg/ml cefotaxim and 50 μg/ml spectinomycin to inhibit the growth of A. tumefaciens and 0.03% Nile blue A (Sigma) to discriminate between fungal colonies and the white color of the membrane. After 5 days of incubation at 28°C, hyphae from visible fungal colonies were transferred to fresh uracil-free SC agar plates, which was repeated three times to obtain candidates for stable transformants. Transformants that could grow on uracil-free SC agar plates but could no longer grow on GY agar plates containing 5-fluoroorotic acid (5-FOA) (0.05%, wt/vol) were regarded as stable transformants (6). Control experiments were performed without acetosyringone. All experiments were carried out in triplicate, and the averages from three separate experiments, which were reproducible within ±10%, are presented in the figures.
Genomic DNA preparation.
M. alpina strains were cultivated in GY medium (glucose 2% and yeast extract 3%) at 28°C for 4 days on a rotary shaker (300 strokes/min). Fungal mycelia were harvested by suction filtration and washed twice with sterile water. Genomic DNA was prepared by the method described previously (19).
Southern blot analysis.
For Southern blot analysis, 10 μg genomic DNA was digested with restriction enzymes overnight. The treated fragments were size fractioned in a 1% agarose gel and then transferred to Hybond N+ membranes (Amersham Biosciences, Buckinghamshire, United Kingdom) using a VacuGene XL Vacuum Blotting System (Amersham Biosciences). Hybridization was performed with a Gene Images Alkphos Direct labeling and detection system (Amersham Bioscience) following the standard procedure.
Analysis of fatty acids and lipids.
Fatty acid analysis was performed basically as described previously (19). Spores of recombinant strains obtained by the ATMT system were inoculated into a 20-ml Erlenmeyer flask containing 4 ml GY medium (glucose, 2%; yeast extract, 1%). All transformants and wild strains were cultivated at 12°C with reciprocal shaking (120 strokes/min) after a 2-day precultivation at 28°C. All experiments were carried out in triplicate, and the averages from three separate experiments, which were reproducible within ±10%, are presented in the figures.
RESULTS
Establishment of ATMT for M. alpina 1S-4. (i) Optimization of ATMT conditions.
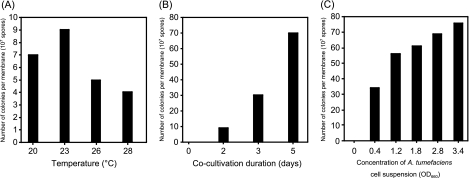
We optimized the conditions of A. tumefaciens C58C1 harboring pBIG3ura5-mediated transformation of M. alpina 1S-4. Preliminary transformation was performed by spreading a mixed suspension of spores and bacteria on sterile nitrocellulose membranes on IM agar plates with or without acetosyringone, which is necessary for the induction of vir genes (23), followed by successive incubation for 2 days at 23°C as described previously (4, 5, 8). After 48 h, the membranes were transferred to SC uracil-free agar plates with Nile blue A (0.04%, wt/vol) and incubated statically at 28°C for 4 days. As a result, some colonies were obtained only with acetosyringone (data not shown). Subsequently, the effects of cocultivation conditions on the efficiency of ATMT of M. alpina 1S-4 were examined. The temperature, cocultivation duration, and concentration of the bacterial cell suspension were optimized. The optimal temperature for cocultivation was 23°C under cocultivation for 2 days (Fig. 3A). The number of colonies also increased depending on the cocultivation duration (Fig. 3B); however, membranes were completely covered by a large number of colonies on selection medium after prolonged cocultivation (over 5 days), which complicated the description of each colony. The number of colonies increased depending on the concentration of the bacterial cell suspension used for cocultivation for 5 days (Fig. 3C).
FIG. 3.
(A) Effect of cocultivation temperature on transformation of M. alpina 1S-4. An Agrobacterium culture at an OD660 of 1.5 was used for cocultivation. The spore suspension (100 μl) and bacterial cell suspension (100 μl) were mixed and then incubated on an IM agar plate containing 200 μM of acetosyringone for 2 days. (B) Effect of cocultivation duration on transformation of M. alpina 1S-4. An Agrobacterium culture at an OD660 of 1.5 was used for cocultivation. The spore suspension (100 μl) and bacterial cell suspension (100 μl) were mixed and then incubated on an IM agar plate containing 200 μΜ of acetosyringone at 23°C. (C) Effect of concentration of A. tumefaciens cell suspension on transformation of M. alpina 1S-4. The spore suspension (100 μl) and bacterial cell suspension (100 μl) at the indicated concentrations (OD660) were mixed and then incubated on an IM agar plate containing 200 μM acetosyringone for 5 days. Other conditions were the same as described in Materials and Methods.
(ii) Mitotic stability.
We have already revealed that stable transformants could not grow on GY agar plates containing 5-FOA but could grow on SC uracil-free agar plates (24). Twenty-seven putative transformants were selected randomly and their stability checked by culturing on GY agar plates containing 5-FOA and on SC uracil-free medium. Furthermore, the mitotic stability of these stable transformants was evaluated by successively subculturing them on GY agar plates for three generations, after which they were transferred to SC uracil-free agar plates. As a result, we obtained 16 mitotically stable transformants out of 27 colonies (>60%). This experiment was carried out in triplicate and showed at least over 60% stable transformants of the total colonies on average. About 70 transformant colonies were obtained with one membrane, resulting in the acquisition of 40 stable transformants per membrane. Ten cocultivated membranes containing 108 spores in total were prepared at once. Thus, the transformation frequency surpassed 400 transformants/108 spores.
(iii) Southern blot analysis.
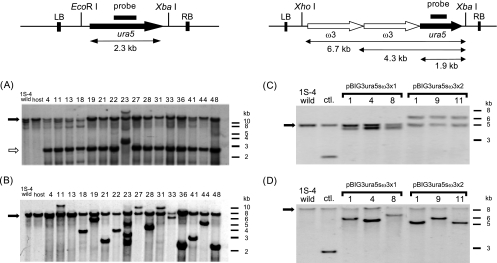
Figure 4 shows T-DNA region integration into chromosomal DNA observed on Southern blot analysis, using genomic DNA from each stable transformant, and probed with the ura5 gene. EcoRI and XbaI recognition sites were located at the upper and lower ends of the ura5 gene cassette, 124 and 81 bp inside of the nick sites of the left and right borders of T-DNA, respectively (Fig. 4A). Almost all transformants showed bands of hybridization corresponding to 2.3 kb, the size of the intact ura5 gene cassette, which suggested that T-DNA was integrated into chromosomal DNA of M. alpina 1S-4 without truncation or disruption. On the other hand, a different, 3.0-kb band was observed for transformant 23. This phenomenon has often been observed with the ATMT system (2, 5, 17). It was considered that integrated T-DNA suffered truncation at the corresponding restriction sites. Subsequently, each prepared chromosomal DNA was digested with both EcoRI and SpeI, the SpeI site of which was absent in the T-DNA region, and hybridized with the same probe to check the copy number and integration location of T-DNA (Fig. 4B). As a result, transformant 23 was considered to have multiple copies of T-DNA due to the observation of four bands.
FIG. 4.
Southern blot analysis of stable transformants. LB, left border; RB, right border. (A) Genomic DNA (10 μg) was digested with EcoRI and XbaI. (B) Genomic DNA (10 μg) was digested with EcoRI and SpeI. (C) Genomic DNA (10 μg) was digested with XhoI and XbaI. (D) Genomic DNA (10 μg) was digested with SpeI and XbaI. Hybridization was performed using a 0.6-kb DNA probe covering the ura5 gene. The white arrow in panel A indicates the size corresponding to an intact 2.3-kb ura5 gene cassette. The endogenous ura5 gene is indicated by black arrows in panels A, B, C, and D. Molecular size markers (kb) are indicated on the right.
Breeding of eicosapentaenoic acid (EPA) producer by ATMT. (i) Southern blot and expression analysis of the ω3-desaturase gene among transformants.
Genomic DNAs were also isolated from pBIG3ura5s transformant 1 (control), pBIG3ura5sω3x1 (no. 1, 4, and 8), pBIG3ura5sω3x2 (no. 1, 9, and 11), and the 1S-4 wild strain, and the copy number and integration location of T-DNA were checked in the same way. As shown in Fig. 4C, 1.9-kb, 4.3-kb, and 6.7-kb signals corresponded to the T-DNA region of each binary vector and integrated into chromosomal DNA without truncation or disruption. Subsequently, genomic DNA was digested with XbaI and SpeI, the restriction site of which was absent in the T-DNA region, and hybridized with the same probe (Fig. 4D). These observations indicated that a single T-DNA was integrated at a random location into chromosomal DNA without truncation or disruption.
(ii) Fatty acid analysis.
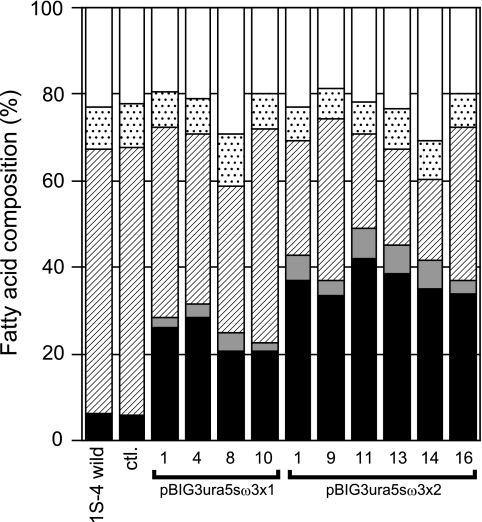
The spores of randomly selected stable transformants overexpressing ω3-desaturase and the wild strain were cultivated to analyze fatty acid composition in 4 ml GY medium for 16 days at 12°C after a 2-day precultivation at 28°C. Transformant 1 with pBIG3ura5s was used as a control. As a result, the contents and productivity (mg/mg dry cells and mg/ml) of n-3 PUFAs increased among all ω3-desaturase-overexpressing transformants (Fig. 5; Table 1). The contents of n-3 PUFAs in pBIG3ura5sω3x1 and pBIG3ura5sω3x2 after 18 days of cultivation were (24.5 ± 2.2)% and (42.1 ± 2.0)% (mean ± standard error), respectively (Fig. 5). The n-3 PUFAs content in pBIG3ura5sω3x2 transformants was approximately 1.7-fold higher than that in pBIG3ura5sω3x1 transformants. In the best transformant (no. 11), the content of EPA, an end product of PUFA synthesized by M. alpina 1S-4, was up to 40% of total fatty acids. On the other hand, AA production was decreased with increasing EPA production, indicating that AA was transformed to EPA by ω3-desaturase.
FIG. 5.
Comparison of fatty acid composition among transformants. Cultivation was performed in 4 ml GY medium with shaking at 120 strokes/min for 2 days at 28°C and for a further 16 days at 12°C. ▪, EPA; shaded bar, other n-3 PUFAs (α-linolenic acid, stearidonic acid, and n-3 eicosatetraenoic acid); (▒), AA; ▩, other n-6 PUFAs (linoleic acid; γ-linolenic acid, and dihomo-γ-linolenic acid); □, other PUFAs.
TABLE 1.
Fatty acid production by transformants
| Strain | Dry cellsa (g/liter) | Total fatty acidsa
|
EPA (20:5) yield
|
||
|---|---|---|---|---|---|
| mg/g | g/liter | mg/g | g/liter | ||
| Mortierella alpina 1S-4 (wild) | 10.6 | 177.7 | 1.89 | 10.9 | 0.12 |
| pBIG3ura5s no. 1 (control) | 10.2 | 143.8 | 1.47 | 8.2 | 0.08 |
| pBIG3ura5sω3x1 no. 1 | 10.7 | 170.1 | 1.82 | 44.2 | 0.47 |
| pBIG3ura5sω3x1 no. 4 | 10.8 | 133.6 | 1.44 | 37.9 | 0.41 |
| pBIG3ura5sω3x1 no. 8 | 10.8 | 153.1 | 1.65 | 31.3 | 0.34 |
| pBIG3ura5sω3x1 no. 10 | 11.2 | 180.8 | 2.02 | 37.4 | 0.42 |
| pBIG3ura5sω3x2 no. 1 | 11.3 | 124.9 | 1.41 | 46.3 | 0.52 |
| pBIG3ura5sω3x2 no. 9 | 11.1 | 137.3 | 1.52 | 45.8 | 0.51 |
| pBIG3ura5sω3x2 no. 11 | 10.3 | 101.5 | 1.04 | 42.8 | 0.44 |
| pBIG3ura5sω3x2 no. 13 | 10.9 | 163.5 | 1.78 | 62.8 | 0.68 |
| pBIG3ura5sω3x2 no. 14 | 11.0 | 172.0 | 1.89 | 60.6 | 0.67 |
| pBIG3ura5sω3x2 no. 16 | 11.5 | 169.4 | 1.94 | 57.0 | 0.65 |
Mycelia were precultured for 2 days at 28°C and then cultured for 16 days at 12°C.
DISCUSSION
In this study, we first used a well-established auxotroph transformation system to evaluate the ATMT system in M. alpina 1S-4. We used spores of ura5− M. alpina 1S-4 as the starting material and the ura5 gene as a selectable marker. Although ATMT has been reported to be applicable to several filamentous fungi, the transformation efficiency varied among fungi due to biological differences among them (16). A. tumefaciens also has the ability to transform not only protoplasts but also intact cells; however, there have been few reports of ATMT systems in zygomycetes. In those studies, protoplasts or germinated conidia were employed as the starting material for ATMT. Thus, it was very important to succeed in using spores as recipient cells for M. alpina transformation to avoid time-consuming processes. One explanation of this success may be that we used a high Agrobacterium suspension concentration and a long cocultivation time compared with general ATMT system conditions.
Previously, we reported the biolistic transformation of M. alpina 1S-4. In that transformation system, a homologous ribosomal DNA region was included in the vector used to obtain stable transformants; however, approximately 90% of transformants remained unstable due to a failure to insert the plasmid into the genome or due to rearrangement of the integrated plasmids (24). The ATMT system reported here was very efficient and useful for the genetic modification of M. alpina 1S-4. We obtained at least 400 stable transformants per 108 spores with the ATMT system. On the other hand, the transformation frequency obtained with the biolistic method was about 30 stable transformants (24). The ATMT system resulted in increased transformation frequency compared with the conventional biolistic method for M. alpina 1S-4. Moreover, although microprojectile bombardment, which employed high pressure to transfer plasmid DNA, often caused physical damage to cells with resulting low growth, this ATMT system transferred the binary vector to host cells under mild conditions, and all transformants showed vigorous growth (1). Most transformants (94%) had a single copy of T-DNA at a random location in chromosomal DNA.
These features of ATMT of M. alpina 1S-4 enabled insertional mutagenesis and further recovery of T-DNA flanking sequences by means of PCR-based techniques or plasmid rescue techniques (3, 13). Moreover, we could analyze a novel promoter of M. alpina 1S-4 accurately due to its single copy. In addition, it has also been reported that ATMT increased homologous recombination frequency to target mutagenesis (gene disruption or gene replacement) (16).
On the other hand, we demonstrated overexpression of the ω3-desaturase gene using the ATMT system and improved the fatty acid composition of M. alpina 1S-4. n-3 fatty acids produced by transformant pBIG3ura5sω3x1 surpassed 30% of total PUFAs on average. When we transformed M. alpina 1S-4 with an ω3 expression cassette by the biolistic method, n-3 fatty acids were approximately 30% regardless of the integrated copy number (data not shown), while n-3 fatty acids produced by transformants pBIG3ura5sω3x2 reached 40%. These observations indicated that ATMT was useful for molecular breeding of M. alpina 1S-4.
Furthermore, most n-3 fatty acids produced by ATMT transformants were composed of EPA (20:5n-3). EPA has attracted considerable attention because of various beneficial physiological effects, anti-inflammatory properties, and prevention of major coronary events (6, 26). At present, fish oil is the only source of EPA but is considered unattractive because it contains substantial amounts of undesirable fatty acids and cholesterol; thus, there is much interest in the development of methods of EPA microbial production.
Previously, we reported that M. alpina was considered as a potential source of EPA for commercial production (22). However, the EPA content of total PUFAs in strain 2O-17 was 13.7% even under optimal conditions (20). We succeeded in producing EPA, with 40% total PUFAs, by M. alpina 1S-4. A pBIG3ura5sω3x2 transformant was cultivated in a 5-liter jar fermentor containing 2.5 liters GY medium at 12°C for 14 days after a 2-day precultivation at 28°C, and 36% of triacylglycerol extracted from mycelia was EPA (data not shown).
Our results demonstrated that ATMT improved the transformation efficiency of M. alpina 1S-4, the mitotic stability of transformants, and the fatty acid composition in M. alpina 1S-4, which indicates that this method constitutes a powerful tool for molecular manipulation of M. alpina 1S-4 to construct an industrial strain library for fatty acid production and to analyze fatty acid metabolism in M. alpina 1S-4.
Acknowledgments
We thank Yasuyuki Kubo (Kyoto Prefectural University, Japan) for providing Agrobacterium tumefaciens C58C1 and binary vector pBIG2RHPH2.
This work was partially supported by grants from the Project for Development of a Technological Infrastructure for Industrial Bioprocesses on R&D of New Industrial Science and Technology Frontiers (to S.S.); the Industrial Technology Research Grant Program (no. 05A07003d to E.S.) of the New Energy and Industrial Technology Development Organization, Japan; Grants-in-Aid for Scientific Research (no. 16688004 to J.O., no. 18208009 to S.S., and no. 19688006 to E.S.); and COE for Microbial-Process Development Pioneering Future Production Systems (to S.S.) from the Ministry of Education, Science, Sports, and Culture, Japan.
Footnotes
Published ahead of print on 6 July 2009.
REFERENCES
- 1.Ando, A., E. Sakuradani, K. Horinaka, J. Ogawa, and S. Shimizu. 2009. Transformation of an oleaginous zygomycete Mortierella alpina 1S-4 with the carboxin resistance gene conferred by mutation of the iron-sulfur subunit of succinate dehydrogenase. Curr. Genet. 55:349-356. [DOI] [PubMed] [Google Scholar]
- 2.Bundock, P., and P. J. Hooykaas. 1996. Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc. Natl. Acad. Sci. USA 93:15272-15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Combier, J. P., D. Melayah, C. Raffier, G. Gay, and R. Marmeisse. 2003. Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in the symbiotic ectomycorrhizal fungus Hebeloma cylindrosporum. FEMS Microbiol. Lett. 220:141-148. [DOI] [PubMed] [Google Scholar]
- 4.Covert, S. F., P. Kapoor, M. H. Lee, A. Briley, and C. J. Nairn. 2001. Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol. Res. 105:259-264. [Google Scholar]
- 5.de Groot, M. J., P. Bundock, P. J. Hooykaas, and A. G. Beijersbergen. 1998. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16:839-842. [DOI] [PubMed] [Google Scholar]
- 6.Endres, S., R. Ghorbani, V. E. Kelley, K. Georgilis, G. Lonnemann, J. W. van der Meer, J. G. Cannon, T. S. Rogers, M. S. Klempner, P. C. Weber, et al. 1989. The effect of dietary supplementation with n-3 polyunsaturated fatty acids on the synthesis of interleukin-1 and tumor necrosis factor by mononuclear cells. N. Engl. J. Med. 320:265-271. [DOI] [PubMed] [Google Scholar]
- 7.Gelvin, S. B. 2000. Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51:223-256. [DOI] [PubMed] [Google Scholar]
- 8.Gouka, R. J., C. Gerk, P. J. J. Hooykaas, P. Bundock, W. Musters, C. T. Verrips, and M. J. A. de Groot. 1999. Transformation of Aspergillus awamori by Agrobacterium tumefaciens-mediated homologous recombination. Nat. Biotechnol. 17:598-601. [DOI] [PubMed] [Google Scholar]
- 9.Guil-Guerrero, J. L. 2007. Stearidonic acid (18:4n-3): metabolism, nutritional importance, medical uses and natural sources. Eur. J. Lipid Sci. Technol. 109:1226-1236. [Google Scholar]
- 10.Jareonkitmongkol, S., H. Kawashima, N. Shirasaka, S. Shimizu, and H. Yamada. 1992. Production of dihomo-γ-linolenic acid by a Δ5-desaturase-defective mutant of Mortierella alpina 1S-4. Appl. Environ. Microbiol. 58:2196-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jareonkitmongkol, S., S. Shimizu, and H. Yamada. 1993. Production of an eicosapentaenoic acid-containing oil by a Δ12 desaturase-defective mutant of Mortierella alpina 1S-4. J. Am. Oil Chem. Soc. 70:119-123. [Google Scholar]
- 12.Kawashima, H., N. Kamada, E. Sakuradani, S. Jareonkitmongkol, K. Akimoto, and S. Shimizu. 1997. Production of 8,11,14,17-cis-eicosatetraenoic acid by Δ5 desaturase-defective mutants of an arachidonic acid-producing fungus, Mortierella alpina. J. Am. Oil Chem. Soc. 74:455-459. [Google Scholar]
- 13.Leclerque, A., H. Wan, A. Abschutz, S. Chen, G. V. Mitina, G. Zimmermann, and H. U. Schairer. 2004. Agrobacterium-mediated insertional mutagenesis (AIM) of the entomopathogenic fungus Beauveria bassiana. Curr. Genet. 45:111-119. [DOI] [PubMed] [Google Scholar]
- 14.Mackenzie, D. A., P. Wongwathanarat, A. T. Carter, and D. B. Archer. 2000. Isolation and use of a homologous histone H4 promoter and a ribosomal DNA region in a transformation vector for the oil-producing fungus Mortierella alpina. Appl. Environ. Microbiol. 66:4655-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattanovich, D., F. Ruker, A. D. Machado, M. Laimer, F. Regner, H. Steinkellner, G. Himmler, and H. Katinger. 1989. Efficient transformation of Agrobacterium spp. by electroporation. Nucleic Acids Res. 17:6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michielse, C. B., P. J. J. Hooykaas, C. A. M. J. J. van den Hondel, and A. F. J. Ram. 2005. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 48:1-17. [DOI] [PubMed] [Google Scholar]
- 17.Michielse, C. B., A. F. Ram, P. J. Hooykaas, and C. A. van den Hondel. 2004. Agrobacterium-mediated transformation of Aspergillus awamori in the absence of full-length VirD2, VirC2, or VirE2 leads to insertion of aberrant T-DNA structures. J. Bacteriol. 186:2038-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakuradani, E., T. Abe, K. Iguchi, and S. Shimizu. 2005. A novel fungal ω3-desaturase with wide substrate specificity from arachidonic acid-producing Mortierella alpina 1S-4. Appl. Microbiol. Biotechnol. 66:648-654. [DOI] [PubMed] [Google Scholar]
- 19.Sakuradani, E., M. Kobayashi, and S. Shimizu. 1999. Δ9-fatty acid desaturase from arachidonic acid-producing fungus: unique gene sequence and its heterologous expression in a fungus, Aspergillus. Eur. J. Biochem. 260:208-216. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu, S., H. Kawashima, Y. Shinmen, K. Akimoto, and H. Yamada. 1988. Production of eicosapentaenoic acid by Mortierella fungi. J. Am. Oil Chem. Soc. 65:1455-1459. [Google Scholar]
- 21.Shimizu, S., and J. Ogawa. 1997. Production of useful fatty acids by microbial processes. Recent Res. Dev. Lipids Res. 1:267-286. [Google Scholar]
- 22.Shimizu, S., Y. Shinmen, H. Kawashima, K. Akimoto, and H. Yamada. 1988. Fungal mycelia as a novel source of eicosapentaenoic acid activation of enzyme(s) involved in eicosapentaenoic acid production at low temperature. Biochem. Biophys. Res. Commun. 150:335-341. [DOI] [PubMed] [Google Scholar]
- 23.Stachel, S. E., E. Messens, M. Vanmontagu, and P. Zambryski. 1985. Identification of the signal molecules produced by wounded plant-cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318:624-629. [Google Scholar]
- 24.Takeno, S., E. Sakuradani, S. Murata, M. Inohara-Ochiai, H. Kawashima, T. Ashikari, and S. Shimizu. 2004. Establishment of an overall transformation system for an oil-producing filamentous fungus, Mortierella alpina 1S-4. Appl. Microbiol. Biotechnol. 65:419-425. [DOI] [PubMed] [Google Scholar]
- 25.Tsuji, G., S. Fujii, N. Fujihara, C. Hirose, S. Tsuge, T. Shiraishi, and Y. Kubo. 2003. Agrobacterium tumefaciens-mediated transformation for random insertional mutagenesis in Colletotrichum lagenarium. J. Gen. Plant Pathol. 69:230-239. [Google Scholar]
- 26.Yokoyama, M., H. Origasa, M. Matsuzaki, Y. Matsuzawa, Y. Saito, Y. Ishikawa, S. Oikawa, J. Sasaki, H. Hishida, H. Itakura, T. Kita, A. Kitabatake, N. Nakaya, T. Sakata, K. Shimada, and K. Shirato. 2007. Effects of eicosapentaenoic acid on major coronary events in hypercholesterolaemic patients (JELIS): a randomised open-label, blinded endpoint analysis. Lancet 369:1090-1098. [DOI] [PubMed] [Google Scholar]
- 27.Zupan, J. R., and P. Zambryski. 1995. Transfer of T-DNA from Agrobacterium to the plant cell. Plant Physiol. 107:1041-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]